Identification of Genomic Regions for Traits Associated with Flowering in Cassava (Manihot esculenta Crantz)
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation, Distribution and Heritability Estimates of Flowering-Associated Traits
2.2. Correlation between Flowering and Flowering-Associated Traits
2.3. Genotyping and SNP Identification
2.4. Population Structure and Kinship
2.5. Linkage Disequilibrium Estimation
2.6. Marker–Trait Association Mapping
2.7. Identification of Candidate Genes
3. Discussion
3.1. Variation in Phenotypic Traits Related to Flowering
3.2. Correlation between Flowering and Flowering-Associated Traits
3.3. Population Structure and Kinship
3.4. Linkage Disequilibrium
3.5. Marker–Trait Association Mapping
3.6. Putative Candidate Genes Linked to Marker Loci for Flowering-Associated Traits
4. Materials and Methods
4.1. Plant Material and Field Trials
4.2. Flowering Traits Evaluated
4.3. Genomic DNA Extraction and Genotyping
4.4. SNP Calling and Annotation
4.5. Statistical Analyses
4.6. Population Structure and Kinship
4.7. Linkage Disequilibrium Estimation
4.8. Marker–Trait Association Mapping
4.9. Identification of Putative Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaur, K.; Ahluwalia, P. Cassava as potential crop for the food and fermentation industry: A review. Int. J. Food Ferment. Technol. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Nanbol, K.K.; Namo, O. The contribution of root and tuber crops to food security: A review. J. Agric. Sci. Technol. B 2019, 9, 221–233. [Google Scholar]
- Li, S.; Cui, Y.; Zhou, Y.; Luo, Z.; Liu, J.; Zhao, M. The industrial applications of cassava: Current status, opportunities and prospects. J. Sci. Food Agric. 2017, 97, 2282–2290. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, H.; Hershey, C.; Iglesias, C.; Zhang, X. Fifty years of a public cassava breeding program: Evolution of breeding objectives, methods, and decision-making processes. Theor. Appl. Genet. 2021, 134, 2335–2353. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, H.; Chareinsuk, R. Excellence in cassava breeding: Perspectives for the future. Crop Breed. Genet. Genom. 2020, 2, e200008. [Google Scholar] [CrossRef]
- Bandeira, E.M.S.; Andrade, L.R.B.; Souza, E.H.; Alves, A.A.C.; de Oliveira, E.J. Reproductive barriers in cassava: Factors and implications for genetic improvement. PLoS ONE 2021, 16, e0260576. [Google Scholar] [CrossRef]
- Cock, J.H.; Connor, D.J. Chapter 19—Cassava. In Crop Physiology Case Histories for Major Crops; Sadras, V.O., Calderini, D.F., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 588–633. [Google Scholar] [CrossRef]
- Alves, A.A.C. Cassava botany and phyiology. In Cassava: Biology, Production and Utilization; Hillocks, R.J., Thresh, J.M., Bellotti, A.C., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 67–89. [Google Scholar]
- Ceballos, H.; Kulakow, P.; Hershey, C. Cassava breeding: Current status, bottlenecks and the potential of biotechnology tools. Trop. Plant Biol. 2012, 5, 73–87. [Google Scholar] [CrossRef]
- Perera, P.I.; Quintero, M.; Dedicova, B.; Kularatne, J.D.; Ceballos, H. Comparative morphology, biology and histology of reproductive development in three lines of Manihot esculenta Crantz (Euphorbiaceae: Crotonoideae). AoB Plants 2013, 5, pls046. [Google Scholar] [CrossRef]
- Silva Souza, L.; Cunha Alves, A.A.; de Oliveira, E.J. Phenological diversity of flowering and fruiting in cassava germplasm. Sci. Hortic. 2020, 265, 109253. [Google Scholar] [CrossRef]
- Ceballos, H.; Kawuki, R.S.; Gracen, V.E.; Yencho, G.C.; Hershey, C.H. Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: A case study for cassava. Theor. Appl. Genet. 2015, 128, 1647–1667. [Google Scholar] [CrossRef]
- McSteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant Biol. 2005, 56, 353–374. [Google Scholar] [CrossRef]
- Gaarslev, N.; Swinnen, G.; Soyk, S. Meristem transitions and plant architecture-learning from domestication for crop breeding. Plant Physiol. 2021, 187, 1045–1056. [Google Scholar] [CrossRef]
- Sun, L.; Nie, T.; Chen, Y.; Yin, Z. From floral induction to blooming: The molecular mysteries of flowering in woody plants. Int. J. Mol. Sci. 2022, 23, 10959. [Google Scholar] [CrossRef]
- Bendahmane, M.; Dubois, A.; Raymond, O.; Bris, M.L. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 2013, 64, 847–857. [Google Scholar] [CrossRef]
- Krishnamurthy, K.V.; Bahadur, B. Genetics of flower development. In Plant Biology and Biotechnology; Bahadur, B., Rajam, M.V., Sahijram, L., Krishnamurthy, K.V., Eds.; Springer: New Delhi, India, 2015; Volume 1, pp. 385–407. [Google Scholar]
- Mallik, M. Flowering control mechanisms in plants and its importance in crop production and breeding. Int. J. Pure Appl. Biosci. 2018, 6, 1033–1038. [Google Scholar] [CrossRef]
- Tokunaga, H.; Quynh, D.T.N.; Anh, N.H.; Nhan, P.T.; Matsui, A.; Takahashi, S.; Tanaka, M.; Anh, N.M.; Dong, N.V.; Ham, L.H.; et al. Field transcriptome analysis reveals a molecular mechanism for cassava flowering in a mountainous environment in SE Asia. Plant Mol. Biol. 2020, 109, 233–248. [Google Scholar] [CrossRef]
- Hyde, P.T.; Setter, T.L. Long-day photoperiod and cool temperature induce flowering in cassava: Expression of signaling genes. Front. Plant Sci. 2022, 13, 973206. [Google Scholar] [CrossRef]
- Adeyemo, O.S.; Hyde, P.T.; Setter, T.L. Identification of FT family genes that respond to photoperiod, temperature and genotype in relation to flowering in cassava (Manihot esculenta, Crantz). Plant Reprod. 2019, 32, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Bull, S.E.; Alder, A.; Barsan, C.; Kohler, M.; Hennig, L.; Gruissem, W.; Vanderschuren, H. FLOWERING LOCUS T triggers early and fertile flowering in glasshouse cassava (Manihot esculenta Crantz). Plants 2017, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Lema, M. Marker Assisted Selection in comparison to conventional plant breeding: Review article. Agric. Res. Technol. Open Access J. 2018, 14, 555914. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, B.; Ma, N.; Liu, X.; Qin, M.; Zhang, Y.; Wang, K.; Guo, N.; Zuo, K.; Liu, X.; et al. Quantitative trait locus mapping and identification of candidate genes controlling flowering time in Brassica napus L. Front. Plant Sci. 2020, 11, 626205. [Google Scholar] [CrossRef]
- Lu, J.; Shi, Y.; Yin, X.; Liu, S.; Liu, C.; Wen, D.; Li, W.; He, X.; Yang, T. The genetic mechanism of sex type, a complex quantitative trait, in Ricinus communis L. Ind. Crops Prod. 2019, 128, 590–598. [Google Scholar] [CrossRef]
- Sun, F.; Liu, P.; Ye, J.; Lo, L.C.; Cao, S.; Li, L.; Yue, G.H.; Wang, C.M. An approach for jatropha improvement using pleiotropic QTLs regulating plant growth and seed yield. Biotechnol. Biofuels 2012, 5, 42. [Google Scholar] [CrossRef]
- Sraphet, S.; Boonchanawiwat, A.; Thanyasiriwat, T.; Thaikert, R.; Whankaew, S.; Smith, D.R.; Boonseng, O.; Lightfoot, D.A.; Triwitayakorn, K. Quantitative trait loci underlying root yield and starch content in an F1 derived cassava population (Manihot esculenta Crantz). J. Agric. Sci. 2016, 155, 569–581. [Google Scholar] [CrossRef]
- Hmwe, N.H.; Sraphet, S.; Srisawad, N.; Smith, D.R.; Triwitayakorn, K. Identification of QTL underlying plant height and first branch height of cassava. Philipp. J. Sci. 2022, 15, 683. [Google Scholar] [CrossRef]
- Tappiban, P.; Smith, D.R.; Triwitayakorn, K.; Bao, J. Recent understanding of starch biosynthesis in cassava for quality improvement: A review. Trends Food Sci. Technol. 2019, 83, 167–180. [Google Scholar] [CrossRef]
