RNA Sequencing Analysis and Verification of Paeonia ostii ‘Fengdan’ CuZn Superoxide Dismutase (PoSOD) Genes in Root Development
Abstract
1. Introduction
2. Results
2.1. Quality Analysis of RNA-seq of Four Root Developmental Stages in P. ostii ‘Fengdan’
2.2. Annotation of Database
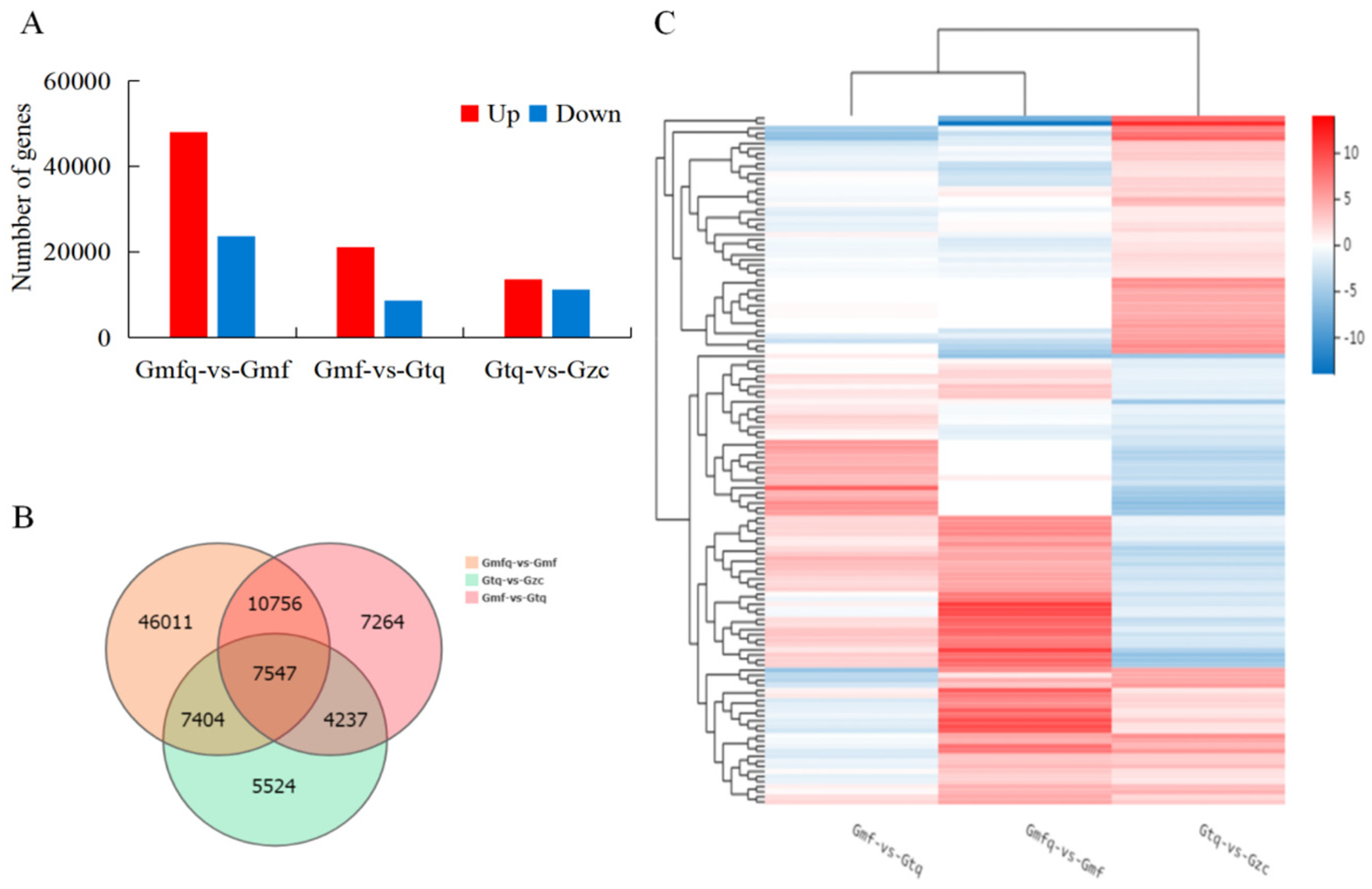
2.3. Identification and Expression of DEGs
2.4. GO Classification and KEGG Enrichment of DEGs
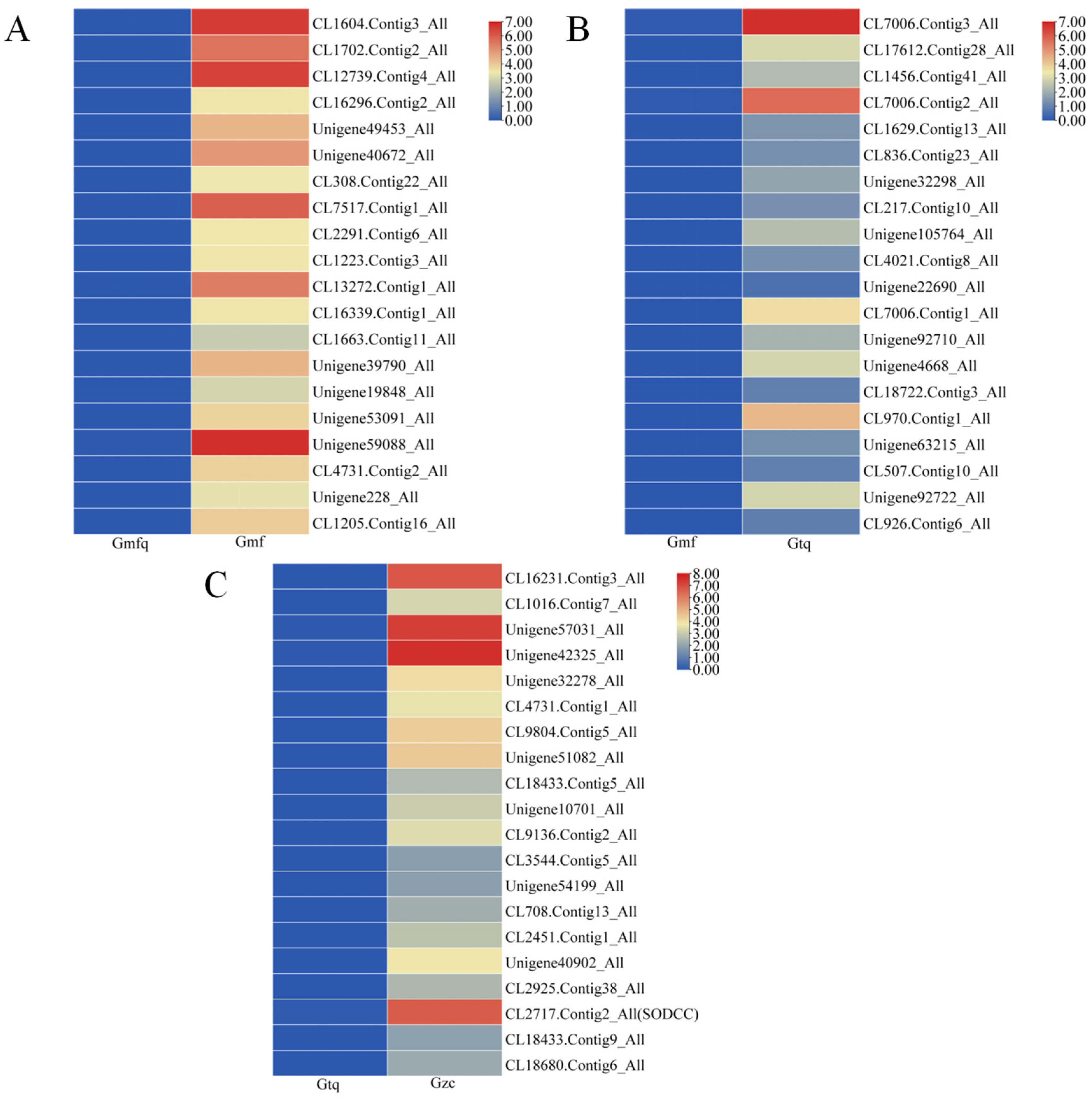
2.5. Analysis of Genes Related to Antioxidant Enzymes
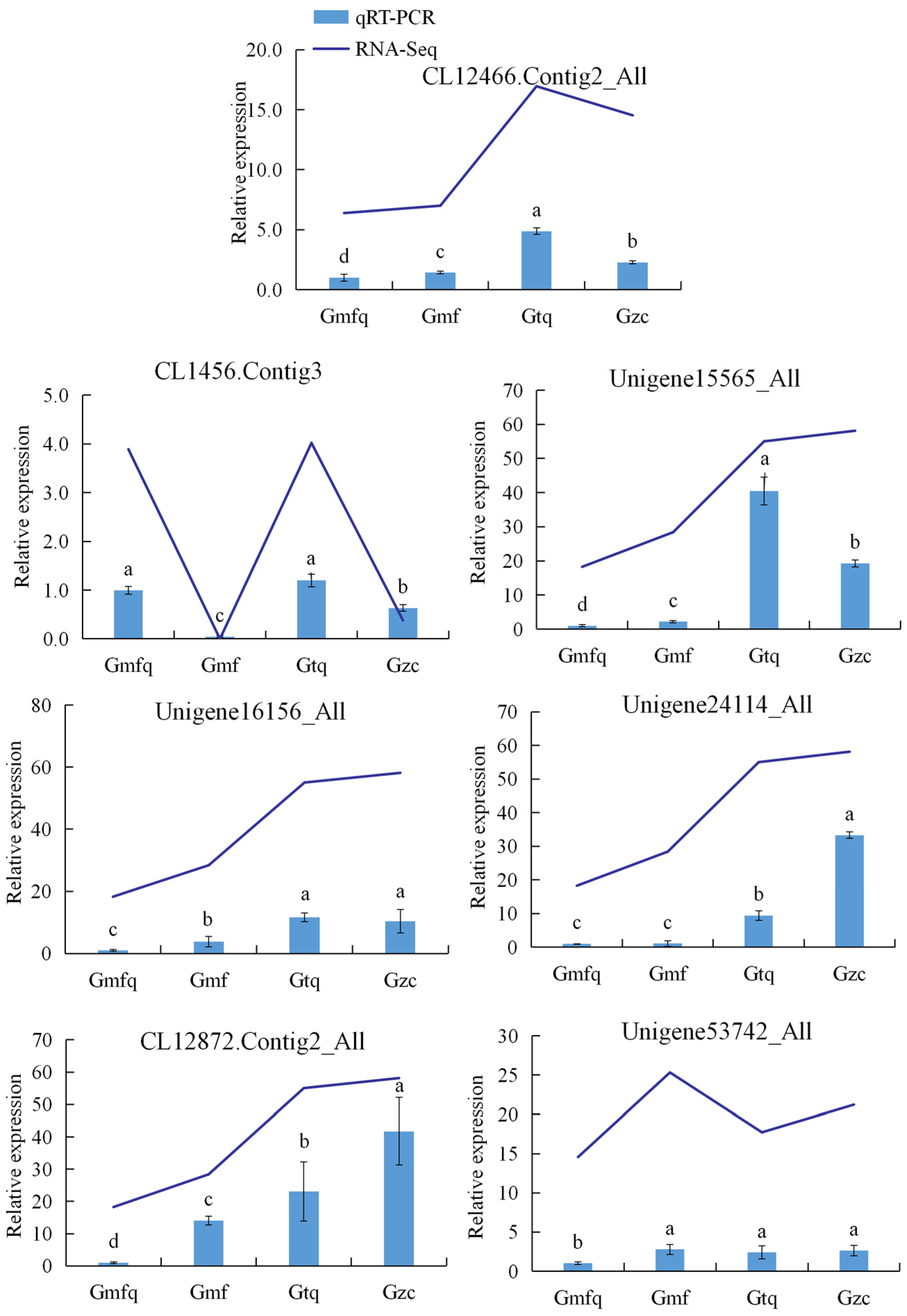
2.6. The Validation of RNA-Seq Data via Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT–PCR)
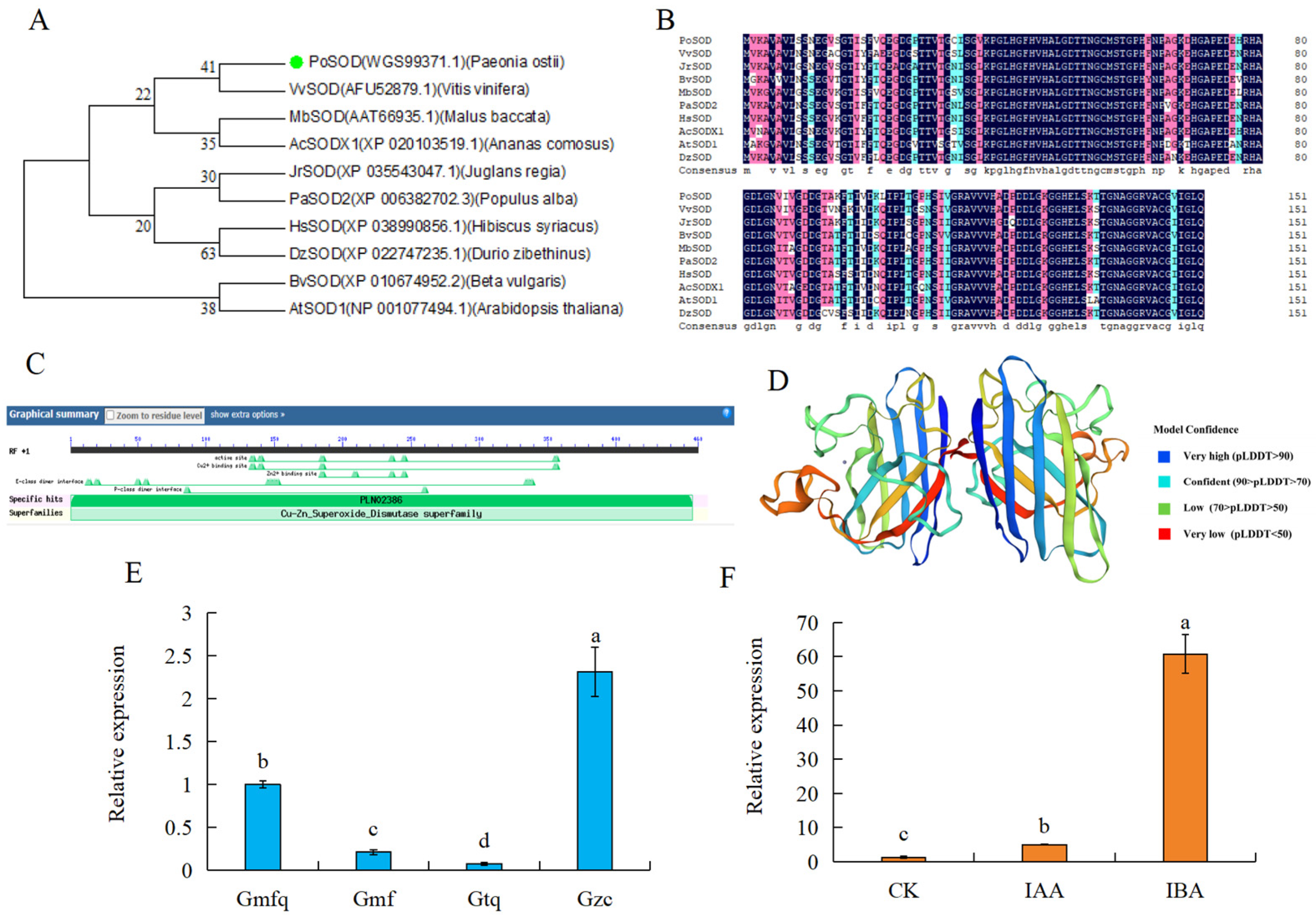
2.7. Cloning and Analysis of PoSOD
2.8. PoSOD Increased the AR Number in P. ostii ‘Fengdan’ Tube Plantlets
2.9. Root Phenotype Analysis of Overexpressing PoSOD in Arabidopsis
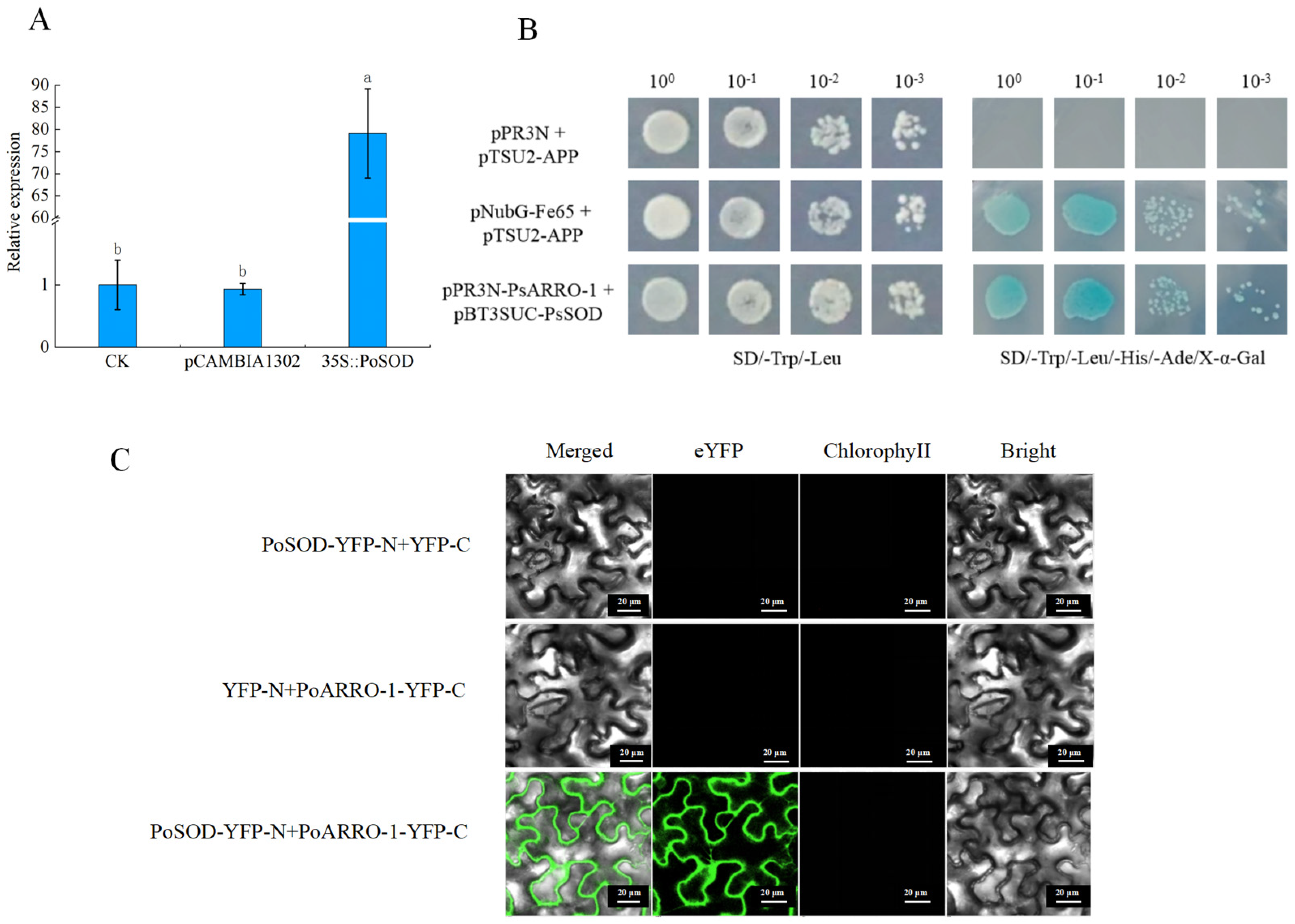
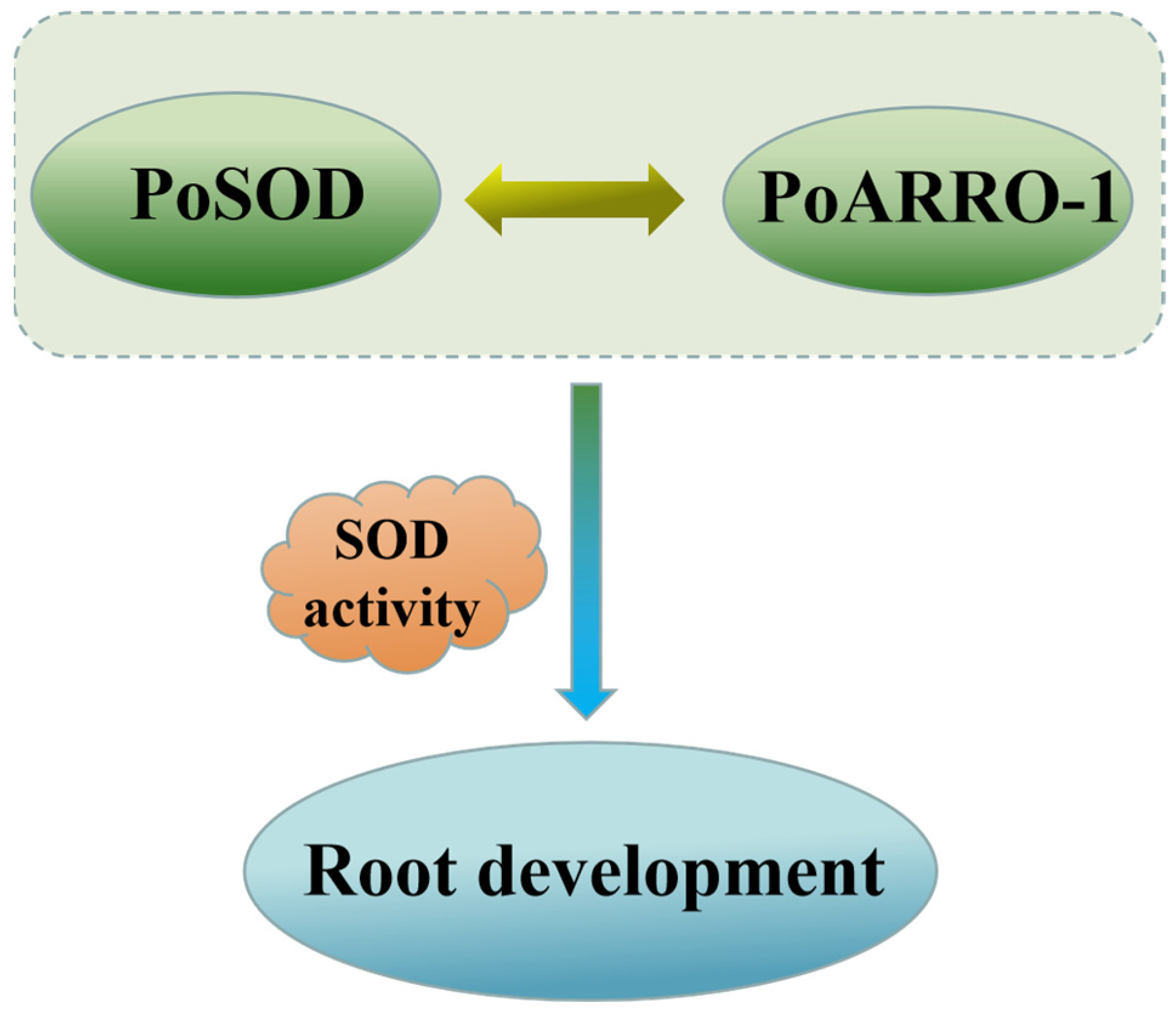
2.10. PoSOD Interacted with PoARRO-1
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction, Library Preparation, Sequencing, Transcriptome Assembly, and Gene Functional Annotation
4.3. Identification and Functional Annotation of DEGs
4.4. The Prediction of the PPI Network
4.5. Isolation of the PoSOD Gene and Bioinformatic Analysis
4.6. P. ostii ‘Fengdan’ Tube Plantlets Treated with Different Substrates
4.7. Transient Expression of PoSOD in P. ostii ‘Fengdan’
4.8. Phenotypic Statistics of Roots in Transgenic PoSOD Arabidopsis
4.9. NBT Staining
4.10. Y2H and BiFC Assays
4.11. qRT-PCR
4.12. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Lakehal, A.; Dob, A.; Novák, O.; Bellini, C. A DAO1-mediated circuit controls auxin and jasmonate crosstalk robustness during adventitious root initiation in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4428. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhry, S.; Kepinski, S. Auxin in root development. Cold Spring Harb. Perspect. Biol. 2022, 14, a039933. