Abstract
Conferring crops with resistance to multiple diseases is crucial for stable food production. Genetic engineering is an effective means of achieving this. The rice receptor-like cytoplasmic kinase BSR1 mediates microbe-associated molecular pattern-induced immunity. In our previous study, we demonstrated that rice lines overexpressing BSR1 under the control of the maize ubiquitin promoter exhibited broad-spectrum resistance to rice blast, brown spot, leaf blight, and bacterial seedling rot. However, unfavorable phenotypes were observed, such as a decreased seed germination rate and a partial darkening of husked rice. Herein, we present a strategy to address these unfavorable phenotypes using an OsUbi7 constitutive promoter with moderate expression levels and a pathogen-inducible PR1b promoter. Rice lines expressing BSR1 under the influence of both promoters maintained broad-spectrum disease resistance. The seed germination rate and coloration of husked rice were similar to those of the wild-type rice.
1. Introduction
Stable crop production is an important agricultural issue because some countries are facing global food shortages owing to population growth, deteriorating security, and global warming. Rice is one of the most important crops worldwide and serves as a staple food for approximately 50% of the world’s population [1]. However, stable rice production is limited by diseases. Approximately 10–30% of the harvest is lost due to blast, one of the seven major crop diseases in the world, caused by the fungus Pyricularia oryzae in rice. However, 10% of the harvest is sufficient to feed 60 million people annually [2]. The bacterial leaf blight caused by Xanthomonas oryzae pv. oryzae (Xoo) is an important rice disease that has caused significant yield losses in Southeast Asian and West African countries [3]. Minimizing these losses would contribute to stabilizing and increasing rice production.
Addressing this problem and enhancing host resistance to these pathogens without relying on pesticides is the most economical and environmentally friendly approach to sustainable food production. Breeding a rice variety with broad-spectrum resistance (BSR) is the safest and most efficient method to achieve this goal. To date, R gene introduction has been the main method used for breeding disease-resistant varieties of rice [4]. However, in many cases, R gene-introduced varieties have only been effective in agricultural production for a few years because of the emergence of new pathogen biotypes that can overcome this gene [5]. In addition, the R gene generally confers resistance to a specific race of the pathogen but does not confer BSR. Furthermore, pyramiding these R genes using conventional breeding methods is labor-intensive, time-consuming, and difficult because of the need to remove linked unfavorable traits. Therefore, we considered using BSR genes as a feasible approach.
To date, several BSR genes have been identified. BSR is classified into species-non-specific (SNS) BSR, which confers resistance against two or more pathogen species, and race-non-specific (RNS) BSR, which confers resistance against two or more races or strains of the same pathogen [6]. SNS BSR is more valuable in agriculture than RNS BSR. Examples of SNS BSR genes include WRKY45 [7] and OsSSI2 [8], which are involved in providing disease resistance to P. oryzae and Xoo via the salicylic acid pathway. WRKY30 [9] and OsACS2 [10] are involved in providing disease resistance to P. oryzae and the sheath blight fungus Rhizoctonia solani through the biosynthesis of jasmonic acid and ethylene, respectively.
BSR1 is an SNS BSR gene, and the BSR1 protein (OsRLCK278) is classified in the RLCK-VII protein family as BIK1 [11], PBL19, and PBL20 [12], which are well known to encode receptor-like cytoplasmic kinases (RLCKs) involved in providing disease resistance in Arabidopsis [13]. We have previously reported the disease resistance mechanism of BSR1 [14,15,16,17].
The BSR1 gene, when highly expressed under the maize ubiquitin promoter (PZmUbi), confers SNS BSR. We have reported that BSR1-overexpressing (OX) rice shows strong resistance to Xoo and Burkholderia glumae, causing bacterial seedling rot, grain rot, P. oryzae, and brown spot fungus Cochliobolus miyabeanus [13,18]. There are few resistance genes for B. glumae and C. miyabeanus, although global warming is expected to increase their propagation in the future [19]. No other genes showed resistance to four or more of these diseases, making the use of BSR1 in breeding desirable. However, because PZmUbi-BSR1 rice plants express high levels of BSR1 throughout the plant body, unfavorable phenotypes, such as reduced seed germination rate and partial darkening of husked rice, have been observed [13,18]. Thus, we used a constitutive promoter with weaker activity than PZmUbi and a pathogen-inducible promoter to generate rice lines expressing BSR1 and evaluated their disease resistance, germination rate, and brown rice color.
2. Results
2.1. Enhancement of BSR1 Expression in Rice and Screening for Bacterial Leaf Blight Resistance
We previously reported rice plants overexpressing BSR1 using a construct (Figure 1a) in which BSR1 cDNA was inserted downstream of the maize ubiquitin promoter (PZmUbi), which is a constitutively high-expressing promoter [13]. PZmUbi-BSR1 transgenic rice lines #5 and #9 exhibited 159- and 130-fold higher expression of BSR1 than the wild-type (WT), respectively, and demonstrated BSR to at least the four pathogens described above [13,18]. However, regarding growth and morphological characteristics, the PZmUbi-BSR1 lines produced partially blackish brown rice (Figure 1b), and the seed germination rate was reduced to approximately 1/3 to 1/4 of the original (Figure 1c), whereas the brown rice color in the WT was white (Figure 1b). We attributed this undesirable trait to the use of a strong promoter and considered that this problem could be resolved by improving the promoter. Therefore, we expressed BSR1 using the rice ubiquitin promoter (POsUbi7) [20], a constitutive promoter with weaker activity than PZmUbi, and the OsPR1b promoter (PPR1b) [21], whose activity was induced by infection with P. oryzae and Xoo.
