Bidirectional Comparisons Revealed Functional Patterns in Interaction between Salmonella enterica and Plants
Abstract
1. Introduction
2. Results
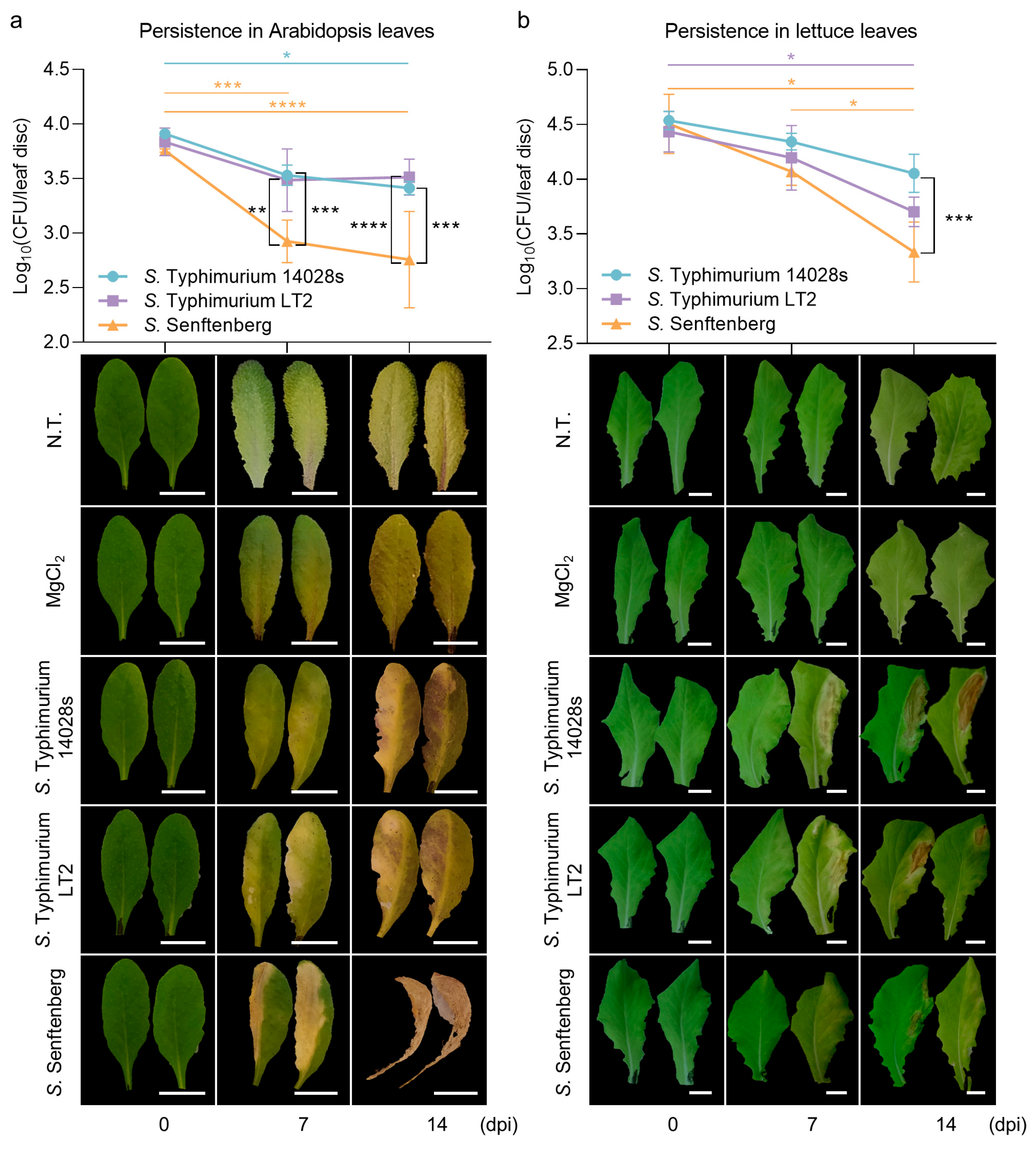
2.1. S. enterica Serovar Typhimurium Strains Persist Better than S. enterica Serovar Senftenberg Strain in Plant Leaves
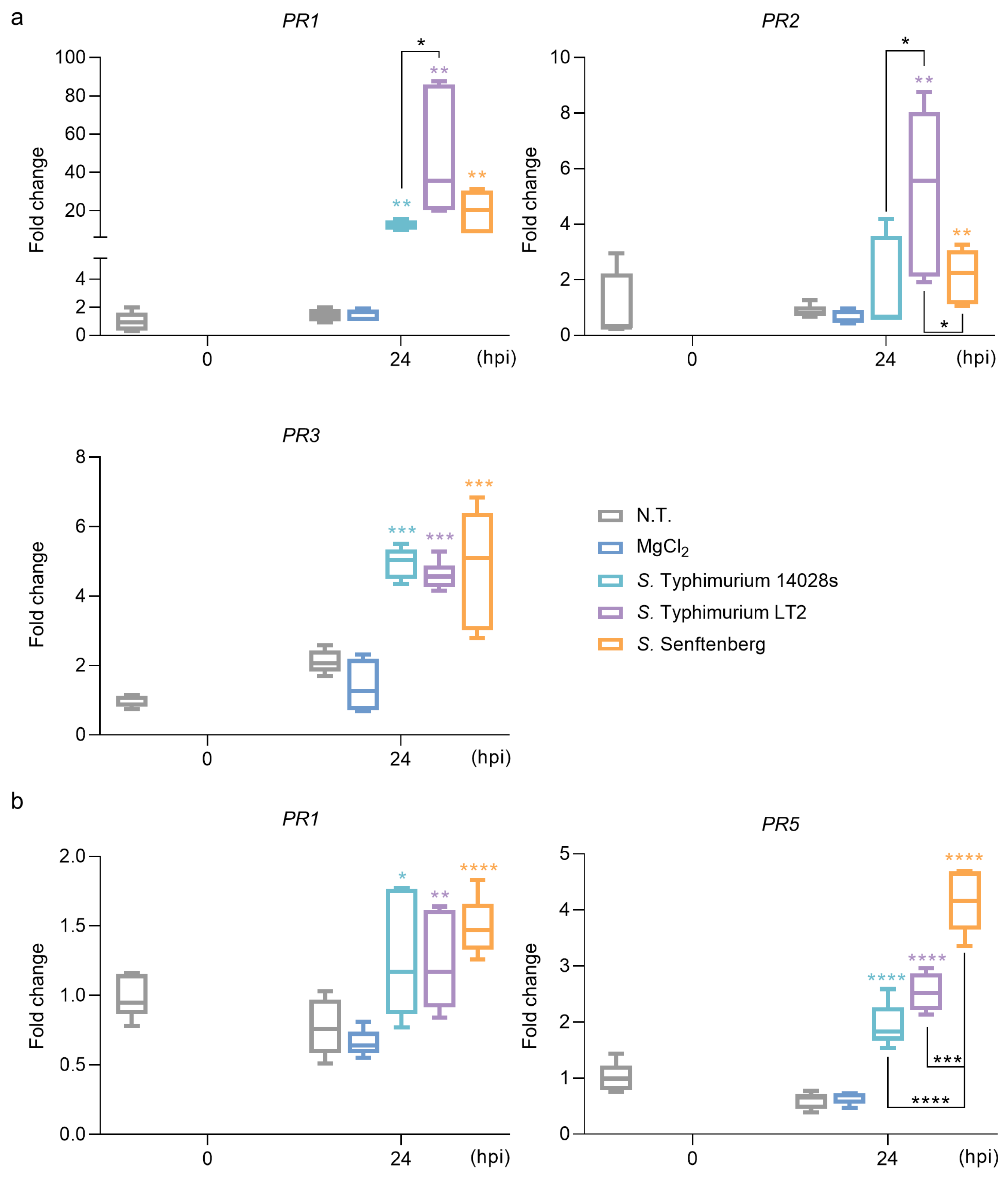