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, X.; Nvsvrot, T.; Huang, L.; Cai, G.; Ding, Y.; Ren, W.; Wang, N. The transcription factor WRKY75 regulates the development of adventitious roots, lateral buds and callus by modulating hydrogen peroxide content in poplar. J. Exp. Bot. 2022, 73, 1483–1498. [Google Scholar] [CrossRef]
- Cavallari, N.; Artner, C.; Benkova, E. Auxin-regulated lateral root organogenesis. Cold Spring Harb. Perspect. Biol. 2021, 13, a039941. [Google Scholar] [CrossRef]
- Kwasniewski, M.; Chwialkowska, K.; Kwasniewska, J.; Kusak, J.; Siwinski, K.; Szarejko, I. Accumulation of peroxidase-related reactive oxygen species in trichoblasts correlates with root hair initiation in barley. J. Plant Physiol. 2013, 170, 185–195. [Google Scholar] [CrossRef]
- Valifard, M.; Le Hir, R.; Müller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef]
- López-Bucio, J.; Ortiz-Castro, R.; Ruíz-Herrera, L.F.; Juárez, C.V.; Hernández-Madrigal, F.; Carreón-Abud, Y.; Martínez-Trujillo, M. Chromate induces adventitious root formation via auxin signalling and SOLITARY-ROOT/IAA14 gene function in Arabidopsis thaliana. Biometals 2015, 28, 353–365. [Google Scholar] [CrossRef]
- da Costa, C.T.; Offringa, R.; Fett-Neto, A.G. The role of auxin transporters and receptors in adventitious rooting of Arabidopsis thaliana pre-etiolated flooded seedlings. Plant Sci. 2020, 290, 110294. [Google Scholar] [CrossRef]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef]
- Yan, Y.H.; Li, J.L.; Zhang, X.Q.; Yang, W.Y.; Wan, Y.; Ma, Y.M.; Zhu, Y.Q.; Peng, Y.; Huang, L.K. Effect of naphthalene acetic acid on adventitious root development and associated physiological changes in stem cutting of Hemarthria compressa. PLoS ONE 2014, 9, e90700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fan, J.; Tan, Q.; Zhao, M.; Zhou, T.; Cao, F. The effects of exogenous hormones on rooting process and the activities of key enzymes of Malus hupehensis stem cuttings. PLoS ONE 2017, 12, e0172320. [Google Scholar] [CrossRef][Green Version]