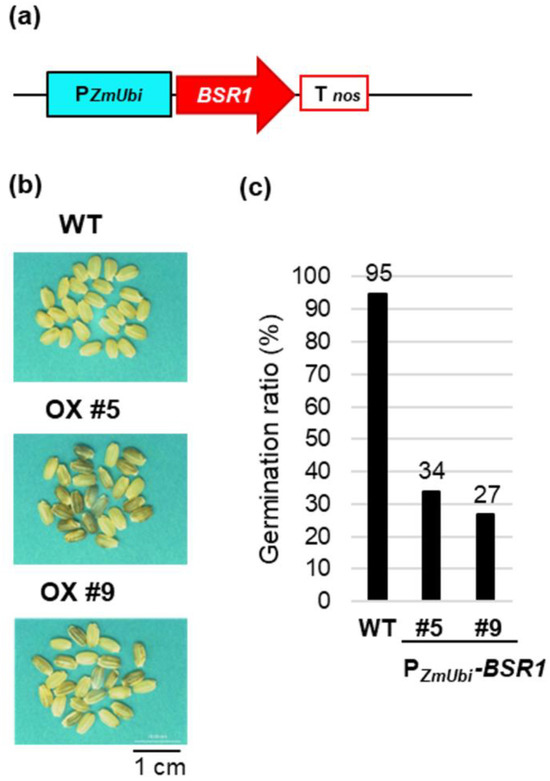
Figure 1.
Undesirable phenotype of BSR1 overexpression by maize ubiquitin promoter in rice seeds: (a) Maize ubiquitin promoter (PZmUbi)-BSR1 construct. Tnos, nos terminator. (b) Grain color of wild-type (WT) PZmUbi-BSR1 (OX) #5 and #9 lines. (c) Germination ratio (%) of WT PZmUbi-BSR1 #5 and #9 lines. n = 116–198.
POsUbi7-BSR1 rice lines were generated using a vector in which BSR1 cDNA was inserted between the POsUbi7 promoter and double terminator (Figure 2a). From approximately 50 T0 plants carrying POsUbi7-BSR1, lines resistant to Xoo race 1 were selected through initial screening, and the expression levels of BSR1 were subsequently analyzed. Next, we verified the resistance of the selected lines in T1 plants and selected nine lines that demonstrated approximately half or less suppression of lesion length compared with the WT. The expression levels of BSR1 and resistance levels to Xoo in the nine selected lines are presented in Figure 2b,c, respectively. Figure S1 presents Xoo resistance in three representative lines. Additionally, since the PZmUbi-BSR1 rice is also resistant to other Xoo races [18], we inoculated race III Xoo to lines #15 and #22 as representatives. Therefore, the lesion lengths were reduced to <1/4 of that in the WT, indicating resistance (Figure S2). Then, the grain color of the T1 seeds from these lines was examined. Three lines (#20, #27, and #46) showed blackish grains (Figure 2b,c, gray bar); however, six lines (#11, #15, #22, #31, #41, and #42) exhibited white grains similar to those of the WT (Figure 2b,c, white bar). The POsUbi7-BSR1 lines showing Xoo-resistant and white grains (Figure 2b, white bar only) had 16.0- to 49.5-fold higher expression of BSR1 than that of WT, which was lower than those of the PZmUbi-BSR1 rice described above. We successfully generated lines with leaf blight resistance and white grains using the OsUbi7 promoter.
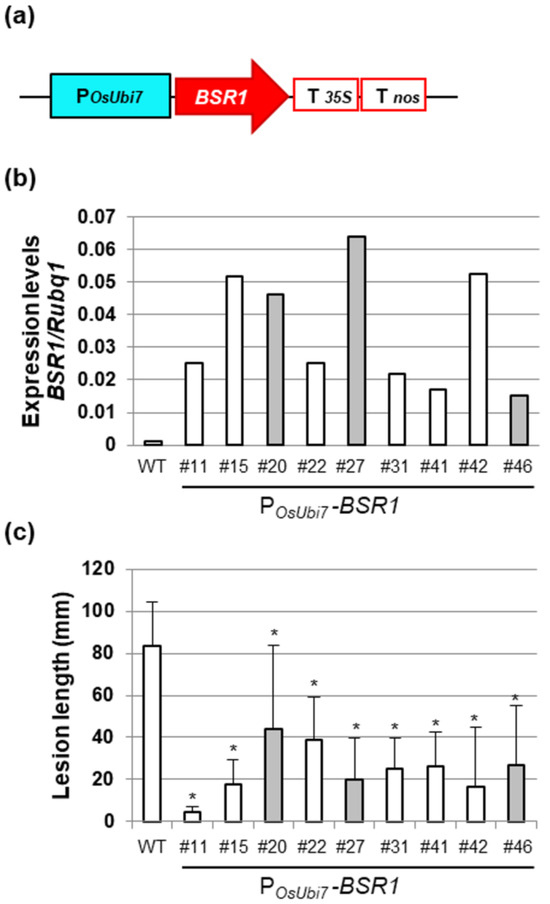
Figure 2.
