A Proposed Saffron Soilless Cultivation System for a Quality Spice as Certified by Genetic Traceability
Abstract
1. Introduction
2. Results
2.1. Field Trials
2.2. Population Genetics
2.2.1. SNPs Identification
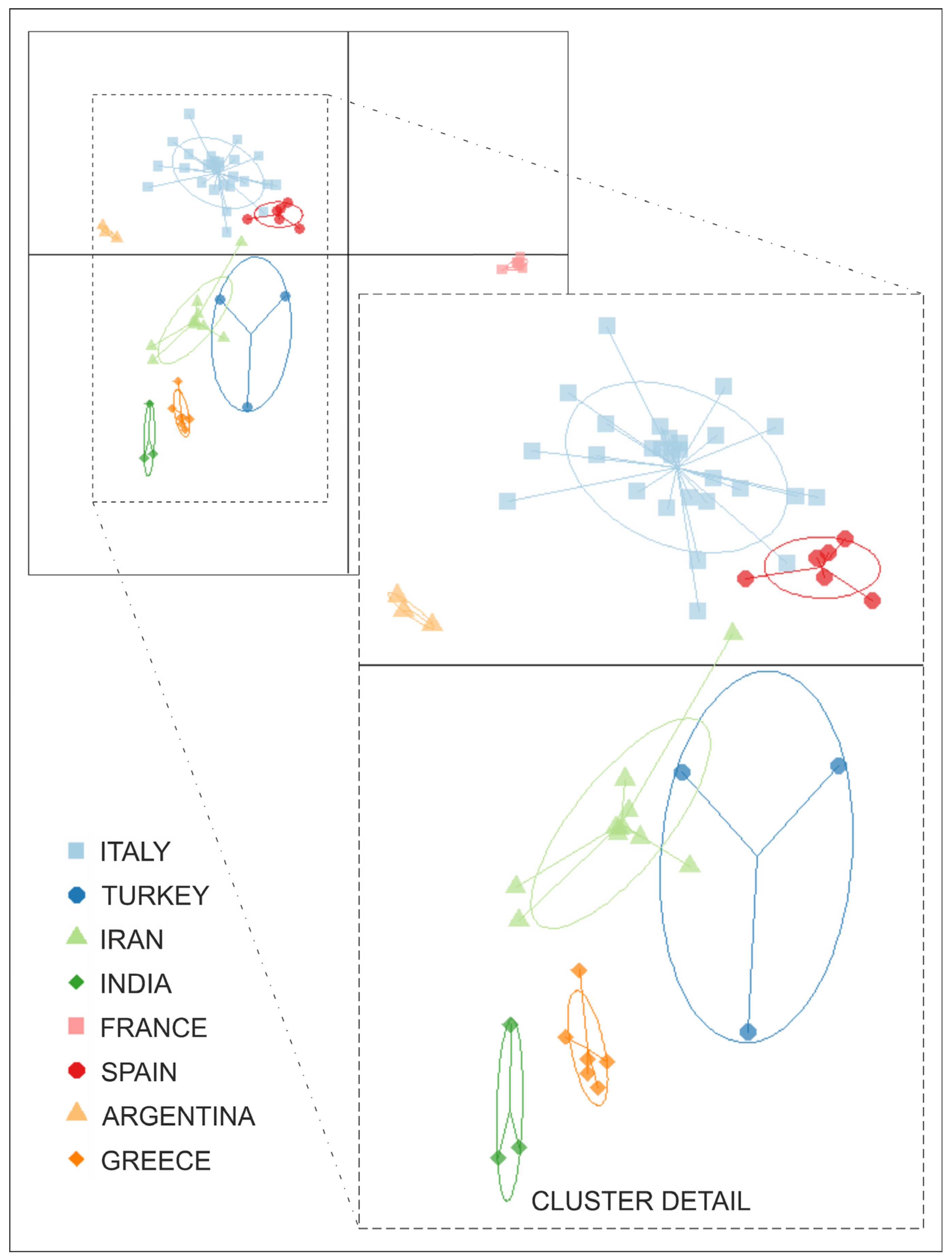
2.2.2. Population Structure and Genetic Variability
2.2.3. Private Alleles
3. Discussion
3.1. Field Trials
3.2. Population Genetics
4. Materials and Methods
4.1. Field Trials
4.1.1. Cultivation Methods
4.1.2. Statistical Analysis
- Yijk is the yield or quality parameter of spice or corms for the i-th treatment (Ti), j-th company (Lj), and k-th year (tk);
- μ is the grand mean;
- Ti is the effect of the i-th treatment;
- Lj is the effect of the j-th location;
- Yk is the effect of the k-th year;
- (TL)ij is the interaction effect between the i-th treatment and the j-th company;
- (TY)ik is the interaction effect between the i-th treatment and the k-th year;
- (LY)jk is the interaction effect between the j-th company and the k-th year;
- (TLY)ijk is the interaction effect between the i-th treatment, the j-th company, and the k-th year;
- eijk is the random experimental error associated with each observation.