- Jin, H.D.; Zheng, C.Y.; Hua, B.; Yu, C.L.; Li, K.Y.; Yu, W.W. Rooting anatomy and physiological related enzyme activities of Torreya grandis cuttings. Acta Agric. Zhejiangensis 2022, 34, 1955–1966. [Google Scholar]
- Xiao, Z.F.; Sheng, P.J.; Xu, Y.F.; Peng, Y.; Jiang, R.; Zheng, B.J.; Li, F. Changes in relevant oxidase activities and carbohydrates during the rooting process of micro-cuttage of Cinnamomum camphora ct. Linalool. J. Yibin Univ. 2022, 22, 62–66. [Google Scholar]
- Irani, S.; Trost, B.; Waldner, M.; Nayidu, N.; Tu, J.; Kusalik, A.J.; Todd, C.D.; Wei, Y.; Bonham-Smith, P.C. Transcriptome analysis of response to Plasmodiophora brassicae infection in the Arabidopsis shoot and root. BMC Genom. 2018, 19, 23. [Google Scholar] [CrossRef]
- Cheng, L.; Jiang, R.; Yang, J.; Xu, X.; Zeng, H.; Li, S. Transcriptome profiling reveals an IAA-regulated response to adventitious root formation in lotus seedling. Z. Naturforschung 2018, 73, 229–240. [Google Scholar]
- Zhai, R.; Feng, Y.; Wang, H.; Zhan, X.; Shen, X.; Wu, W.; Zhang, Y.; Chen, D.; Dai, G.; Yang, Z.; et al. Transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genom. 2013, 14, 19. [Google Scholar] [CrossRef]
- Song, L.; Prince, S.; Valliyodan, B.; Joshi, T.; Maldonado dos Santos, J.V.; Wang, J.; Lin, L.; Wan, J.; Wang, Y.; Xu, D.; et al. Genome-wide transcriptome analysis of soybean primary root under varying water-deficit conditions. BMC Genom. 2016, 17, 57. [Google Scholar] [CrossRef]
- Fu, Z.Z.; Xu, M.L.; Wang, H.J.; Wang, E.Q.; Li, Y.M.; Wang, L.M.; Gao, J.; Zhang, Z.; Yuan, X.; Zhang, H.C. Analysis of the transcriptome and related physiological indicators of tree peony (Paeonia suffruticosa Andr.) plantlets before and after rooting in vitro. Plant Cell Tissue Organ Cult. 2021, 147, 529–543. [Google Scholar] [CrossRef]
- Li, S.W.; Leng, Y.; Shi, R.F. Transcriptomic profiling provides molecular insights into hydrogen peroxide-induced adventitious rooting in mung bean seedlings. BMC Genom. 2017, 18, 188. [Google Scholar] [CrossRef]