Generation and screening of POsUBi7-BSR1 lines: (a) Rice Ubi7 promoter (POsUbi7)-BSR1 construct. T35S, CaMV35S terminator; Tnos, nos terminator. (b) BSR1 expression levels in POsUbi7-BSR1 T0 lines. (c) Disease resistance to Xanthomonas oryzae pv. oryzae (T7174, race I) in POsUbi7-BSR1 T1 lines. Lesion lengths in POsUbi7-BSR1 plants were significantly lower than those in WT plants (* p < 0.05 by Dunnett’s test). Values are mean ± standard deviation (n = 4–14). Gray bars in (b,c) indicate that blackish grains were included, whereas white bars indicate that only white grains were included.
PPR1b-BSR1 rice lines were generated using a vector in which BSR1 cDNA was inserted between the PPR1b promoter and the double terminator, as illustrated in Figure 3a. From approximately 30 T0 plants carrying PPR1b-BSR1, lines resistant to Xoo race 1 were selected through initial screening. By confirming the resistance of the selected lines in T1 plants, six lines that exhibited approximately half or less suppression of lesion length compared with the WT were identified (Figure 3b). The grain color of the T1 seeds from these lines was then examined, and all six lines showed a white grain color (Figure 3b, white bar) similar to that of the WT.
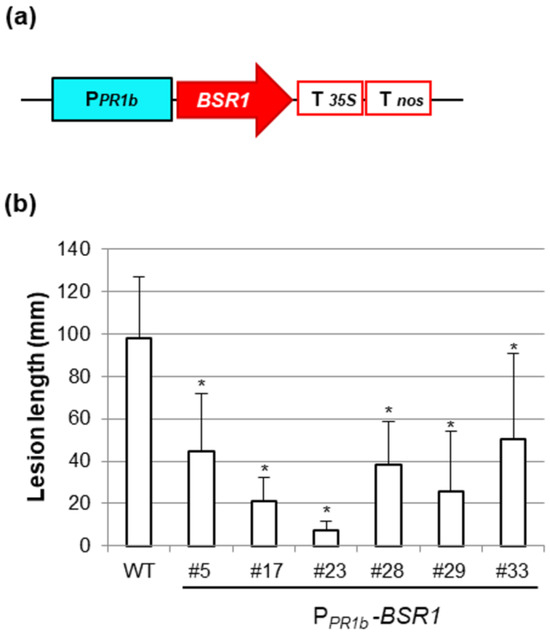
Figure 3.
Generation and screening of POsPR1b-BSR1 lines: (a) Rice PR1b promoter (PPR1b)-BSR1 construct. T35S, CaMV35S terminator; Tnos, nos terminator. (b) Disease resistance to Xanthomonas oryzae pv. oryzae (T7174) in PPR1b-BSR1 T1 lines. Lesion lengths in PPR1b-BSR1 plants were significantly lower than those in WT plants (* p < 0.05 by Dunnett’s test). Values are mean ± standard deviation (n = 3–9). White bars indicate that only white grains were included.
2.2. Other Disease Resistance in POsUbi7-BSR1 Lines
The T4 generation of POsUbi7-BSR1 lines #22 and #42 was used as a representative for subsequent disease resistance tests. Overexpression of BSR1 was confirmed before disease resistance evaluation (Figure S3). First, the disease resistance to B. glumae was investigated. Although the WT plants succumbed to the infection, displaying 0% survival, lines #22 and #42 exhibited 100% survival, indicating resistance to bacterial seedling rot (Figure 4a,b). Subsequently, disease resistance to the fungus P. oryzae, which causes rice blast, was assessed. Lines #22 and #42 demonstrated robust resistance, with <1/8 and <1/3 of the lesions observed in comparison with the WT, respectively (Figure 4c,d). Subsequently, disease resistance to the fungus C. miyabeanus, which causes brown spots, was evaluated. Lines #22 and #42 showed strong resistance, with lesion numbers < 1/2 and <1/9, respectively, compared with the WT (Figure 4e,f and Figure S4). Overall, the POsUbi7-BSR1 lines (#22 and #42) maintained resistance to at least four pathogens, similar to the PZmUbi-BSR1 lines. Additionally, the descendants of lines #22 and #42 displayed resistance to panicle blast (Figure S5).