- Garcia-Oliveira, A.L.; Kimata, B.; Kasele, S.; Kapinga, F.; Masumba, E.; Mkamilo, G.; Sichalwe, C.; Bredeson, J.V.; Lyons, J.B.; Shah, T.; et al. Genetic analysis and QTL mapping for multiple biotic stress resistance in cassava. PLoS ONE 2020, 15, e0236674. [Google Scholar] [CrossRef] [PubMed]
- Challa, S.; Neelapu, N.R.R. Genome-Wide Association Studies (GWAS) for abiotic stress tolerance in plants. In Biochemical, Physiological and Molecular Avenues for Combating Abiotic Stress Tolerance in Plants; Academic Press: Vienna, Austria, 2018; pp. 135–150. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Kostova, D.; Bulut, M.; Fernie, A.R. Genome-wide association studies: Assessing trait characteristics in model and crop plants. Cell. Mol. Life Sci. 2021, 78, 5743–5754. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Togninalli, M.; Seren, U.; Meng, D.; Fitz, J.; Nordborg, M.; Weigel, D.; Borgwardt, K.; Korte, A.; Grimm, D.G. The AraGWAS Catalog: A curated and standardized Arabidopsis thaliana GWAS catalog. Nucleic Acids Res. 2018, 46, D1150–D1156. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, E.; Kocher, T.; Filiault, D.L.; Nordborg, M. Revisiting a GWAS peak in Arabidopsis thaliana reveals possible confounding by genetic heterogeneity. Heredity 2021, 127, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Morinaka, Y.; Wang, F.; Huang, P.; Takehara, S.; Hirai, T.; Ito, A.; Koketsu, E.; Kawamura, M.; Kotake, K.; et al. GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc. Natl. Acad. Sci. USA 2019, 116, 21262–21267. [Google Scholar] [CrossRef] [PubMed]
- Ruanjaichon, V.; Khammona, K.; Thunnom, B.; Suriharn, K.; Kerdsri, C.; Aesomnuk, W.; Yongsuwan, A.; Chaomueang, N.; Thammapichai, P.; Arikit, S.; et al. Identification of gene associated with sweetness in corn (Zea mays L.) by genome-wide association study (GWAS) and development of a functional SNP marker for predicting sweet corn. Plants 2021, 10, 1239. [Google Scholar] [CrossRef] [PubMed]
- Tomkowiak, A.; Bocianowski, J.; Wolko, L.; Adamczyk, J.; Mikolajczyk, S.; Kowalczewski, P.L. Identification of markers associated with yield traits and morphological features in maize (Zea mays L.). Plants 2019, 8, 330. [Google Scholar] [CrossRef]
- Zhou, H.; Xiao, X.; Asjad, A.; Han, D.; Zheng, W.; Xiao, G.; Huang, Y.; Zhou, Q. Integration of GWAS and transcriptome analyses to identify SNPs and candidate genes for aluminum tolerance in rapeseed (Brassica napus L.). BMC Plant Biol. 2022, 22, 130. [Google Scholar] [CrossRef]
- Jaiswal, V.; Gupta, S.; Gahlaut, V.; Muthamilarasan, M.; Bandyopadhyay, T.; Ramchiary, N.; Prasad, M. Genome-Wide Association Study of major agronomic traits in foxtail millet (Setaria italica L.) using ddRAD sequencing. Sci. Rep. 2019, 9, 5020. [Google Scholar] [CrossRef]
- do Carmo, C.D.; e Sousa, M.B.; Brito, A.C.; de Oliveira, E.J. Genome-wide association studies for waxy starch in cassava. Euphytica 2020, 216, 82. [Google Scholar] [CrossRef]
- Rabbi, I.Y.; Udoh, L.I.; Wolfe, M.; Parkes, E.Y.; Gedil, M.A.; Dixon, A.; Ramu, P.; Jannink, J.L.; Kulakow, P. Genome-wide association mapping of correlated traits in cassava: Dry matter and total carotenoid content. Plant Genome 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Ikeogu, U.N.; Akdemir, D.; Wolfe, M.D.; Okeke, U.G.; Chinedozi, A.; Jannink, J.L.; Egesi, C.N. Genetic correlation, genome-wide association and genomic prediction of portable NIRS predicted carotenoids in cassava roots. Front. Plant Sci. 2019, 10, 1570. [Google Scholar] [CrossRef]