- Song, Y.L.; Shang, W.Q.; Wang, Z.; He, S.L.; Meng, X.Y.; Shi, L.Y.; Shen, Y.X.; He, D.; Lou, X.Y.; Sun, Y.K. Transcriptome analysis and genes function verification of root development of Paeonia suffruticosa under sandy loam cultivation. Phyton-Int. J. Exp. Bot. 2022, 91, 2791–2812. [Google Scholar] [CrossRef]
- Wen, S.S.; Miao, D.P.; Cui, H.Y.; Li, S.G.; Gu, Y.N.; Jia, R.R.; Leng, Y.F. Physiology and transcriptomic analysis of endogenous hormones regulating in vitro adventitious root formation in tree peony. Sci. Hortic. 2023, 318, 112122. [Google Scholar] [CrossRef]
- Gao, J.; Xue, J.; Xue, Y.; Liu, R.; Ren, X.; Wang, S.; Zhang, X. Transcriptome sequencing and identification of key callus browning-related genes from petiole callus of tree peony (Paeonia suffruticosa cv. Kao) cultured on media with three browning inhibitors. Plant Physiol. Biochem. 2020, 149, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.S.; Cheng, F.Y.; Zhong, Y.; Wang, X.; Li, L.Z.M.; Zhang, X.Y.; Qiu, J.M. Efficient protocols for the micropropagation of tree peony (Paeonia suffruticosa ‘Jin Pao Hong’, P. suffruticosa ‘Wu Long Peng Sheng’, and P. x lemoinei ‘High Noon’) and application of arbuscular mycorrhizal fungi to improve plantlet establishment. Sci. Hortic. 2016, 201, 10–17. [Google Scholar] [CrossRef]
- Meng, X.Y.; Wang, Z.; He, S.L.; Shi, L.Y.; Song, Y.L.; Lou, X.Y.; He, D. Endogenous hormone levels and activities of IAA-modifying enzymes during adventitious rooting of tree peony cuttings and grafted scions. Hortic. Environ. Biotechnol. 2019, 60, 187–197. [Google Scholar] [CrossRef]
- Shang, W.Q.; Wang, Z.; He, S.L.; He, D.; Dong, N.L.; Guo, Y. Changes of endogenous IAA and related activities during rooting of Paeonia suffruticosa in vitro. J. Northwest Univ. (Nat. Sci. Ed.) 2021, 49, 129–136. [Google Scholar]
- Shang, W.Q.; Wang, Z.; He, S.L.; He, D.; Liu, Y.P.; Fu, Z.Z. Research on the relationship between phenolic acids and rooting of tree peony (Paeonia suffruticosa) plantlets in vitro. Sci. Hortic. 2017, 224, 53–60. [Google Scholar] [CrossRef]