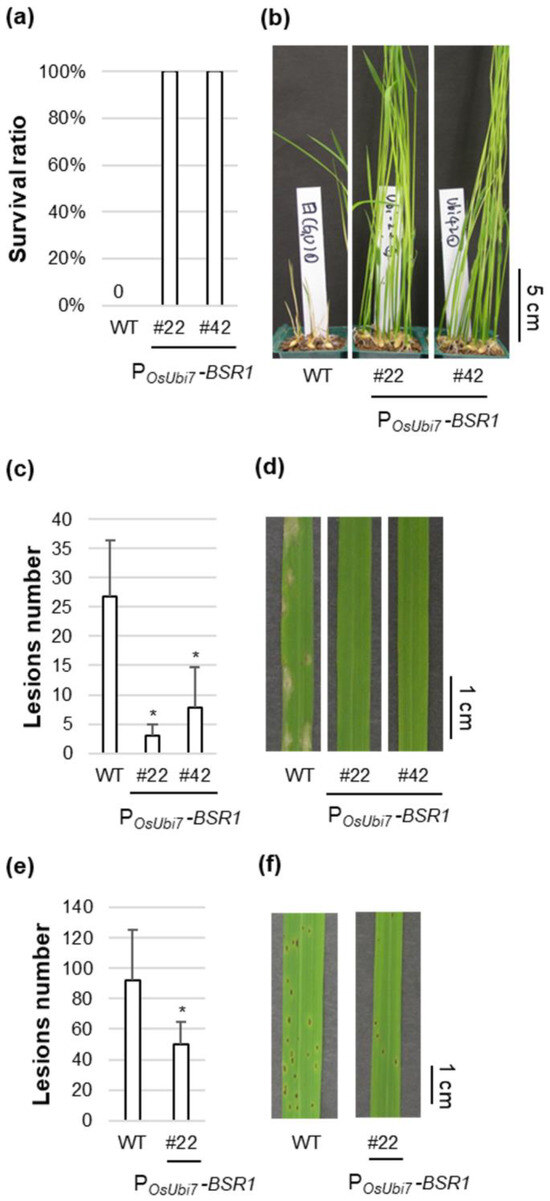
Figure 4.
Disease resistance in the representative POsUBi7-BSR1 lines: (a,b) Disease resistance to Burkholderia glumae in POsUbi7-BSR1 lines. (a) Pre-germinated T4 seeds of POsUbi7-BSR1 and WT lines were inoculated with B. glumae. The inoculum concentration was OD520 = 0.0004. Survival ratio was calculated 8 d post-inoculation (n = 9–20). Tests were repeated three times with similar results. (b) Photographs of B. gluma-infected shoots in POsUbi7-BSR1 and WT lines 8 d post-inoculation. (c,d) Disease resistance to Pyricularia oryzae in POsUbi7-BSR1 lines. (c) Lesion numbers on P. oryzae-infected T4 leaves in POsUbi7-BSR1 and WT lines 6 d post-inoculation. The inoculum concentration was 1.3 × 105 conidia/mL. Lesion numbers in POsUbi7-BSR1 plants were significantly lower than those in WT plants (* p < 0.05 by Dunnett’s test). Values are mean ± standard deviation (n = 3–7). (d) Photographs of leaves infected with P. oryzae in POsUbi7-BSR1 and WT 7 d post-inoculation. (e,f) Disease resistance to Cochliobolus miyabeanus in POsUbi7-BSR1 line. (e) Lesion numbers on C. miyabeanus-infected T4 leaves in POsUbi7-BSR1 and WT lines 4 d post-inoculation. The inoculum concentration was 1.4 × 104 conidia/mL. Lesion numbers in POsUbi7-BSR1 plants were significantly lower than in WT plants (* p < 0.05 by t-test). Values are mean ± standard deviation (n = 6–7). (f) Photographs of leaves infected with C. miyabeanus in POsUbi7-BSR1 and WT lines 4 d post-inoculation.
2.3. Resistance to Other Diseases in PPR1b-BSR1 Lines
The progenies of PPR1b-BSR1 lines #23 and #28 were used as representatives for disease resistance tests. First, disease resistance to B. glumae was investigated. The WT plants succumbed to the infection, displaying 0% survival. Lines #23 and #28 exhibited 95 and 35% survival, respectively, indicating their resistance to bacterial seedling rot (Figure 5a,b and Figure S6). Subsequently, the disease resistance to P. oryzae was evaluated. Lines #23 and #28 demonstrated strong resistance, with lesion numbers < 1/4 and <1/5, respectively, compared with the WT (Figure 5c,d). Third, disease resistance to C. miyabeanus was assessed. Lines #23 and #28 displayed strong resistance, with lesion numbers < 1/5 and <1/2 of that of the WT, respectively (Figure 5e,f). In summary, the PPR1b-BSR1 lines (#23 and #28) maintained resistance to at least four pathogens, similar to the PZmUbi-BSR1 and POsUbi7-BSR1 lines.

Figure 5.
Disease resistance in the representative PPR1b-BSR1 lines. (a,b) Disease resistance to Burkholderia glumae in PPR1b-BSR1 line. (a) Pre-germinated T3 seeds of PPR1b-BSR1 and WT lines were inoculated with B. glumae. The inoculum concentration was OD520 = 0.004. The survival ratio was calculated 7 d post-inoculation (n = 20). Tests were repeated three times with similar results. (b) Photographs of B. glumae-infected shoots in PPR1b-BSR1 and WT 7 d post-inoculation. (c,d) Disease resistance to Pyricularia oryzae in PPR1b-BSR1 lines. (c) Lesion numbers on P. oryzae-infected T1 leaves in PPR1b-BSR1 and WT lines 6 d post-inoculation. The inoculum concentration was 1.3 × 105 conidia/mL. Lesion numbers in PPR1b-BSR1 plants were significantly lower than those in WT plants 6 d post-inoculation (* p < 0.05 by Dunnett’s test). Values are mean ± standard deviation (n = 4–7). (d) Photographs of leaves infected with P. oryzae in PPR1b-BSR1 and WT 7 d post-inoculation. (e,f) Disease resistance to Cochliobolus miyabeanus in PPR1b-BSR1 lines. (e) Lesion number on C. miyabeanus-infected T1 leaves in PPR1b-BSR1 and WT lines 4 d post-inoculation. The inoculum concentration was 1.4 × 104 conidia/mL. Lesion numbers in PPR1b-BSR1 plants were significantly lower than in wild-type plants (* p < 0.05 by Dunnett’s test). Values are means ± standard deviation (n = 3–6). (f) Photographs of leaves infected with C. miyabeanus in PPR1b-BSR1 and WT lines 4 d post-inoculation.