4.2. Population Genetics
4.2.1. Plant Material
4.2.2. Genomic Library Preparation, Sequencing, and Genetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nemati, Z.; Harpke, D.; Gemicioglu, A.; Kerndorff, H.; Blattner, F.R. Saffron (Crocus sativus) Is an Autotriploid That Evolved in Attica (Greece) from Wild Crocus cartwrightianus. Mol. Phylogenet. Evol. 2019, 136, 14–20. [Google Scholar] [CrossRef]
- Gresta, F.; Lombardo, M.; Siracusa, L.; Ruberto, G. Saffron, an Alternative Crop for Sustainable Agricultural Systems. A Review. Agron. Sustain. Dev. 2008, 28, 95–112. [Google Scholar] [CrossRef]
- Winterhalter, P.; Straubinger, M. Saffron—Renewed Interest in an Ancient Spice. Food Rev. Int. 2000, 16, 39–59. [Google Scholar] [CrossRef]
- Basker, D.; Negbi, M. Uses of Saffron I. Econ. Bot. 1983, 37, 228–236. [Google Scholar] [CrossRef]
- Abu-Izneid, T.; Rauf, A.; Khalil, A.A.; Olatunde, A.; Khalid, A.; Alhumaydhi, F.A.; Aljohani, A.S.M.; Sahab Uddin, M.; Heydari, M.; Khayrullin, M.; et al. Nutritional and Health Beneficial Properties of Saffron (Crocus sativus L.): A Comprehensive Review. Crit. Rev. Food Sci. Nutr. 2022, 62, 2683–2706. [Google Scholar] [CrossRef]
- Ghaffari, S.; Roshanravan, N. Saffron; An Updated Review on Biological Properties with Special Focus on Cardiovascular Effects. Biomed. Pharmacother. 2019, 109, 21–27. [Google Scholar] [CrossRef]
- Thachil, A.F.; Mohan, R.; Bhugra, D. The Evidence Base of Complementary and Alternative Therapies in Depression. J. Affect. Disord. 2007, 97, 23–35. [Google Scholar] [CrossRef]
- Shahnoushi, N.; Abolhassani, L.; Kavakebi, V.; Reed, M.; Saghaian, S. Economic Analysis of Saffron Production. In Saffron: Science, Technology and Health; Elsevier: Amsterdam, The Netherlands, 2020; pp. 337–356. ISBN 9780128186381. [Google Scholar]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the King of Spices: An Overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Condurso, C.; Cincotta, F.; Tripodi, G.; Verzera, A. Bioactive Volatiles in Sicilian (South Italy) Saffron: Safranal and Its Related Compounds. J. Essent. Oil Res. 2017, 29, 221–227. [Google Scholar] [CrossRef]
- D’Archivio, A.A.; Giannitto, A.; Maggi, M.A.; Ruggieri, F. Geographical Classification of Italian Saffron (Crocus sativus L.) Based on Chemical Constituents Determined by High-Performance Liquid-Chromatography and by Using Linear Discriminant Analysis. Food Chem. 2016, 212, 110–116. [Google Scholar] [CrossRef]