- He, D.; Li, R.; Ji, S.; Wu, J.; Wang, Z.; Liu, Y.P.; He, S.L. Cloning and expression analysis of adventitious rooting related gene PsARRO-1 of tree peony. Plant Physiol. J. 2014, 50, 1151–1158. [Google Scholar]
- Sun, Y.K.; Shang, W.Q.; Yuan, J.; Wang, Z.; He, S.L.; Song, Y.L.; Wang, J.G. Functional analysis of PsARRO-1 in root development of Paeonia suffruticosa. Horticulturae 2022, 8, 903. [Google Scholar] [CrossRef]
- Wang, Z.; Su, G.; He, S.; Shi, L.; He, D.; Shang, W.; Yang, D. Effects of root pruning on adventitious root formation, enzyme activities, and hormone levels in Paeonia suffruticosa ‘Fengdanbai’ seedlings. Hortic. Sci. Technol. 2021, 39, 10–22. [Google Scholar] [CrossRef]
- Guo, M.; Pan, Y.M.; Gao, Z.M. First report of medical tree peony root rot caused by fusarium solani in Tongling, China. Plant Dis. 2012, 96, 909. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.A.; Agbévénou, K.; Herrbach, V.; Gough, C.; Bensmihen, S. Abscisic acid promotes pre-emergence stages of lateral root development in Medicago truncatula. Plant Signal. Behav. 2015, 10, e977741. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Giehl, R.F.H.; von Wirén, N. Nutrient-hormone relations: Driving root plasticity in plants. Mol. Plant 2022, 15, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Owusu Adjei, M.; Xiang, Y.; He, Y.; Zhou, X.; Mao, M.; Liu, J.; Hu, H.; Luo, J.; Zhang, H.; Feng, L.; et al. Adventitious root primordia formation and development in the stem of Ananas comosus var. bracteatus slip. Plant Signal. Behav. 2021, 16, 1949147. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, C.; Li, C.; Zhao, Z.; Wei, L.; Liu, Z.; Hu, D.; Liao, W. Nitric oxide is involved in hydrogen sulfide-induced adventitious rooting in tomato (Solanum lycopersicum). Funct. Plant Biol. 2022, 49, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.J.; Park, S.U.; Kim, S.E.; Lim, Y.H.; Ji, C.Y.; Kim, Y.H.; Kim, H.S.; Kwak, S.S. Overexpression of IbLfp in sweetpotato enhances the low-temperature storage ability of tuberous roots. Plant Physiol. Biochem. 