- Esuma, W.; Herselman, L.; Labuschagne, M.T.; Ramu, P.; Lu, F.; Baguma, Y.; Buckler, E.S.; Kawuki, R.S. Genome-wide association mapping of provitamin A carotenoid content in cassava. Euphytica 2016, 212, 97–110. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Rabbi, I.Y.; Egesi, C.; Hamblin, M.; Kawuki, R.; Kulakow, P.; Lozano, R.; Carpio, D.P.; Ramu, P.; Jannink, J.L. Genome-wide association and prediction reveals genetic architecture of cassava mosaic disease resistance and prospects for rapid genetic improvement. Plant Genome 2016, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nandudu, L.; Kawuki, R.; Ogbonna, A.; Kanaabi, M.; Jannink, J.-L. Genetic dissection of cassava brown streak disease in a genomic selection population. Front. Plant Sci. 2023, 13, 5627. [Google Scholar] [CrossRef] [PubMed]
- Kayondo, S.I.; Pino Del Carpio, D.; Lozano, R.; Ozimati, A.; Wolfe, M.; Baguma, Y.; Gracen, V.; Offei, S.; Ferguson, M.; Kawuki, R.; et al. Genome-wide association mapping and genomic prediction for CBSD resistance in Manihot esculenta. Sci. Rep. 2018, 8, 1549. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Silva, P.P.; Sousa, M.B.e.; de Oliveira, E.J.; Morgante, C.V.; de Oliveira, C.R.S.; Vieira, S.L.; Borel, J.C. Genome-wide association study of drought tolerance in cassava. Euphytica 2021, 217, 60. [Google Scholar] [CrossRef]
- Diack, O.; Kanfany, G.; Gueye, M.C.; Sy, O.; Fofana, A.; Tall, H.; Serba, D.D.; Zekraoui, L.; Berthouly-Salazar, C.; Vigouroux, Y.; et al. GWAS unveils features between early- and late-flowering pearl millets. BMC Genom. 2020, 21, 777. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Cairns, J.E.; Babu, R.; Gowda, M.; Makumbi, D.; Magorokosho, C.; Zhang, A.; Liu, Y.; Wang, N.; Hao, Z.; et al. Genome-wide association mapping and genomic prediction analyses reveal the genetic architecture of grain yield and flowering time under drought and heat stress conditions in maize. Front. Plant Sci. 2018, 9, 1919. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.A.; Habyarimana, E.; Karaköy, T.; Baloch, F.S. Genetic dissection of days to flowering via genome-wide association studies in Turkish common bean germplasm. Physiol. Mol. Biol. Plants 2021, 27, 1609–1622. [Google Scholar] [CrossRef]
- Raggi, L.; Caproni, L.; Carboni, A.; Negri, V. Genome-wide association study reveals candidate genes for flowering time variation in common bean (Phaseolus vulgaris L.). Front. Plant Sci. 2019, 10, 962. [Google Scholar] [CrossRef]
- Sabag, I.; Morota, G.; Peleg, Z. Genome-wide association analysis uncovers the genetic architecture of tradeoff between flowering date and yield components in sesame. BMC Plant Biol. 2021, 21, 549. [Google Scholar] [CrossRef]
- Xu, L.; Hu, K.; Zhang, Z.; Guan, C.; Chen, S.; Hua, W.; Li, J.; Wen, J.; Yi, B.; Shen, J.; et al. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res. 2016, 23, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Akhatar, J.; Goyal, A.; Kaur, N.; Atri, C.; Mittal, M.; Singh, M.P.; Kaur, R.; Rialch, I.; Banga, S.S. Genome wide association analyses to understand genetic basis of flowering and plant height under three levels of nitrogen application in Brassica juncea (L.) Czern & Coss. Sci. Rep. 2021, 11, 4278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, Q.; Cregan, P.B.; Nelson, R.L.; Wang, X.; Wu, J.; Jiang, G.L. Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genom. 2015, 16, 217. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, X.; Lu, C.; Ye, J.; Zou, M.; Lu, K.; Feng, S.; Pei, J.; Liu, C.; Zhou, X.; et al. Genome-wide association studies of 11 agronomic traits in cassava (Manihot esculenta Crantz). Front. Plant Sci. 2018, 9, 503. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.A.; Iacuone, S.; Li, S.F.; Parish, R.W. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 2011, 23, 2209–2224. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Yang, W.; He, Y.; Amasino, R.M. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 2007, 19, 2975–2987. [Google Scholar] [CrossRef]