- Giupponi, L.; Leoni, V.; Sala, S.; Giorgi, A.; Bertoni, D. Saffron Growing in Italy: A Sustainable Secondary Activity for Farms in Hilly and Sub-Mountain Areas. Int. J. Agric. Sustain. 2023, 21, 2270263. [Google Scholar] [CrossRef]
- Giupponi, L.; Ceciliani, G.; Leoni, V.; Panseri, S.; Pavlovic, R.; Lingua, G.; Di Filippo, A.; Giorgi, A. Quality Traits of Saffron Produced in Italy: Geographical Area Effect and Good Practices. J. Appl. Bot. Food Qual. 2019, 92, 336–342. [Google Scholar] [CrossRef]
- del Campo, C.P.; Carmona, M.; Maggi, L.; Kanakis, C.D.; Anastasaki, E.G.; Tarantilis, P.A.; Polissiou, M.G.; Alonso, G.L. Effects of Mild Temperature Conditions during Dehydration Procedures on Saffron Quality Parameters. J. Sci. Food Agric. 2010, 90, 719–725. [Google Scholar] [CrossRef]
- Gregory, M.J.; Menary, R.C.; Davies, N.W. Effect of Drying Temperature and Air Flow on the Production and Retention of Secondary Metabolites in Saffron. J. Agric. Food Chem. 2005, 53, 5969–5975. [Google Scholar] [CrossRef]
- Negbi, M. SAFFRON: Crocus sativus L.; Harwood Academic Publishers: Reading, UK, 1999. [Google Scholar]
- Temperini, O.; Rea, R.; Temperini, A.; Colla, G.; Rouphael, Y. Evaluation of Saffron (Crocus sativus L.) Production in Italy: Effects of the Age of Saffron Fields and Plant Density. J. Food Agric. Environ. 2009, 7, 19–23. [Google Scholar]
- Cardone, L.; Candido, V.; Castronuovo, D.; Perniola, M.; Cicco, N. Comparing Annual and Biennial Crop Cycle on the Growth, Yield and Quality of Saffron Using Three Corm Dimensions. Sci. Hortic. 2021, 288, 110393. [Google Scholar] [CrossRef]
- Souret, F.F.; Weathers, P.J. The Growth of Saffron (Crocus sativus L.) in Aeroponics and Hydroponics. J. Herbs Spices Med. Plants 2000, 7, 25–35. [Google Scholar] [CrossRef]
- Maggio, A.; Raimondi, G.; Martino, A.; De Pascale, S. Soilless cultivation of saffron in mediterranean environment. Acta Hortic. 2006, 718, 515–522. [Google Scholar] [CrossRef]
- Lakhiar, I.A.; Gao, J.; Syed, T.N.; Chandio, F.A.; Buttar, N.A. Modern Plant Cultivation Technologies in Agriculture under Controlled Environment: A Review on Aeroponics. J. Plant Interact. 2018, 13, 338–352. [Google Scholar] [CrossRef]
- Caser, M.; Demasi, S.; Victorino, Í.M.M.; Donno, D.; Faccio, A.; Lumini, E.; Bianciotto, V.; Scariot, V. Arbuscular Mycorrhizal Fungi Modulate the Crop Performance and Metabolic Profile of Saffron in Soilless Cultivation. Agronomy 2019, 9, 232. [Google Scholar] [CrossRef]