2021, 167, 577–585. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Dvořák, P.; Krasylenko, Y.; Ovečka, M.; Basheer, J.; Zapletalová, V.; Šamaj, J.; Takáč, T. In vivo light-sheet microscopy resolves localisation patterns of FSD1, a superoxide dismutase with function in root development and osmoprotection. Plant Cell Environ. 2021, 44, 68–87. [Google Scholar] [CrossRef]
- Chen, H.; Lee, J.; Lee, J.M.; Han, M.; Emonet, A.; Lee, J.; Jia, X.; Lee, Y. MSD2, an apoplastic Mn-SOD, contributes to root skotomorphogenic growth by modulating ROS distribution in Arabidopsis. Plant Sci. 2022, 317, 111192. [Google Scholar] [CrossRef]
- Lin, K.H.; Sei, S.C.; Su, Y.H.; Chiang, C.M. Overexpression of the Arabidopsis and winter squash superoxide dismutase genes enhances chilling tolerance via ABA-sensitive transcriptional regulation in transgenic Arabidopsis. Plant Signal. Behav. 2019, 14, 1685728. [Google Scholar] [CrossRef]
- Sun, J.J.; Bi, S.W.; Jin, S.S.; Wang, Y.G.; Chen, H.L.; Ruoyi, N.; Ma, M.Y.; Quan, J.E. Influence of various growth modulators on the rooting, enzymatic activity, and nutrient content of Catalpa Bignonioides (bigeoniacece). Pak. J. Bot. 2024, 56, 75–84. [Google Scholar] [CrossRef]
- Rout, G.R. Effect of auxins on adventitious root development from single node cuttings of Camellia sinensis (L.) Kuntze and associated biochemical changes. Plant Growth Regul. 2006, 48, 111–117. [Google Scholar] [CrossRef]
- Voß, U.; Wilson, M.H.; Kenobi, K.; Gould, P.D.; Robertson, F.C.; Peer, W.A.; Lucas, M.; Swarup, K.; Casimiro, I.; Holman, T.J.; et al. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat. Commun. 2015, 6, 7641. [Google Scholar] [CrossRef]