- Bak, S.; Tax, F.E.; Feldmann, K.A.; Galbraith, D.W.; Feyereisen, R. CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in arabidopsis. Plant Cell 2001, 13, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Distefano, A.M.; Setzes, N.; Cascallares, M.; Fiol, D.F.; Zabaleta, E.; Pagnussat, G.C. Roles of cytochromes P450 in plant reproductive development. Int. J. Dev. Biol. 2021, 65, 187–194. [Google Scholar] [CrossRef]
- Burko, Y.; Geva, Y.; Refael-Cohen, A.; Shleizer-Burko, S.; Shani, E.; Berger, Y.; Halon, E.; Chuck, G.; Moshelion, M.; Ori, N. From organelle to organ: ZRIZI MATE-Type transporter is an organelle transporter that enhances organ initiation. Plant Cell Physiol. 2011, 52, 518–527. [Google Scholar] [CrossRef]
- Xia, C.; Wang, Y.J.; Li, W.Q.; Chen, Y.R.; Deng, Y.; Zhang, X.Q.; Chen, L.Q.; Ye, D. The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis. Plant J. 2010, 63, 189–202. [Google Scholar] [CrossRef]
- Ren, D.; Rao, Y.; Yu, H.; Xu, Q.; Cui, Y.; Xia, S.; Yu, X.; Liu, H.; Hu, H.; Xue, D.; et al. MORE FLORET1 encodes a MYB transcription factor that regulates spikelet development in rice. Plant Physiol. 2020, 184, 251–265. [Google Scholar] [CrossRef]
- Edstam, M.M.; Edqvist, J. Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant. 2014, 152, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Lebel-Hardenack, S.; Ye, D.; Koutnikova, H.; Saedler, H.; Grant, S.R. Conserved expression of a TASSELSEED2 homolog in the tapetum of the dioecious Silene latifolia and Arabidopsis thaliana. Plant J. 1997, 12, 515–526. [Google Scholar] [CrossRef]
- Zhao, H.; Guan, X.; Xu, Y.; Wang, Y. Characterization of novel gene expression related to glyoxal oxidase by agro-infiltration of the leaves of accession Baihe-35-1 of Vitis pseudoreticulata involved in production of H2O2 for resistance to Erysiphe necator. Protoplasma 2013, 250, 765–777. [Google Scholar] [CrossRef]
- Wang, R.; Liu, X.; Liang, S.; Ge, Q.; Li, Y.; Shao, J.; Qi, Y.; An, L.; Yu, F. A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis. J. Exp. Bot. 2015, 66, 6327–6343. [Google Scholar] [CrossRef]
- Wei, K.; Zhong, X. Non-specific lipid transfer proteins in maize. BMC Plant Biol. 2014, 14, 281. [Google Scholar] [CrossRef]
- Taneva, K.; Bozhanova, V.; Petrova, I. Variability, heritability and genetic advance of some grain quality traits and grain yield in durum wheat genotypes. Bulg. J. Agric. Sci. 2019, 25, 288–295. [Google Scholar]
- Favour, E.; Emeka, N.; Chiedozie, E.; Bunmi, O.; Emmanuel, O. Genetic variability, heritability and variance components of some yield and yield related traits in second backcross population (BC2) of cassava. Afr. J. Plant Sci. 2017, 11, 185–189. [Google Scholar] [CrossRef]
- Rodrmguez, E.P.B.; Morante, N.; Salazar, S.; Hyde, P.T.; Setter, T.L.; Kulakow, P.; Aparicio, J.S.; Zhang, X. Flower-inducing technology facilitates speed breeding in cassava. Front. Plant Sci. 2023, 14, 1172056. [Google Scholar] [CrossRef]
- Aquilino, L.; Pariyo, A.; Baguma, Y.; Edema, R.; Gibson, P.; Bisikwa, J. Genotypes by environment effect on flowering and seed set in cassava, Manihot esculenta Crantz in Uganda. J. Food Nutr. Agric. 2021, 4, 26–37. [Google Scholar] [CrossRef]
- Ren, D.; Cai, X.; Lin, Q.; Ye, H.; Teng, J.; Li, J.; Ding, X.; Zhang, Z. Impact of linkage disequilibrium heterogeneity along the genome on genomic prediction and heritability estimation. Genet. Sel. Evol. 2022, 54, 47. [Google Scholar] [CrossRef] [PubMed]
- Leelawijitkul, S.; Kongsil, P.; Kittipadakul, P.; Juntawong, P. Correlation between relative gene expression patterns of two Flowering locus T (MeFT1 and MeFT2) in cassava leaf and flowering traits under different flowering induction conditions. Pak. J. Biol. Sci. 2022, 25, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Pineda, M.; Morante, N.; Salazar, S.; Cuásquer, J.; Hyde, P.T.; Setter, T.L.; Ceballos, H. Induction of earlier flowering in cassava through extended photoperiod. Agronomy 2020, 10, 1273. [Google Scholar] [CrossRef]