- Zsombik, L.; Hanász, A.; Sipos, T.; Basal, O.; Magyar-Tábori, K. Seedling Morphology of Different Wheat Genotypes at Early Stages under Hydrocultural Conditions. Acta Agrar. Debreceniensis 2021, 1, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Kour, K.; Gupta, D.; Gupta, K.; Dhiman, G.; Juneja, S.; Viriyasitavat, W.; Mohafez, H.; Islam, M.A. Smart-Hydroponic-Based Framework for Saffron Cultivation: A Precision Smart Agriculture Perspective. Sustainability 2022, 14, 1120. [Google Scholar] [CrossRef]
- Chourak, Y.; Belarbi, E.H.; da Cunha-Chiamolera, T.P.L.; Guil-Guerrero, J.L.; Carrasco, G.; Urrestarazu, M. Effect of Macronutrient Conditions and Electrical Conductivity on the Quality of Saffron Grown in Soilless Culture Systems. J. Soil Sci. Plant Nutr. 2022, 22, 4449–4457. [Google Scholar] [CrossRef]
- Dewir, Y.H.; Alsadon, A.; Ibrahim, A.; El-Mahrouk, M. Effects of Growing Substrate, Mode of Nutrient Supply, and Saffron Corm Size on Flowering, Growth, Photosynthetic Competence, and Cormlet Formation in Hydroponics. Horttechnology 2022, 32, 234–240. [Google Scholar] [CrossRef]
- Alavi-Siney, S.M.; Saba, J.; Siahpirani, A.F.; Nasiri, J. ISSR-Assisted Spatial Genetic Structure, Population Admixture, and Biodiversity Estimates across Locally Adopted Saffron Ecotypes from 18 Different Provenances of Iran. J. Appl. Res. Med. Aromat. Plants 2023, 35, 100467. [Google Scholar] [CrossRef]
- Alavi-Siney, S.M.; Saba, J.; Nasiri, J. Genetic Variability and Population Genetic Structure in Autotriploid Saffron Using Allelic Phenotypes of Microsatellite Markers. Sci. Hortic. 2022, 299, 111043. [Google Scholar] [CrossRef]
- Mir, M.A.; Mansoor, S.; Sugapriya, M.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Deciphering Genetic Diversity Analysis of Saffron (Crocus sativus L.) Using RAPD and ISSR Markers. Saudi J. Biol. Sci. 2021, 28, 1308–1317. [Google Scholar] [CrossRef]
- Busconi, M.; Colli, L.; Sánchez, R.A.; Santaella, M.; De-Los-Mozos Pascual, M.; Santana, O.; Roldán, M.; Fernández, J.A. AFLP and MS-AFLP Analysis of the Variation within Saffron Crocus (Crocus sativus L.) Germplasm. PLoS ONE 2015, 10, e0123434. [Google Scholar] [CrossRef]
- Siracusa, L.; Gresta, F.; Avola, G.; Albertini, E.; Raggi, L.; Marconi, G.; Lombardo, G.M.; Ruberto, G. Agronomic, Chemical and Genetic Variability of Saffron (Crocus sativus L.) of Different Origin by LC-UV-Vis-DAD and AFLP Analyses. Genet. Resour. Crop Evol. 2013, 60, 711–721. [Google Scholar] [CrossRef]
- Rochette, N.C.; Rivera-Colón, A.G.; Catchen, J.M. Stacks 2: Analytical Methods for Paired-End Sequencing Improve RADseq-Based Population Genomics. Mol. Ecol. 2019, 28, 4737–4754. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2024. Available online: https://www.r-project.org/ (accessed on 18 November 2024).