- Hernández-Vega, J.C.; Langford, S.; Hurtado, D.A.; Cady, B.; Kayanja, G.; Okwara, N.; Mauriello, A.; Alkio, M.; Colón-Carmona, A. Detoxification of phenanthrene in Arabidopsis thaliana involves a Dioxygenase for Auxin Oxidation 1 (AtDAO1). J. Biotechnol. 2021, 342, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Dobrev, P.I.; Pěnčík, A.; Hošek, P.; Vondráková, Z.; Filepová, R.; Malínská, K.; Brunoni, F.; Helusová, L.; Moravec, T.; et al. DIOXYGENASE FOR AUXIN OXIDATION 1 catalyzes the oxidation of IAA amino acid conjugates. Plant Physiol. 2021, 187, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, N.M.N.; Chandrasekara, B.C.H.W.M.; Bandaranayake, P.C.G. A CTAB protocol for obtaining high-quality total RNA from cinnamon (Cinnamomum zeylanicum Blume). 3 Biotech 2021, 11, 201. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Moran, Y.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Neogy, A.; Garg, T.; Kumar, A.; Dwivedi, A.K.; Singh, H.; Singh, U.; Singh, Z.; Prasad, K.; Jain, M.; Yadav, S.R. Genome-Wide Transcript Profiling Reveals an Auxin-Responsive Transcription Factor, OsAP2/ERF-40, Promoting Rice Adventitious Root Development. Plant Cell Physiol. 2019, 60, 2343–2355. [Google Scholar] [CrossRef]
- Xu, Y.F.; Shang, W.Q.; Li, L.; Song, Y.L.; Wang, G.; Shi, L.Y.; Shen, Y.X.; Sun, Y.K.; He, S.L.; Wang, Z. Transcriptome landscape analyses of the regulatory network for zygotic embryo development in Paeonia ostii. Int. J. Mol. Sci. 2023, 24, 10715. [Google Scholar] [CrossRef]
- Gascuel, O.; Steel, M. Neighbor-joining revealed. Mol. Biol. Evol. 2006, 23, 1997–2000. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Bagos, P.G.; Tsirigos, K.D.; Plessas, S.K.; Liakopoulos, T.D.; Hamodrakas, S.J. Prediction of signal peptides in archaea. Protein Eng. Des. Sel. 2009, 22, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Gupta, S.C.; Chauhan, P.S.; Lata, C. An OsNAM gene plays important role in root rhizobacteria interaction in transgenic Arabidopsis through abiotic stress and phytohormone crosstalk. Plant Cell Rep. 2021, 40, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Huang, Y.Y.; Xin, J.L.; He, C.T.; Yang, Z.Y. A novel microRNA IamiR-4-3p from water spinach (Ipomoea aquatica Forsk.) increased Cd uptake and translocation in Arabidopsis thaliana. Environ. Sci. Pollut. Res. Int. 2022, 29, 41375–41385. [Google Scholar] [CrossRef] [PubMed]