- Ochoa, A.; Storey, J.D. Estimating FST and kinship for arbitrary population structures. PLoS Genet. 2021, 17, e1009241. [Google Scholar] [CrossRef] [PubMed]
- Meisner, J.; Albrechtsen, A. Inferring population structure and admixture proportions in low-depth NGS data. Genetics 2018, 210, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ralph, P. Local PCA shows how the effect of population structure differs along the genome. Genetics 2019, 211, 289–304. [Google Scholar] [CrossRef]
- Bangarwa, S.K.; Kumawat, R.; Barupal, H.l.; Kumar, A.; Yadav, M.K. Association mapping in crops plants. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 1313–1325. [Google Scholar] [CrossRef]
- Vos, P.G.; Paulo, M.J.; Voorrips, R.E.; Visser, R.G.; van Eck, H.J.; van Eeuwijk, F.A. Evaluation of LD decay and various LD-decay estimators in simulated and SNP-array data of tetraploid potato. Theor. Appl. Genet. 2017, 130, 123–135. [Google Scholar] [CrossRef]
- de Albuquerque, H.Y.G.; do Carmo, C.D. Genetic diversity of Manihot esculenta Crantz germplasm based on SNP markers. Ann. Appl. Biol. 2018, 173, 271–284. [Google Scholar] [CrossRef]
- Santos, C.S.D.; Sousa, M.B.; Brito, A.C.; de Oliveira, L.A.; Carvalho, C.W.P.; de Oliveira, E.J. Genome-wide association study of cassava starch paste properties. PLoS ONE 2022, 17, e0262888. [Google Scholar] [CrossRef]
- Rabbi, I.Y.; Kayondo, S.I.; Bauchet, G.; Yusuf, M.; Aghogho, C.I.; Ogunpaimo, K.; Uwugiaren, R.; Smith, I.A.; Peteti, P.; Agbona, A.; et al. Genome-wide association analysis reveals new insights into the genetic architecture of defensive, agro-morphological and quality-related traits in cassava. Plant Mol. Biol. 2022, 109, 195–213. [Google Scholar] [CrossRef]
- Simko, I.; Haynes, K.G.; Jones, R.W. Assessment of linkage disequilibrium in potato genome with single nucleotide polymorphism markers. Genetics 2006, 173, 2237–2245. [Google Scholar] [CrossRef] [PubMed]
- D’Hoop, B.B.; Paulo, M.J.; Kowitwanich, K.; Sengers, M.; Visser, R.G.; van Eck, H.J.; van Eeuwijk, F.A. Population structure and linkage disequilibrium unravelled in tetraploid potato. Theor. Appl. Genet. 2010, 121, 1151–1170. [Google Scholar] [CrossRef] [PubMed]
- Braulio, J.; Cloutier, S. Association mapping in plant genomes. In Genetic Diversity in Plants; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Mulugeta, B.; Tesfaye, K.; Ortiz, R.; Johansson, E.; Hailesilassie, T.; Hammenhag, C.; Hailu, F.; Geleta, M. Marker-trait association analyses revealed major novel QTLs for grain yield and related traits in durum wheat. Front. Plant Sci. 2022, 13, 1009244. [Google Scholar] [CrossRef] [PubMed]
- Boonchanawiwat, A.; Sraphet, S.; Boonseng, O.; Lightfoot, D.A.; Triwitayakorn, K. QTL underlying plant and first branch height in cassava (Manihot esculenta Crantz). Field Crops Res. 2011, 121, 343–349. [Google Scholar] [CrossRef]
- Crouch, D.J.M.; Bodmer, W.F. Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc. Natl. Acad. Sci. USA 2020, 117, 18924–18933. [Google Scholar] [CrossRef] [PubMed]
- Vance, G.; Shackelford, T.K. Polygenic Inheritance. In Encyclopedia of Animal Cognition and Behavior; Vonk, J., Shackelford, T., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–2. [Google Scholar] [CrossRef]