- Jombart, T.; Collins, C. A Tutorial for Discriminant Analysis of Principal Components (DAPC) Using Adegenet 2.1.6; MRC Centre for Outbreak Analysis and Modelling: London, UK, 2022. [Google Scholar]
- Alonso, G.L.; Arghittu, A.; Astrka, K.; Betza, T.; Cambae Conada, M.; Cilloco, M.T.; Pitzalis, P. White Book, Saffron in Europe. Problems and Strategies for Improving the Quality and Strengthen Competitiveness; Alexandros-LLC: Chicago, IL, USA, 2007. [Google Scholar]
- Khajeh-Hosseini, M.; Fallahpour, F. Emerging Innovation in Saffron Production. In Saffron: Science, Technology and Health; Elsevier: Amsterdam, The Netherlands, 2020; pp. 205–216. ISBN 9780128186381. [Google Scholar]
- Shahrokhi, M.B.; Rahimi, H.; Rashed, M.H. Chapter 6—Saffron Pests, Diseases, and Weeds. In Saffron (Crocus sativus). Production and Processing; Kafi, M., Koocheki, A., Rashed, M.H., Nassiri, M., Eds.; Science Publishers: Enfield, NH, USA, 2006. [Google Scholar]
- Bazoobandi, M.; Rahimi, H.; Karimi-Shahri, M.R. Saffron Crop Protection. In Saffron: Science, Technology and Health; Woodhead Publishing: Cambridge, UK, 2020; pp. 169–185. [Google Scholar] [CrossRef]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Molina, R.V.; Renau-Morata, B.; Nebauer, S.G.; Candido, V. Crocus sativus L. Ecotypes from Mediterranean Countries: Phenological, Morpho-Productive, Qualitative and Genetic Traits. Agronomy 2021, 11, 551. [Google Scholar] [CrossRef]
- Rubio-Moraga, A.; Castillo-Lápez, R.; Gámez-Gámez, L.; Ahrazem, O. Saffron Is a Monomorphic Species as Revealed by RAPD, ISSR and Microsatellite Analyses. BMC Res. Notes 2009, 2, 189. [Google Scholar] [CrossRef] [PubMed]
- Busconi, M.; Wischnitzki, E.; Del Corvo, M.; Colli, L.; Soffritti, G.; Stagnati, L.; Fluch, S.; Sehr, E.M.; de los Mozos Pascual, M.; Fernández, J.A. Epigenetic Variability Among Saffron Crocus (Crocus sativus L.) Accessions Characterized by Different Phenotypes. Front. Plant Sci. 2021, 12, 642631. [Google Scholar] [CrossRef] [PubMed]
- Nemati, Z.; Mardi, M.; Majidian, P.; Zeinalabedini, M.; Pirseyedi, S.M.; Bahadori, M. Saffron (Crocus sativus L.), a Monomorphic or Polymorphic Species? Span. J. Agric. Res. 2014, 12, 753–762. [Google Scholar] [CrossRef]
- Anabat, M.M.; Riahi, H.; Sheidai, M.; Koohdar, F. Population Genetic Study and Barcoding in Iran Saffron (Crocus sativus L.). Ind. Crops Prod. 2020, 143, 111915. [Google Scholar] [CrossRef]
- Villa, C.; Costa, J.; Meira, L.; Oliveira, M.B.P.P.; Mafra, I. Exploiting DNA Mini-Barcodes as Molecular Markers to Authenticate Saffron (Crocus sativus L.). Food Control 2016, 65, 21–31. [Google Scholar] [CrossRef]
- Huang, W.J.; Li, F.F.; Liu, Y.J.; Long, C.L. Identification of Crocus sativus and Its Adulterants from Chinese Markets by Using DNA Barcoding Technique. Iran. J. Biotechnol. 2015, 13, 36–42. [Google Scholar] [CrossRef]
- Marconi, G.; Capomaccio, S.; Comino, C.; Acquadro, A.; Portis, E.; Porceddu, A.; Albertini, E. Methylation Content Sensitive Enzyme DdRAD (MCSeEd): A Reference-Free, Whole Genome Profiling System to Address Cytosine/Adenine Methylation Changes. Sci. Rep. 2019, 9, 14864. [Google Scholar] [CrossRef]