2.4. Improvement of Undesirable Traits Detected in the Seeds of PZmUbi-BSR1 Lines
As mentioned above, the two POsUbi7-BSR1 and two PPR1b-BSR1 lines exhibited resistance to the four major diseases, mirroring the disease resistance observed in the PZmUbi-BSR1 lines. The grains of the T4 homo-seeds of the two POsUbi7-BSR1 lines and T3 homo-seeds of the two PPR1b-BSR1 lines were white (Figure 6). Additionally, the issue of reduced germination in PZmUbi-BSR1 seeds was addressed, as these seeds demonstrated a 100% germination rate, similar to that of the WT (Figure 6). As described above, the successful utilization of OsUbi7 and OsPR1b promoters preserved resistance to the four major diseases, effectively improved grain color, and resolved germination concerns in the seeds.
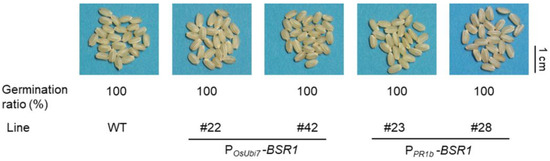
Figure 6.
Improved seed phenotypes in the POsUbi7-BSR1 and PPR1b-BSR1 lines. Photographs of dehusked grains from T4 seeds of the POsUbi7-BSR1 lines, T3 seeds of the PPR1b-BSR1 lines, and WT. The grain color of these transgenic lines was white, as in the WT. The germination ratios (%) of these lines were similar to WT. n = 25.
3. Discussion
When BSR1 was expressed using the rice constitutive promoter POsUbi7 with moderate expression levels and the pathogen-inducible promotor PPR1b to adjust the expression level and timing in rice, both rice plants exhibited sufficient resistance to the four major diseases (rice blast, brown spot, bacterial leaf blight, and bacterial seedling rot), the PZmUbi-BSR1 rice plants did as well. This indicates that disease resistance can be conferred by appropriate expression of BSR1 without using PZmUbi. These promoters have been used to improve the growth of PZmUbi-WRKY45 rice plants with blast and bacterial leaf blight resistance [20,21]. In this study, we have shown that these promoters are useful for improving unfavorable phenotypes (such as the blackish color of brown rice and low germination rate) by overexpressing BSR1. Since the POsUbi7-BSR1 and PPR1b-BSR1 lines are transgenic, they were grown in a small, isolated greenhouse, where exact yield comparisons are not feasible. Although no significant differences were observed compared to the WT, field evaluation is essential for accurate yield assessment.
BSR1-OX rice plants with PZmUbi, which exhibit strong resistance to leaf blight and blast disease, show browning at the infection site [13,18]. In the present study, BSR1-expressing rice plants with POsUBi7 and PPR1b showed browning at the infection site, especially when infected with leaf blight, but this was not as remarkable as that in the case of PZmUbi. We have previously reported that BSR1 is involved in chitin-, peptidoglycan-, and lipopolysaccharide-triggered ROS production and defense responses and ROS production is enhanced in BSR1-OX rice [14,16]. Furthermore, it has been reported that BSR1 is involved in plant responses to OsPeps and damage-associated molecular patterns (DAMPs), and these responses are enhanced in BSR1-OX rice [17]. Thus, the browning observed at the pathogen infection may be caused by cell death due to excessive ROS production by the recognition of microbe-associated molecular patterns (MAMPs) or DAMPs.
In PZmUbi-BSR1 rice plants, overexpression of BSR1 caused a blackish grain color and reduced the seed germination rate, as described above (Figure 1). According to the Rice Expression Profile Database [22] (https://ricexpro.dna.affrc.go.jp/ (accessed on 11, March, 2024)), BSR1 is transcribed at certain levels in leaves and roots at different growth stages and in developing panicles (young anthers, pistils, lemmas, palea, ovaries, embryos, and endosperms). Studies reported that ROS is involved in almost all the stages of growth, development, and differentiation and that they play important roles in programmed cell death and signal transduction [23,24,25]. Therefore, BSR1, which is expressed during ovary, embryo, and endosperm development after fertilization, may contribute to ROS generation in response to signals from ligands other than MAMPs and DAMPs in WT rice. Alternatively, it may respond to the MAMP signals produced by endophytes. Therefore, the overexpression of BSR1 with PZmUbi may induce the production of large amounts of ROS in the endosperm and embryo, causing partial darkening of the endosperm and cell death of the embryo, resulting in a reduced germination rate. In contrast, the improvement in brown rice color and germination rate in POsUbi7-BSR1 and PPR1b-BSR1 rice seeds, similar to those in WT seeds, may be attributed to the moderate control of ROS production by adjusting BSR1 expression to an appropriate level. The upstream signals of BSR1 during panicle and seed development have not yet been elucidated; however, future studies could be conducted to uncover the ligands involved.