- Kumar, S.; Kirk, C.; Deng, C.H.; Shirtliff, A.; Wiedow, C.; Qin, M.; Wu, J.; Brewer, L. Marker-trait associations and genomic predictions of interspecific pear (Pyrus) fruit characteristics. Sci. Rep. 2019, 9, 9072. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, M.; Cavalleri, A.; Chandler, J.W.; Colombo, L. Auxin and flower development: A blossoming field. Cold Spring Harb. Perspect. Biol. 2021, 13, a039974. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kamanga, B.M.; Zhang, W.; Xu, Y.; Xu, L. Research progress of aldehyde oxidases in plants. PeerJ 2022, 10, e13119. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, L.; Liu, D.; Ma, S.; Dai, Y.; Zhang, X.; Wang, Y.; Hu, T.; Xiao, M.; Zhou, Y.; et al. Identification and expression of the Multidrug and Toxic Compound Extrusion (MATE) gene family in Capsicum annuum and Solanum tuberosum. Plants 2020, 9, 1448. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef]
- Ozimati, A.; Kawuki, R.; Esuma, W.; Kayondo, I.S.; Wolfe, M.; Lozano, R.; Rabbi, I.; Kulakow, P.; Jannink, J.L. Training population optimization for prediction of cassava brown streak disease resistance in West African clones. G3 Genes Genomes Genet. 2018, 8, 3903–3913. [Google Scholar] [CrossRef] [PubMed]
- Ozimati, A.; Kawuki, R.; Esuma, W.; Kayondo, S.I.; Pariyo, A.; Wolfe, M.; Jannink, J.L. Genetic variation and trait correlations in an East African cassava breeding population for genomic selection. Crop Sci. 2019, 59, 460–473. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Merk, H.L.; Yarnes, S.C.; Van Deynze, A.; Tong, N.; Menda, N.; Mueller, L.A.; Mutschler, M.A.; Loewen, S.A.; Myers, J.R.; Francis, D.M. Trait diversity and potential for selection indices based on variation among regionally adapted processing tomato germplasm. J. Am. Soc. Hortic. Sci. 2012, 137, 427–437. [Google Scholar] [CrossRef]
- Lloyd-Smith, J.O. Maximum likelihood estimation of the negative binomial dispersion parameter for highly overdispersed data, with applications to infectious diseases. PLoS ONE 2007, 2, e180. [Google Scholar] [CrossRef]
- Towers, S. Negative Binomial Likelihood Fits for Overdispersed Count Data. 2018. Available online: https://sherrytowers.com/2018/04/10/negative-binomial-likelihood-fits-for-overdispersed-count-data/ (accessed on 20 November 2023).
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- The Alliance of Genome Resources Consortium. Alliance of Genome Resources Portal: Unified model organism research platform. Nucleic Acids Res. 2020, 48, D650–D658. [Google Scholar] [CrossRef]
- Baguma, J.K.; Mukasa, S.B.; Nuwamanya, E.; Alicai, T.; Omongo, C.; Hyde, P.T.; Setter, T.L.; Ochwo-Ssemakula, M.; Esuma, W.; Kanaabi, M.; et al. Flowering and fruit-set in cassava under extended red-light photoperiod supplemented with plant-growth regulators and pruning. BMC Plant Biol. 2023, 23, 335. [Google Scholar] [CrossRef]
- Oluwasanya, D.; Esan, O.; Hyde, P.T.; Kulakow, P.; Setter, T.L. Flower development in cassava Is feminized by cytokinin, while proliferation is stimulated by anti-ethylene and pruning: Transcriptome responses. Front. Plant Sci. 2021, 12, 666266. [Google Scholar] [CrossRef]
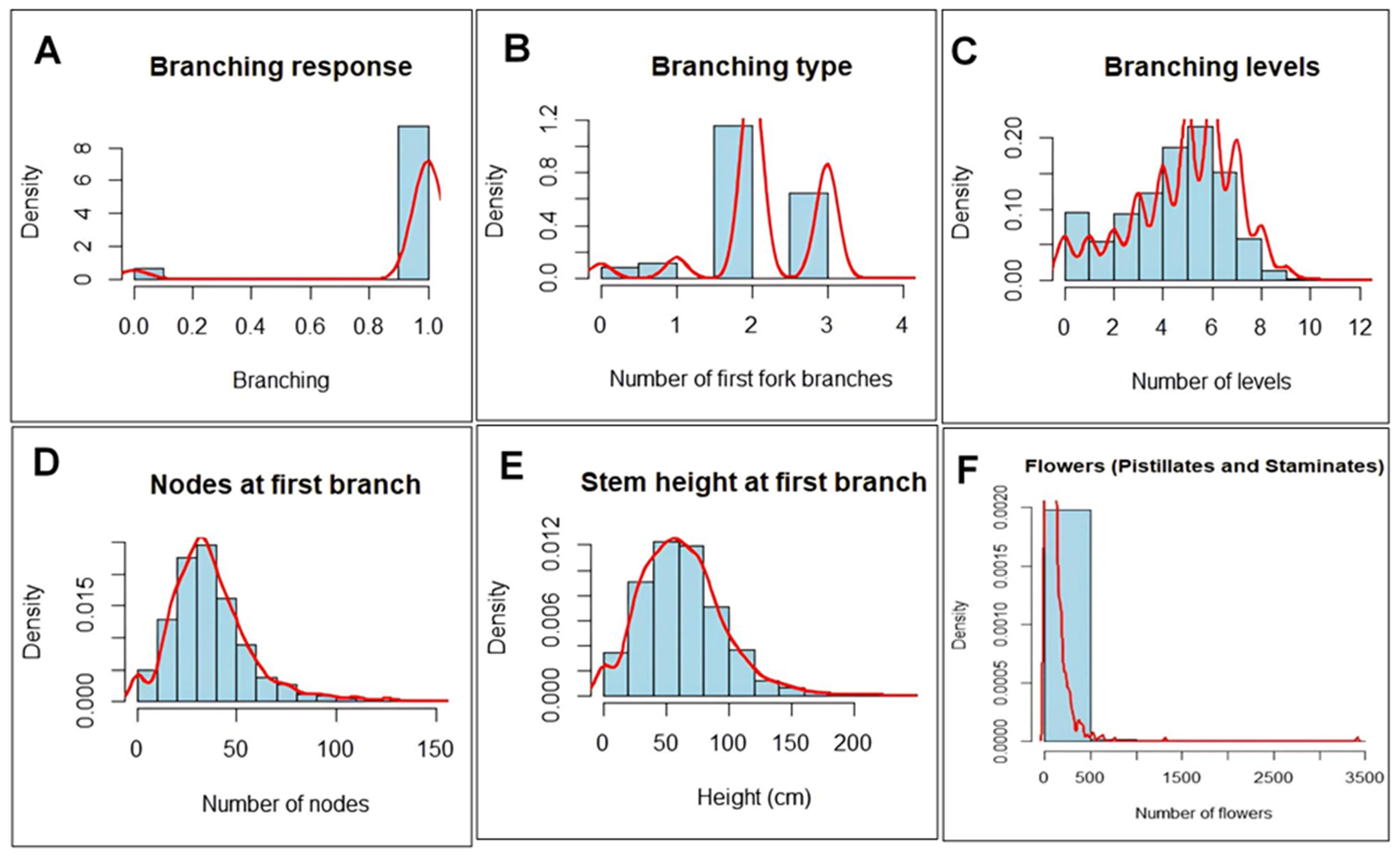
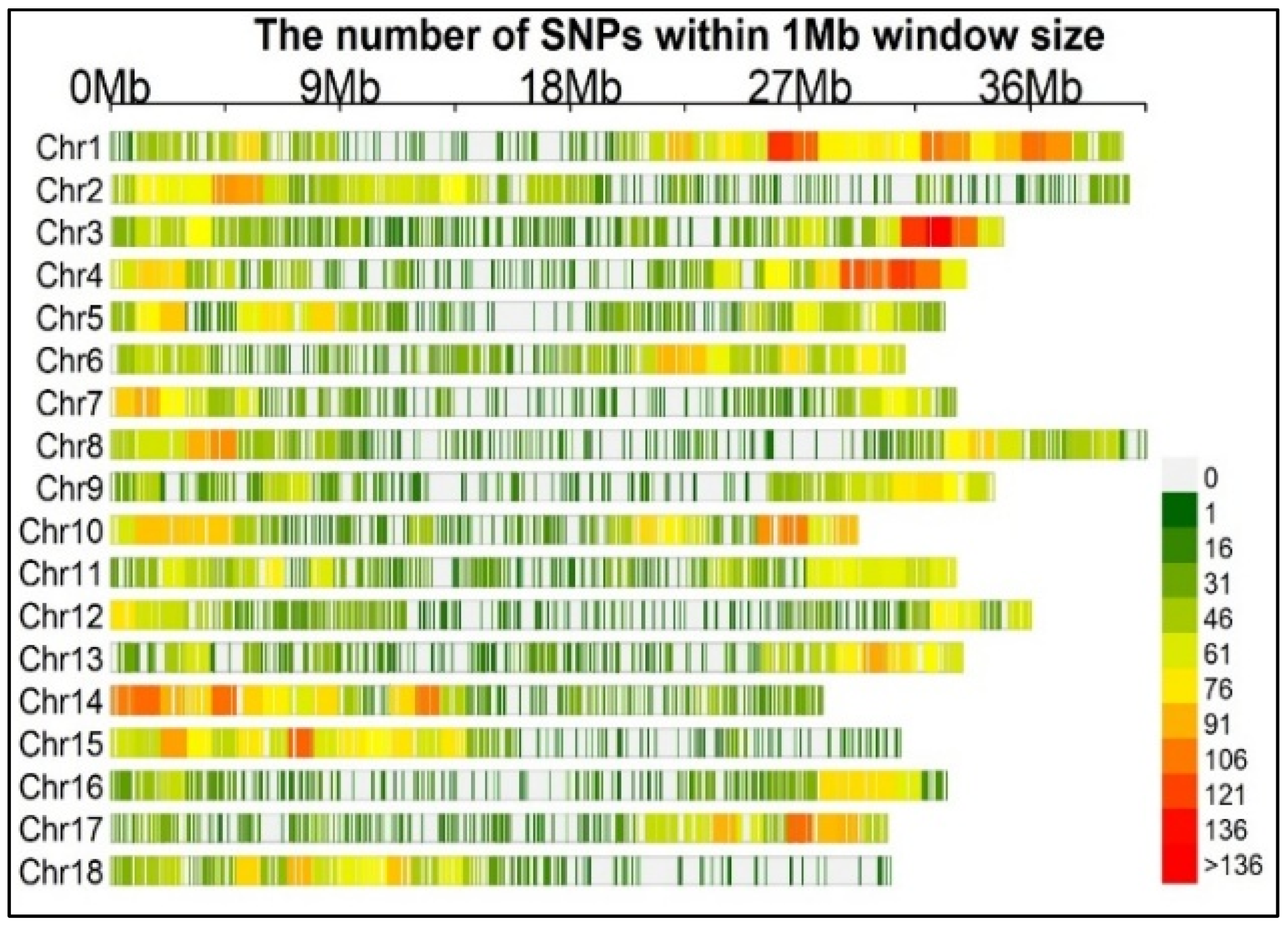

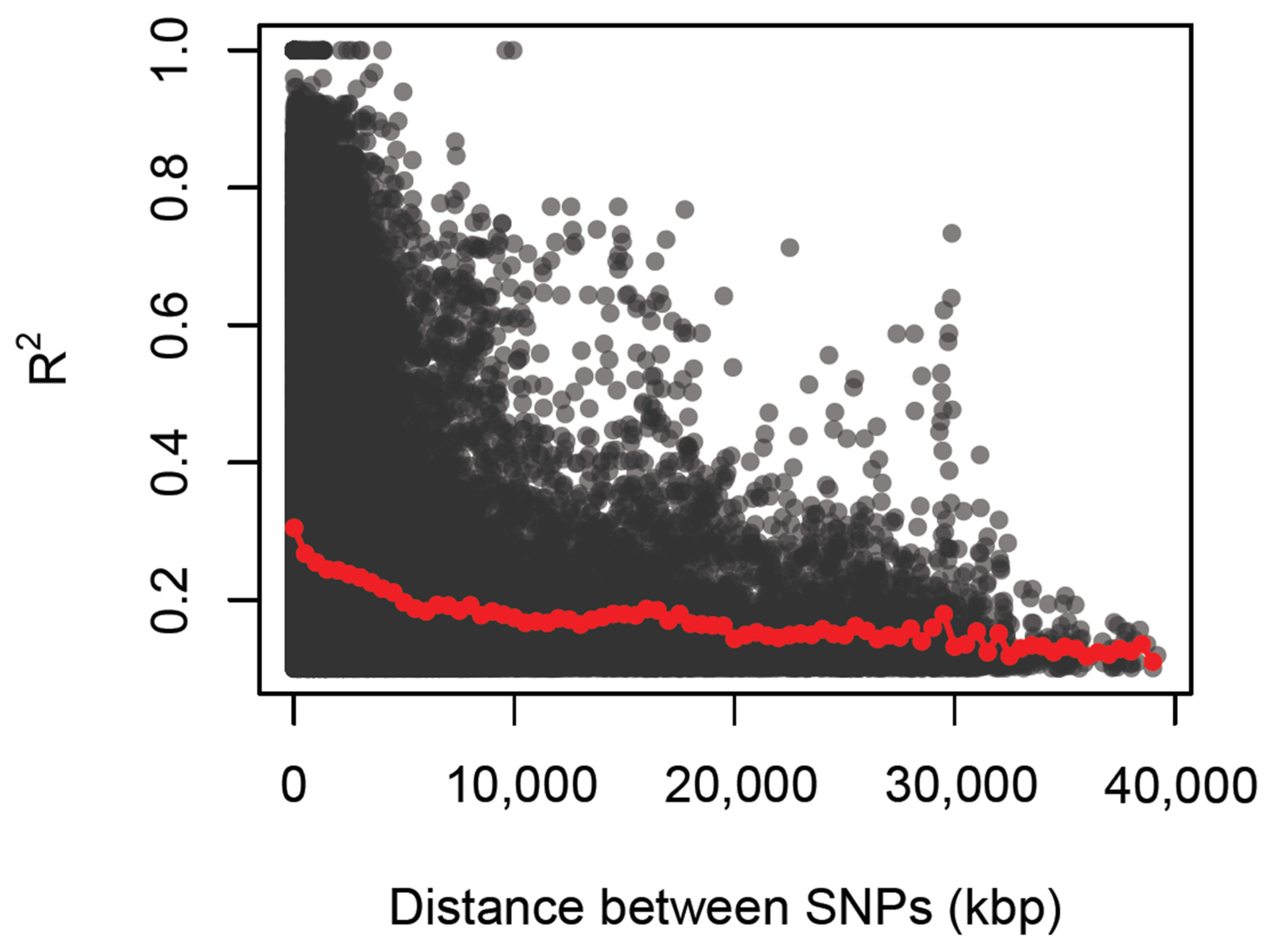
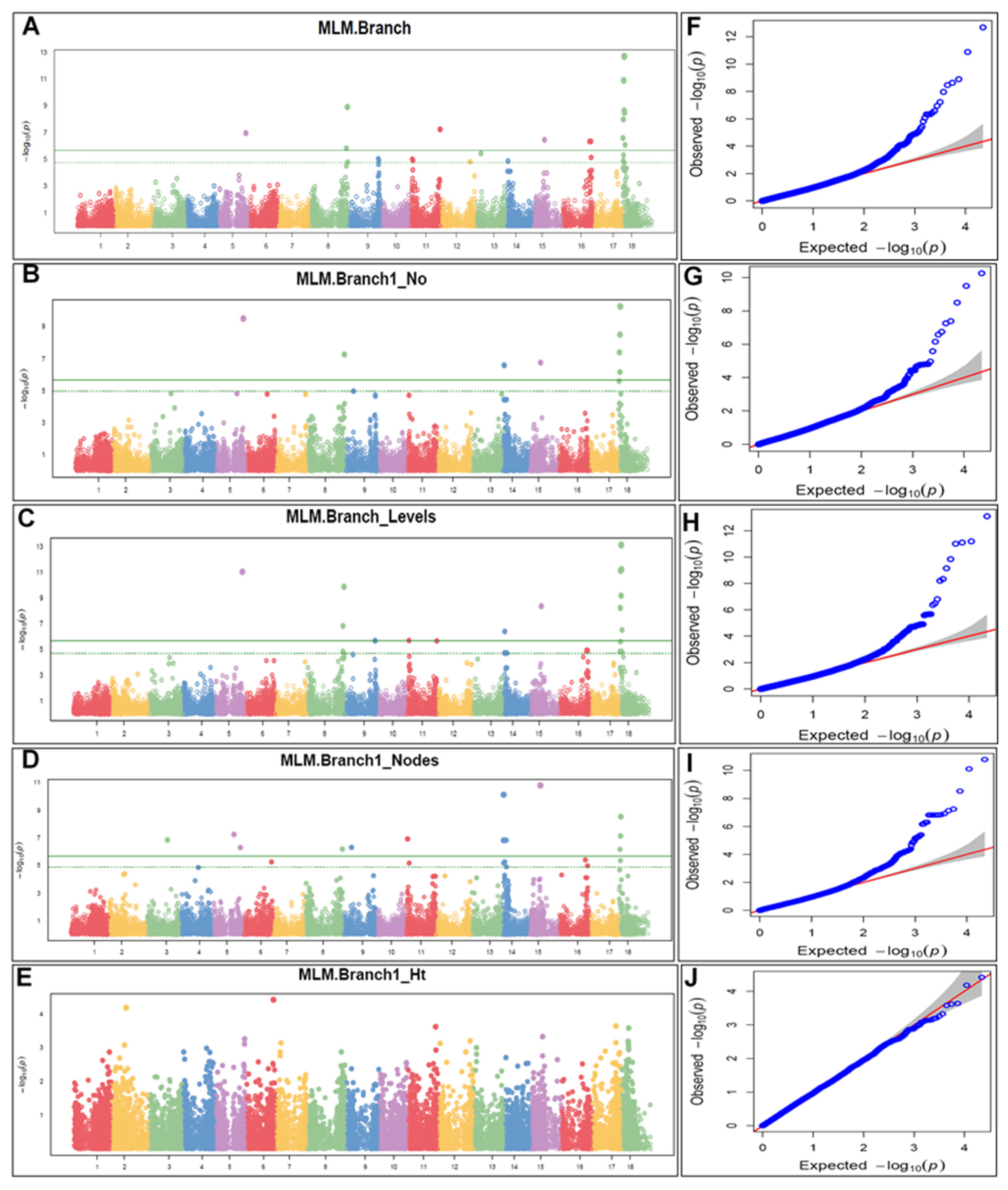
| Variables | Branching | Branch Type | Branching Levels (Number) | Nodes (Number) | Stem Height (cm) | Pistillates (Number) | Staminates (Number) |
|---|---|---|---|---|---|---|---|
| Observations (n) | 1565 | 1565 | 1565 | 1547 | 1562 | 801 | 801 |
| Mean | 0.93 | 2.19 | 4.86 | 36.90 | 63.06 | 2.09 | 34.63 |
| Skewedness | - | + | - | + | + | + | + |
| SEM | 0.01 | 0.02 | 0.05 | 0.51 | 0.85 | 0.30 | 8.51 |
| CI (0.95) | 0.01 | 0.04 | 0.11 | 1.01 | 1.67 | 0.59 | 16.72 |
| Variance | 0.06 | 0.51 | 4.60 | 407.04 | 1119.56 | 39.00 | 31,506.04 |
| SD | 0.25 | 0.71 | 2.15 | 20.18 | 33.46 | 6.25 | 177.50 |
| CV (%) | 26.87 | 32.72 | 44.16 | 54.66 | 53.08 | 2.98 | 5.13 |
| Heritability, H2 | 0.34 | 0.53 | 0.38 | 0.42 | 0.60 | 0.83 | 0.00 |
| SNP heritability, h2 | 0.25 | 0.00 | 0.19 | 0.01 | 0.35 | - | - |