- Chong, Z.; Ruan, J.; Wu, C.I. Rainbow: An Integrated Tool for Efficient Clustering and Assembling RAD-Seq Reads. Bioinformatics 2012, 28, 2732–2737. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]




| (a) | |||||||
|---|---|---|---|---|---|---|---|
| Spice Yield (g/m2) | Flowers (No./m2) | Flowering (No. of Days) | |||||
| Source of Variation | Df | MS | MS | MS | |||
| Rep (location year) | 72 | 0.258 | - | 1075.8 | 5.5 | ||
| Treatments | 2 | 6.163 | * | 7002.8 | ns | 126.9 | * |
| Locations | 7 | 2.750 | ns | 32,595.8 | ns | 1049.8 | * |
| Years | 2 | 17.337 | *** | 105,236.0 | *** | 444.4 | *** |
| Treatments × location | 14 | 0.744 | ns | 10,740.3 | ** | 43.3 | ns |
| Treatments × year | 4 | 0.866 | *** | 3555.2 | *** | 11.2 | * |
| Location × year | 14 | 2.146 | *** | 22,505.0 | *** | 335.0 | *** |
| Treatments × location × year | 27 | 0.365 | *** | 3511.0 | *** | 26.1 | *** |
| Error | 140 | 0.054 | 573.3 | 4.5 | |||
| Total | 282 | ||||||
| (b) | |||||||
| Corm Number (m2) | Corm Yield (g/m2) | Corm Weight (g) | |||||
| Source of Variation | Df | MS | MS | MS | |||
| Rep (location year) | 63 | 340.5 | - | 145,089.0 | - | 3.88 | - |
| Treatments | 2 | 9838.3 | ns | 9,581,795.4 | ns | 694.82 | ** |
| Locations | 6 | 40,258.5 | ns | 10,440,362.8 | ns | 534.37 | ns |
| Years | 2 | 22,248.5 | *** | 16,512,813.3 | *** | 381.23 | *** |
| Treatments × location | 12 | 100,062.1 | ns | 3,831,779.7 | ns | 93.86 | ns |
| Treatments × year | 4 | 6292.4 | *** | 1,663,513.8 | *** | 13.49 | * |
| Location × year | 12 | 23,973.8 | *** | 4,027,401.94 | *** | 337.24 | *** |
| Treatments × location × year | 20 | 5419.4 | *** | 2,412,028.3 | *** | 57.76 | *** |
| Error | 112 | 380.6 | 162,896.0 | 4.20 | |||
| Total | 233 | ||||||
| Spice Yield (g/m2) 1 | Flowers (No./m2) | Flowering Length (days) | Bulbs (No./m2) | Bulbs (g/m2) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Treatments | ||||||||||
| Bin–farm soil | 1.150 | B | 121.9 | 18.2 | B | 136.7 | 1919.2 | |||
| Bin–organic soil | 1.377 | A | 138.5 | 19.4 | B | 129.4 | 2576.5 | |||
| Field (control) | 0.849 | C | 126.2 | 22.2 | A | 121.1 | 2295.2 | |||
| Locations | ||||||||||
| Alfonsi | 0.898 | 102.6 | 31.7 | A | 90.0 | 2293.3 | ||||
| Fontanelle | 0.781 | 84.1 | 12.6 | E | 101.2 | 1930.1 | ||||
| Mazzuoli | 1.372 | 165.1 | 17.5 | D | 187.3 | 2116.1 | ||||
| Porta Sole | 1.373 | 161.5 | 22.1 | B | 63.6 | 1947.9 | ||||
| Ro_Lo | 0.927 | 106.2 | 18.2 | CD | 114.4 | 1878.1 | ||||
| Venturi | 1.546 | 164.7 | 19.1 | C | 131.0 | 2177.1 | ||||
| Vinerbi | 1.263 | 129.1 | 17.5 | D | - | - | ||||
| Zafferano and Dintorni | 0.849 | 110.0 | 20.2 | C | 118.3 | 1768.7 | ||||
| Years | ||||||||||
| 2018 | 1.405 | A | 150.3 | A | 20.0 | A | 145.9 | A | 2882.8 | A |
| 2019 | 1.340 | B | 146.2 | A | 21.0 | A | 134.4 | B | 1926.0 | B |
| 2020 | 0.632 | C | 90.1 | B | 18.9 | B | 107.0 | C | 1982.1 | B |
| Moisture (%) | Bitterness | Aromatic Strength | Coloring Strength | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of Variation | Df | MS | Df | MS | Df | MS | Df | MS | ||||