- Porco, S.; Pěnčík, A.; Rashed, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef]
- Jambunathan, N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol. Biol. 2010, 639, 292–298. [Google Scholar] [PubMed]
- Bai, Z.; Zhang, J.; Ning, X.; Guo, H.; Xu, X.; Huang, X.; Wang, Y.; Hu, Z.; Lu, C.; Zhang, L.; et al. A Kinase-Phosphatase-Transcription factor module regulates adventitious root emergence in Arabidopsis root-hypocotyl junctions. Mol. Plant 2020, 13, 1162–1177. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Song, Y.; Wang, Z.; Shi, L.; Yu, S.; Xu, Y.; Wang, G.; He, D.; Jiang, L.; Shang, W.; et al. RNA Sequencing Analysis and Verification of Paeonia ostii ‘Fengdan’ CuZn Superoxide Dismutase (PoSOD) Genes in Root Development. Plants 2024, 13, 421. https://doi.org/10.3390/plants13030421
Wang J, Song Y, Wang Z, Shi L, Yu S, Xu Y, Wang G, He D, Jiang L, Shang W, et al. RNA Sequencing Analysis and Verification of Paeonia ostii ‘Fengdan’ CuZn Superoxide Dismutase (PoSOD) Genes in Root Development. Plants. 2024; 13(3):421. https://doi.org/10.3390/plants13030421
Chicago/Turabian StyleWang, Jiange, Yinglong Song, Zheng Wang, Liyun Shi, Shuiyan Yu, Yufeng Xu, Guiqing Wang, Dan He, Liwei Jiang, Wenqian Shang, and et al. 2024. "RNA Sequencing Analysis and Verification of Paeonia ostii ‘Fengdan’ CuZn Superoxide Dismutase (PoSOD) Genes in Root Development" Plants 13, no. 3: 421. https://doi.org/10.3390/plants13030421
APA StyleWang, J., Song, Y., Wang, Z., Shi, L., Yu, S., Xu, Y., Wang, G., He, D., Jiang, L., Shang, W., & He, S. (2024). RNA Sequencing Analysis and Verification of Paeonia ostii ‘Fengdan’ CuZn Superoxide Dismutase (PoSOD) Genes in Root Development. Plants, 13(3), 421. https://doi.org/10.3390/plants13030421