2.2. S. enterica Strains Regulate the Expression of PR Genes
2.3. Tomato Plants Modulate Their Responses to S. Typhimurium 14028s
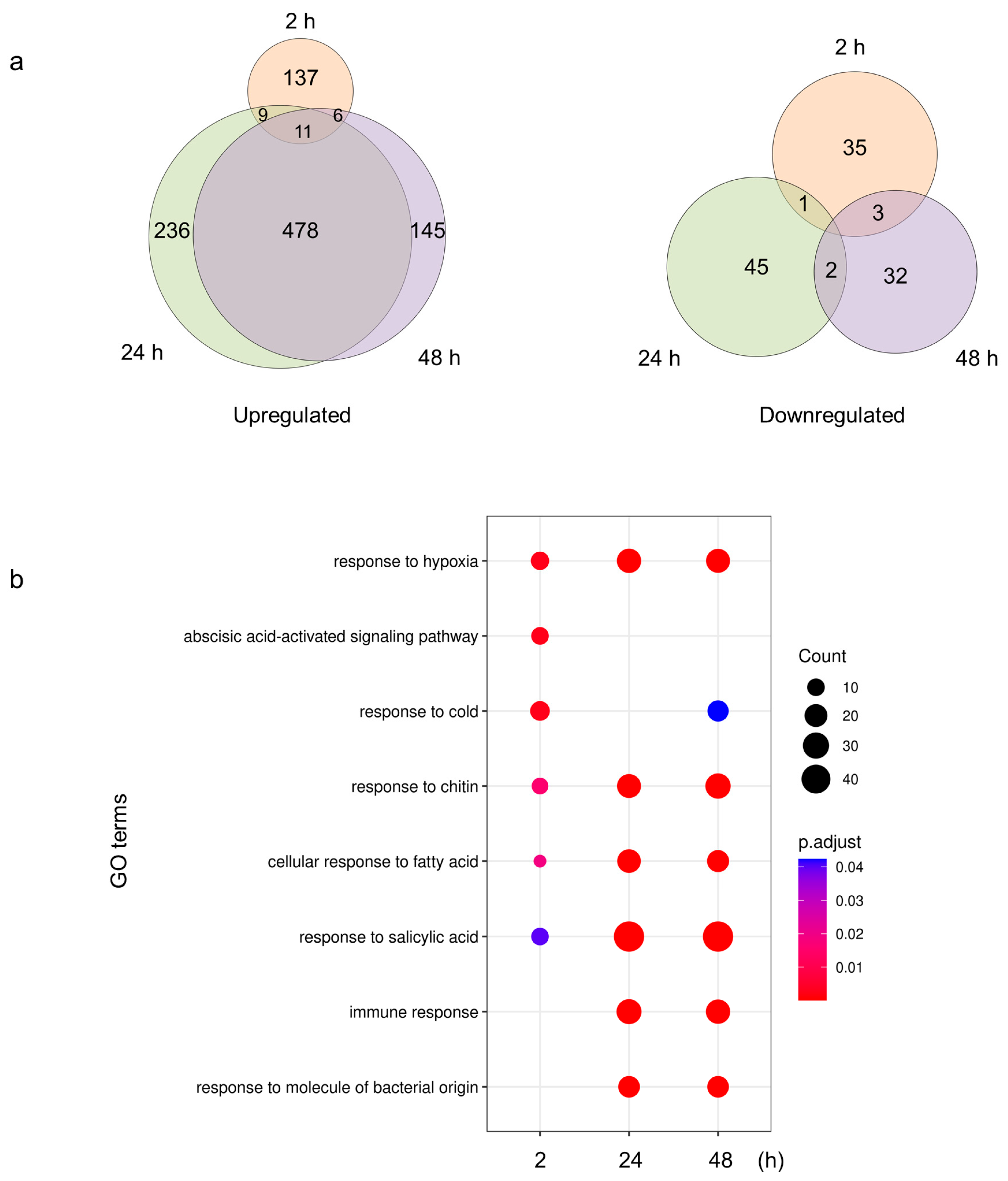
2.4. Plants Respond to S. Typhimurium 14028s with Metabolic and Immunological Adjustments
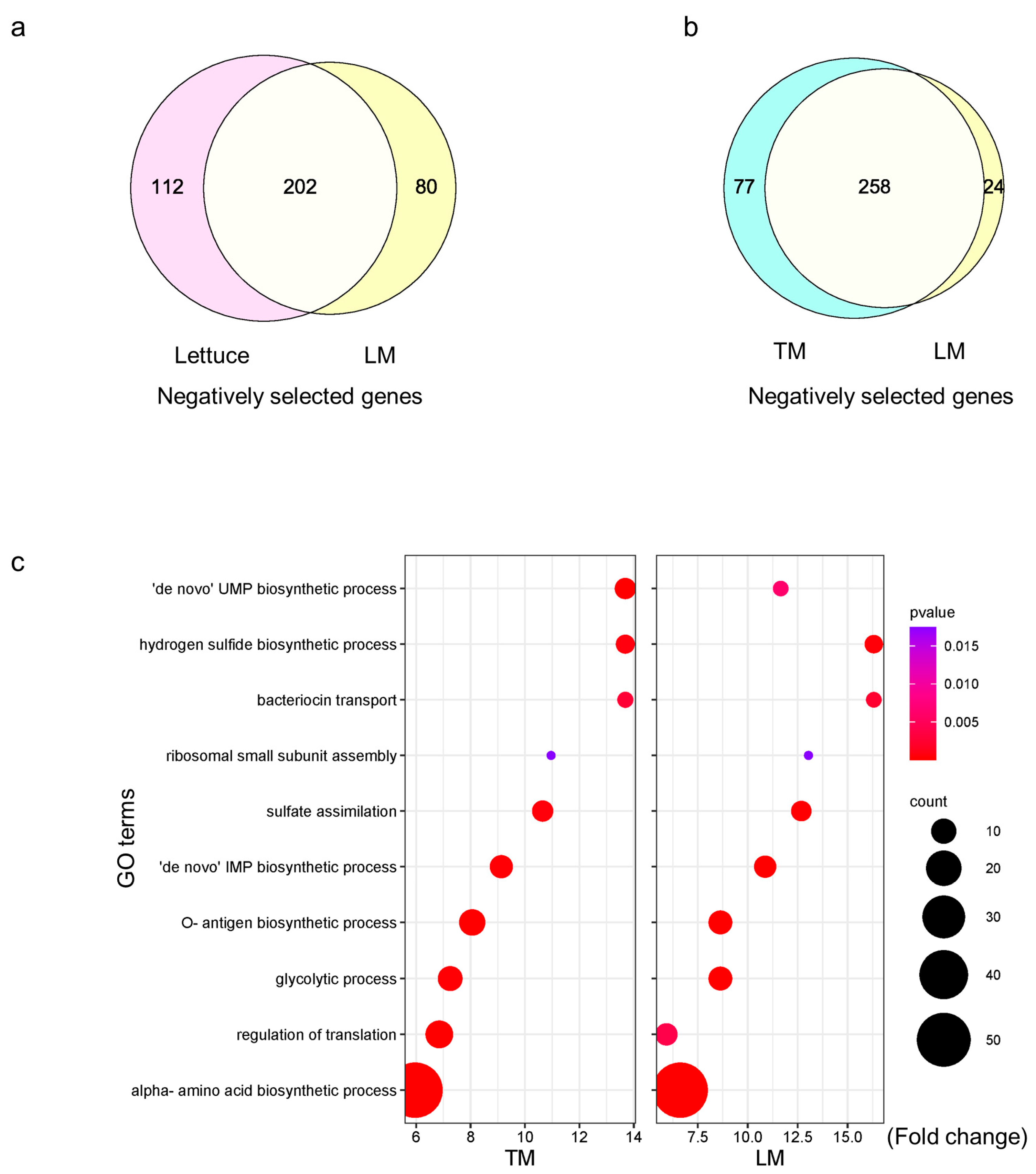
2.5. S. Typhimurium 14028s Adapts to Plant Leaf-Mimicking Media with Cellular Component Biosynthesis and Adjusted Metabolism
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Medium Recipes, and Culture Conditions
4.2. Plant Cultivation
4.3. Salmonella Persistence Assay
4.4. RT-qPCR Assay
4.5. Tomato Transcriptome Assay
4.6. Salmonella Tn-Seq Library Assay
4.7. Comparison of Plant Transcriptomes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silva, C.; Calva, E.; Maloy, S. One Health and Food-Borne Disease: Salmonella Transmission between Humans, Animals, and Plants. Microbiol. Spectr. 2014, 2, OH-0020-2013. [Google Scholar] [CrossRef]
- CDC. Reports of Selected Salmonella Outbreak Investigations. Available online: https://www.cdc.gov/salmonella/outbreaks.html (accessed on 18 August 2023).