| Significance | * | *** | *** | *** | * | * | *** |
| Variable | Sums of Squares | Mean Squares | F Value | Significance | ||||
|---|---|---|---|---|---|---|---|---|
| Acc | Loc | Acc | Loc | Acc | Loc | Acc | Loc | |
| Branching | 57.143 | 0.634 | 0.12841 | 0.63371 | 3.533 | 17.4353 | *** | *** |
| Branch type | 435.27 | 0.96 | 0.9781 | 0.9603 | 3.2537 | 3.1945 | *** | ns |
| Branching levels | 5021.7 | 367.3 | 11.28 | 367.26 | 9.733 | 316.7597 | *** | *** |
| Nodes (at 1st branch) | 280,458 | 16954 | 630.2 | 16,954 | 2.4511 | 65.9356 | *** | *** |
| Stem height (1st branch) (cm) | 755,806 | 4734 | 1698 | 4734 | 2.2902 | 6.3829 | *** | * |
| Trait | SNP | Ch | Gene ID | Description | Function | Reference Species | References |
|---|---|---|---|---|---|---|---|
| Branch, Branch1_No., Branch_Levels | S18_1832353 | 18 | Manes.18G016700 | aldehyde oxidase GLOX | Catalyzes the oxidation of aldehydes to the corresponding carboxylates; involved in anther development and plays a role in tapetum and pollen development | A. thaliana Vitis pseudoreticulata (Chinese wild grapevine) | [60,69] |
| Manes.18G016725 | lysine-specific histone demethylase 1 homolog 3 | Promotion of the floral transition | A. thaliana | [61] | |||
| Manes.18G016200 | cytochrome P450 83B1 | Functions in auxin homeostasis and plant reproductive development | A. thaliana | [62,63] | |||
| S5_29309724 | 5 | Manes.05G186700 | protein DETOXIFICATION 48 | Could be involved in specifying the lateral organ initiation rate | A. thaliana | [64,70] | |
| S15_11747301 | 15 | Manes.15G140500 | eukaryotic translation initiation factor 3 subunit I | Negatively regulates translation during flower development | A. thaliana | [65] | |
| Manes.15G140900 | myb family transcription factor MOF1 | Transcriptional repressor that plays a role in the regulation of organ identity and spikelet meristem determinacy | Oryza sativa subsp. japonica (rice) | [66] | |||
| Manes.15G140300 | non-specific lipid-transfer protein 4.1 | Lipid transfer protein involved in seed and ovule maturation and development, probably by regulating fatty acid homeostasis during suberin and sporopollenin biosynthesis or deposition | Hordeum vulgare (barley); A. thaliana; maize | [67,71] | |||
| Manes.15G140600 | short-chain dehydrogenase reductase ATA1 | May play a role in tapetum development | A. thaliana | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baguma, J.K.; Mukasa, S.B.; Nuwamanya, E.; Alicai, T.; Omongo, C.A.; Ochwo-Ssemakula, M.; Ozimati, A.; Esuma, W.; Kanaabi, M.; Wembabazi, E.; et al. Identification of Genomic Regions for Traits Associated with Flowering in Cassava (Manihot esculenta Crantz). Plants 2024, 13, 796. https://doi.org/10.3390/plants13060796
Baguma JK, Mukasa SB, Nuwamanya E, Alicai T, Omongo CA, Ochwo-Ssemakula M, Ozimati A, Esuma W, Kanaabi M, Wembabazi E, et al. Identification of Genomic Regions for Traits Associated with Flowering in Cassava (Manihot esculenta Crantz). Plants. 2024; 13(6):796. https://doi.org/10.3390/plants13060796
Chicago/Turabian StyleBaguma, Julius K., Settumba B. Mukasa, Ephraim Nuwamanya, Titus Alicai, Christopher Abu Omongo, Mildred Ochwo-Ssemakula, Alfred Ozimati, Williams Esuma, Michael Kanaabi, Enoch Wembabazi, and et al. 2024. "Identification of Genomic Regions for Traits Associated with Flowering in Cassava (Manihot esculenta Crantz)" Plants 13, no. 6: 796. https://doi.org/10.3390/plants13060796
APA StyleBaguma, J. K., Mukasa, S. B., Nuwamanya, E., Alicai, T., Omongo, C. A., Ochwo-Ssemakula, M., Ozimati, A., Esuma, W., Kanaabi, M., Wembabazi, E., Baguma, Y., & Kawuki, R. S. (2024). Identification of Genomic Regions for Traits Associated with Flowering in Cassava (Manihot esculenta Crantz). Plants, 13(6), 796. https://doi.org/10.3390/plants13060796