The BSR1 gene has also been introduced into plants other than rice and has been shown to confer disease resistance. In Arabidopsis, the overexpression of BSR1 under the 35S promoter containing two enhancers resulted in disease resistance to three pathogens, including the bacterium Pst DC3000 as well as fungi C. higginsianum and R. solani [13,26]. When overexpressed in various plants under the same promoter as in Arabidopsis, BSR1 confers disease resistance to Pst DC3000 and R. solani in tomatoes and to R. solani in torenia. BSR1-overexpressed sugarcane is resistant to one of the most serious diseases caused by the fungus Sporisorium scitamineum [26]. When BSR1 was appropriately expressed in these plants, the transgenic plants exhibited normal growth and morphology similar to that of the WT and retained disease resistance, although a dwarf phenotype was observed in sugarcane with the highest expression level of BSR1 [26]. By selecting appropriate promoters and fine-tuning the expression level of BSR1, it would be possible to generate plants with broad-spectrum disease resistance without compromising the growth in other crops as well as in rice.
Recently, POsUbi7-BSR1 rice was also reported to confer resistance to the chewing herbivore Mythimna loreyi Duponchel (Lepodoptera: Noctuidae), a rice pest [17]. As described above, by optimizing the expression level of BSR1 using a suitable promoter such as OsUbi7, we were able to generate rice plants with BSR to four major diseases and one pest while minimizing the adverse effects of overexpression. The next step will be to determine whether these plants are effective against a wide range of pathogens and pests. Another question is whether similar traits can be maintained in a field. We cannot exclude the possibility that unfavorable phenotypes may occur due to somaclonal mutations derived from callus cultures during the generation process of transgenic rice when released into the field environment. We also cannot exclude the possibility of other problems arising from the accumulation of mutations over several generations. In such cases, it may be necessary to produce a larger number of transgenic rice plants and reduce the effects of cultural mutations through multiple backcrossings. Using a leaf-specific promoter may be effective because undesirable traits of BSR1 were found in the seed.
4. Materials and Methods
4.1. Plasmid Construction and Transformation
The PPR1b-BSR1 plasmid was constructed as follows: Briefly, the WRKY45 cDNA of the PPR1b:WRKY45:TT plasmid [21] was replaced with BSR1 cDNA. In detail, the BSR1 cDNA (AK070024; Os09t0533600-01) provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences was first amplified by PCR using the following primers: 5′-TTGATTAACTAAGCTTGTGCGTGCGTGCGTGCTTGC-3′ and 5′-TGATTTCAGCGGATCTCGTCTCTGTGTCTCTCTTT-3′. Subsequently, the PPR1b:WRKY45:TT plasmid was digested with BamHI and partially digested with HindIII to excise the WRKY45 cDNA from the vector, and the amplified BSR1 cDNA was incorporated in place of the WRKY45 cDNA using an In-Fusion HD Cloning Kit w/Cloning Enhancer (Takara Bio, Tokyo, Japan). Transgenic rice lines were obtained from Nipponbare using the resulting plasmid via the Agrobacterium-mediated method [27].
Construction of the POsUbi7-BSR1 plasmid and generation of POsUbi7-BSR1 rice lines have been previously reported briefly [17]. In detail, the BSR1 cDNA was amplified by PCR using the following primers: 5′-GCAAAAGAAGAAGCTGTGCGTGCGTGCGTGCTTGC-3′ and 5′-TGATTTCAGCGGATCTGCTCTCTGTGTCTCTCTTT-3′. Subsequently, the P OsUbi7:WRKY45:TT plasmid was digested with BamHI and partially digested with HindIII to excise the WRKY45 cDNA from the vector. The WRKY45 cDNA was then replaced with amplified BSR1 cDNA using an In-Fusion HD Cloning Kit with Cloning Enhancer.
4.2. RNA Extraction and Quantitative Real-Time (qRT)-PCR Analysis
Total RNA was extracted and purified from rice leaves using the Sepasol-RNA Super G reagent (Nacalai Tesque, Kyoto, Japan) according to the manufacturer’s protocol. First-strand cDNAs were synthesized from equal amounts of total RNA in a volume of 5 μL using the PrimeScript RT Reagent Kit (Takara, Tokyo, Japan), according to the manufacturer’s protocol. The Thermal Cycler Dice TP800 system (Takara) and Kapa SYBR FAST qPCR kit (Kapa Biosystems, Cape Town, South Africa) were used for qRT-PCR analysis according to the manufacturer’s instructions. Primers used for qRT-PCR were as follows: for BSR1, 5′-CCGGGACTTCAAAGCATCTAAC-3′ and 5′-TGTTGGTCCCTCCCTTGCT-3′; for Rubq1, 5′-GGAGCTGCTGCTGTTCTAGG-3′ and 5′-TTCAGACACCATCAAACCAGA-3′, serving as an internal control. BSR1 transcript levels were normalized to the endogenous rice reference gene Rubq1. The relative expression level of each gene was calculated using the 2−ΔΔCt expression ratio, which corrects for gene-specific PCR amplification efficiencies [28].
Overexpression of BSR1 cDNA was confirmed by qRT-PCR analysis of independent T0 plants from the POsUbi7-BSR1 lines, and their progenies were subsequently utilized in further experiments.