| Rep (location) | 15 | 1.32 | 15 | 118.8 | 15 | 14.1 | 15 | 880.5 | ||||
| Treatments | 2 | 0.94 | * | 2 | 30.2 | ns | 2 | 14.7 | ns | 2 | 203.4 | ns |
| Locations | 7 | 1.70 | *** | 7 | 81.0 | * | 7 | 20.4 | ns | 7 | 327.3 | ns |
| Treatments × location | 14 | 0.34 | ns | 14 | 39.5 | ns | 14 | 16.9 | ns | 14 | 27.08 | ns |
| Error | 26 | 0.27 | 26 | 31.8 | 26 | 10.3 | 26 | 327.7 | ||||
| Total | 64 | 64 | 64 | 64 | ||||||||
| Moisture (%) 1 | Bitterness | Aromatic Strength | Aromatic Strength | |||||
|---|---|---|---|---|---|---|---|---|
| Treatments | ||||||||
| Bin–organic soil (BO) | 7.24 | B | 93.98 | 28.02 | 252.8 | |||
| Bin–farm soil (BF) | 7.55 | A | 91.79 | 28.69 | 246.5 | |||
| Field (control) | 7.56 | A | 91.94 | 29.75 | 248.5 | |||
| Locations | ||||||||
| Alfonsi | 7.96 | A | 85.25 | B | 30.72 | A | 230.3 | |
| Fontanelle | 7.56 | AB | 99.20 | A | 28.67 | E | 264.5 | |
| Mazzuoli | 7.66 | AB | 90.78 | B | 28.00 | D | 252.2 | |
| Porta Sole | 7.43 | AB | 92.89 | AB | 28.11 | B | 248.3 | |
| Ro_Lo | 7.70 | AB | 97.00 | A | 28.72 | CD | 248.3 | |
| Venturi | 7.14 | B | 92.56 | AB | 29.67 | C | 247.7 | |
| Vinerbi | 6.58 | C | 92.56 | AB | 30.89 | D | 250.4 | |
| Zafferano and Dintorni | 7.58 | AB | 91.38 | AB | 25.78 | C | 252.2 | |
| Farm Name | Location | Latitude | Longitude | Altitude (m asl) |
|---|---|---|---|---|
| Alfonsi | Nocera Umbra | 43.1153 | 12.7698 | 520 |
| Fontanelle | Marsciano | 42.9070 | 12.3390 | 180 |
| Mazzuoli | Città della Pieve | 42.9546 | 12.0039 | 508 |
| Porta Sole | Perugia | 43.1280 | 12.4866 | 200 |
| Ro_Lo | Foligno | 42.9544 | 12.6995 | 234 |
| Venturi | Gualdo Tadino | 43.2288 | 12.7809 | 536 |
| Vinerbi | Città della Pieve | 42.9546 | 12.0039 | 508 |
| Zafferano and Dintorni | Sant’Anatolia di Narco | 42.7337 | 12.8355 | 328 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mariani, A.; Marconi, G.; Ferradini, N.; Bocchini, M.; Lorenzetti, S.; Chiorri, M.; Russi, L.; Albertini, E. A Proposed Saffron Soilless Cultivation System for a Quality Spice as Certified by Genetic Traceability. Plants 2025, 14, 51. https://doi.org/10.3390/plants14010051
Mariani A, Marconi G, Ferradini N, Bocchini M, Lorenzetti S, Chiorri M, Russi L, Albertini E. A Proposed Saffron Soilless Cultivation System for a Quality Spice as Certified by Genetic Traceability. Plants. 2025; 14(1):51. https://doi.org/10.3390/plants14010051
Chicago/Turabian StyleMariani, Alessandro, Gianpiero Marconi, Nicoletta Ferradini, Marika Bocchini, Silvia Lorenzetti, Massimo Chiorri, Luigi Russi, and Emidio Albertini. 2025. "A Proposed Saffron Soilless Cultivation System for a Quality Spice as Certified by Genetic Traceability" Plants 14, no. 1: 51. https://doi.org/10.3390/plants14010051
APA StyleMariani, A., Marconi, G., Ferradini, N., Bocchini, M., Lorenzetti, S., Chiorri, M., Russi, L., & Albertini, E. (2025). A Proposed Saffron Soilless Cultivation System for a Quality Spice as Certified by Genetic Traceability. Plants, 14(1), 51. https://doi.org/10.3390/plants14010051