4.3. Plant Materials and Culture
Rice (Oryza sativa L. cv. Nipponbare) was used as the WT control. Seeds from T1 to T4 of the POsUbi7-BSR1 and PPR1b-BSR1 lines were sown on half-strength MS medium (Wako, Osaka, Japan) containing 3% sucrose, 0.4% Gelrite (Wako), and hygromycin B (30 μg/mL; Wako), and the hygromycin-resistant seedlings were selected on this medium. WT seeds were sown and grown on the same medium without hygromycin B. WT and hygromycin-resistant transgenic seedlings were then transplanted into pots containing soil (Bonsol no. 2; Sumitomo Kagaku Kougyo, Osaka, Japan) and grown in a greenhouse at 27–30 °C, as previously described [19].
4.4. Pathogens and Pathogen Cultures
The bacterial isolates used in this study were T7174 (MAFF311018, race I) and T7133 (MAFF311020, race III) from Xanthomonas oryzae pv. oryzae (Xoo), and AZ8204 (MAFF301682) from Burkholderia glumae. Additionally, the fungal isolates were Kyu89-246 (MAFF101506, race 003.0) from Pyricularia oryzae and H11-42-1 from Cochliobolus miyabeanus. Culture procedures for these pathogens were conducted as previously described [18].
4.5. Bacterial and Fungal Pathogen Resistance Assay
For the Xoo resistance tests, a suspension of Xoo adjusted to OD600 = 0.3 in water was inoculated onto seedlings of WT and transgenic lines at the 5- to 10-leaf stage using the clip and dip method, as previously described [18]. For B. glumae resistance assays, pre-germinated seeds of WT and transgenic lines were soaked in a suspension of B. glumae adjusted to OD520 = 0.0004–0.004 and subjected to vacuum treatment following previously described protocols [18]. For P. oryzae resistance assays, conidia of a compatible race 003 (isolate Kyu89-246) were suspended in 0.01% Tween 20 at a density of 1.3 × 105/mL and sprayed onto seedlings of WT and transgenic lines at the 5-leaf stage, as previously described [18]. For C. miyabeanus resistance assays, a suspension of C. miyabeanus conidia adjusted to 1.4–5 × 104/mL was sprayed onto seedlings of WT and transgenic lines at the 7-leaf stage, following previously established procedures [18].
4.6. Brown Rice Color Evaluation and Germination Test
The brown rice color of WT and transgenic rice seeds (25 seeds each) was visually evaluated. For the germination test, sterilized brown rice was sown in half-strength MS medium without hygromycin B. After 3 d, seed germination was examined, and the germination ratio was calculated.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants13081138/s1, Figure S1: Disease resistance to Xanthomonas oryzae pv. oryzae (T7174, race I) in POsUbi7-BSR1 T1 lines; Figure S2: Disease resistance to Xanthomonas oryzae pv. oryzae (T7133, race III) in POsUbi7-BSR1 T1 lines; Figure S3: BSR1 expression levels in POsUbi7-BSR1 T4 lines; Figure S4: Disease resistance to Cochliobolus miyabeanus in POsUbi7-BSR1 T4 line; Figure S5: Disease resistance to panicle blast caused by Pyricularia oryzae (isolate Kyu89-246) in the descendants of the POsUbi7-BSR1 #22 and #42 lines; Figure S6: Disease resistance to Burkholderia glumae in PPR1b-BSR1 T2 lines; Method S1: Evaluation of panicle blast resistance [29,30,31].
Author Contributions
Conceptualization, H.T. and M.M.; formal analysis, S.M., H.I. and H.S.; methodology, S.G.; resources, S.G. and H.T.; funding acquisition, M.M.; project administration, M.M.; writing-original draft, S.M., H.I. and M.M.; writing-review and editing, H.T. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the JSPS KAKENHI, grant number JP20H02953, and Genomics-based Technology for Agricultural Improvement, grant number GMO-1006a (Ministry of Agriculture, Forestry, and Fisheries of Japan).
Data Availability Statement
Data are contained within the article.
Acknowledgments
We thank Lois Ishizaki, Naoko Minami, and Sachiko Ohashi (Institute of Agrobiological Sciences, NARO, Tsukuba, Japan) for their support with overall technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.-L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed]
- Skamnioti, P.; Gurr, S.J. Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Li, M.; Wang, X.; Xu, Z.; Wu, K.; Sun, Q.; Du, H.; Younas, M.U.; Zhang, Y.; Feng, Z. Identification of Elite R-Gene Combinations against Blast Disease in Geng Rice Varieties. Int. J. Mol. Sci. 2023, 24, 3984. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.-R.; Pan, H. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.; Ning, Y.; He, Z.; Wang, G.-L. Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Annu. Rev. Plant Biol. 2020, 71, 575–603. [Google Scholar] [CrossRef] [PubMed]
- Shimono, M.; Koga, H.; Akagi, A.; Hayashi, N.; Goto, S.; Sawada, M.; Kurihara, T.; Matsushita, A.; Sugano, S.; Jiang, C.J.; et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 2012, 13, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.J.; Shimono, M.; Maeda, S.; Inoue, H.; Mori, M.; Hasegawa, M.; Sugano, S.; Takatsuji, H. Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant Microbe Interact. 2009, 22, 820–829. [Google Scholar] [CrossRef]
- Peng, X.; Hu, Y.; Tang, X.; Zhou, P.; Deng, X.; Wang, H.; Guo, Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 2012, 236, 1485–1498. [Google Scholar] [CrossRef]
- Helliwell, E.E.; Wang, Q.; Yang, Y. Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 2013, 11, 33–42. [Google Scholar] [CrossRef]
- Veronese, P.; Nakagami, H.; Bluhm, B.; Abuqamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.; Hirt, H.; Mengiste, T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef]
- Rao, S.; Zhou, Z.; Miao, P.; Bi, G.; Hu, M.; Wu, Y.; Feng, F.; Zhang, X.; Zhou, J.M. Roles of Receptor-Like Cytoplasmic Kinase VII Members in Pattern-Triggered Immune Signaling. Plant Physiol. 2018, 177, 1679–1690. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Maeda, S.; Sugano, S.; Ohtake, M.; Hayashi, N.; Ichikawa, T.; Kondou, Y.; Kuroda, H.; Horii, Y.; Matsui, M.; et al. Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 2011, 9, 466–485. [Google Scholar] [CrossRef]
- Kanda, Y.; Yokotani, N.; Maeda, S.; Nishizawa, Y.; Kamakura, T.; Mori, M. The receptor-like cytoplasmic kinase BSR1 mediates chitin-induced defense signaling in rice cells. Biosci. Biotechnol. Biochem. 2017, 81, 1497–1502. [Google Scholar] [CrossRef]
- Sugano, S.; Maeda, S.; Hayashi, N.; Kajiwara, H.; Inoue, H.; Jiang, C.J.; Takatsuji, H.; Mori, M. Tyrosine phosphorylation of a receptor-like cytoplasmic kinase, BSR1, plays a crucial role in resistance to multiple pathogens in rice. Plant J. 2018, 96, 1137–1147. [Google Scholar] [CrossRef]
- Kanda, Y.; Nakagawa, H.; Nishizawa, Y.; Kamakura, T.; Mori, M. Broad-Spectrum Disease Resistance Conferred by the Overexpression of Rice RLCK BSR1 Results from an Enhanced Immune Response to Multiple MAMPs. Int. J. Mol. Sci. 2019, 20, 5523. [Google Scholar] [CrossRef]
- Kanda, Y.; Shinya, T.; Maeda, S.; Mujiono, K.; Hojo, Y.; Tomita, K.; Okada, K.; Kamakura, T.; Galis, I.; Mori, M. BSR1, a Rice Receptor-like Cytoplasmic Kinase, Positively Regulates Defense Responses to Herbivory. Int. J. Mol. Sci. 2023, 24, 10395. [Google Scholar] [CrossRef]
- Maeda, S.; Hayashi, N.; Sasaya, T.; Mori, M. Overexpression of BSR1 confers broad-spectrum resistance against two bacterial diseases and two major fungal diseases in rice. Breed. Sci. 2016, 66, 396–406. [Google Scholar] [CrossRef][Green Version]
- Mizobuchi, R.; Fukuoka, S.; Tsushima, S.; Yano, M.; Sato, H. QTLs for Resistance to Major Rice Diseases Exacerbated by Global Warming: Brown Spot, Bacterial Seedling Rot, and Bacterial Grain Rot. Rice 2016, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Sasakura-Shimoda, F.; Suetsugu, M.; Selvaraj, M.G.; Hayashi, N.; Yamazaki, M.; Ishitani, M.; Shimono, M.; Sugano, S.; Matsushita, A.; et al. Development of disease-resistant rice by optimized expression of WRKY45. Plant Biotechnol. J. 2015, 13, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Sasakura-Shimoda, F.; Yamazaki, M.; Hayashi, N.; Suetsugu, M.; Ochiai, H.; Takatsuji, H. Development of disease-resistant rice by pathogen-responsive expression of WRKY45. Plant Biotechnol. J. 2016, 14, 1127–1138. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef]
- Bethke, P.C.; Jones, R.L. Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J. 2001, 25, 19–29. [Google Scholar] [CrossRef]
- Gapper, C.; Dolan, L. Control of plant development by reactive oxygen species. Plant Physiol. 2006, 141, 341–345. [Google Scholar] [CrossRef]
- Kimura, S.; Kaya, H.; Kawarazaki, T.; Hiraoka, G.; Senzaki, E.; Michikawa, M.; Kuchitsu, K. Protein phosphorylation is a prerequisite for the Ca2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2012, 1823, 398–405. [Google Scholar] [CrossRef]
- Maeda, S.; Ackley, W.; Yokotani, N.; Sasaki, K.; Ohtsubo, N.; Oda, K.; Mori, M. Enhanced Resistance to Fungal and Bacterial Diseases Due to Overexpression of BSR1, a Rice RLCK, in Sugarcane, Tomato, and Torenia. Int. J. Mol. Sci. 2023, 24, 3644. [Google Scholar] [CrossRef]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N., Jr. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T.; et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Hayashi, N. The Panicle Blast Resistance Mechanism of qPbm11 in the Rice Cultivar Miyazaki-mochi is Independent from that of Pb1. Jpn. Agric. Res. Q. JARQ 2019, 53, 289–293. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).