- Islam, M.; Morgan, J.; Doyle, M.P.; Phatak, S.C.; Millner, P.; Jiang, X. Persistence of Salmonella enterica Serovar Typhimurium on Lettuce and Parsley and in Soils on Which They Were Grown in Fields Treated with Contaminated Manure Composts or Irrigation Water. Foodborne Pathog. Dis. 2004, 1, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, A.; Yaron, S. Transfer of Salmonella enterica Serovar Typhimurium from Contaminated Irrigation Water to Parsley Is Dependent on Curli and Cellulose, the Biofilm Matrix Components. J. Food Prot. 2009, 72, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.M.; Harris, L.J.; Beuchat, L.R. Survival and Recovery of Escherichia coli O157:H7, Salmonella, and Listeria monocytogenes on Lettuce and Parsley as Affected by Method of Inoculation, Time between Inoculation and Analysis, and Treatment with Chlorinated Water. J. Food Prot. 2004, 67, 1092–1103. [Google Scholar] [CrossRef] [PubMed]
- Jechalke, S.; Schierstaedt, J.; Becker, M.; Flemer, B.; Grosch, R.; Smalla, K.; Schikora, A. Salmonella Establishment in Agricultural Soil and Colonization of Crop Plants Depend on Soil Type and Plant Species. Front. Microbiol. 2019, 10, 967. [Google Scholar] [CrossRef]
- Takeuchi, K.; Matute, C.M.; Hassan, A.N.; Frank, J.F. Comparison of the Attachment of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella Typhimurium, and Pseudomonas fluorescens to Lettuce Leaves. J. Food Prot. 2000, 63, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Zarkani, A.A.; Schierstaedt, J.; Becker, M.; Krumwiede, J.; Grimm, M.; Grosch, R.; Jechalke, S.; Schikora, A. Salmonella adapts to plants and their environment during colonization of tomatoes. FEMS Microbiol. Ecol. 2019, 95, fiz152. [Google Scholar] [CrossRef] [PubMed]
- Marvasi, M.; Cox, C.E.; Xu, Y.; Noel, J.T.; Giovannoni, J.J.; Teplitski, M. Differential regulation of Salmonella typhimurium genes involved in O-antigen capsule production and their role in persistence within tomato fruit. Mol. Plant Microbe Interact. 2013, 26, 793–800. [Google Scholar] [CrossRef]
- Gu, G.; Hu, J.; Cevallos-Cevallos, J.M.; Richardson, S.M.; Bartz, J.A.; van Bruggen, A.H. Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS ONE 2011, 6, e27340. [Google Scholar] [CrossRef]
- Klerks, M.M.; Franz, E.; van Gent-Pelzer, M.; Zijlstra, C.; van Bruggen, A.H. Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 2007, 1, 620–631. [Google Scholar] [CrossRef]
- Tan, M.S.; White, A.P.; Rahman, S.; Dykes, G.A. Role of Fimbriae, Flagella and Cellulose on the Attachment of Salmonella Typhimurium ATCC 14028 to Plant Cell Wall Models. PLoS ONE 2016, 11, e0158311. [Google Scholar] [CrossRef] [PubMed]
- Montano, J.; Rossidivito, G.; Torreano, J.; Porwollik, S.; Sela Saldinger, S.; McClelland, M.; Melotto, M. Salmonella enterica Serovar Typhimurium 14028s Genomic Regions Required for Colonization of Lettuce Leaves. Front. Microbiol. 2020, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Kroupitski, Y.; Pinto, R.; Brandl, M.T.; Belausov, E.; Sela, S. Interactions of Salmonella enterica with lettuce leaves. J. Appl. Microbiol. 2009, 106, 1876–1885. [Google Scholar] [CrossRef] [PubMed]
- Kroupitski, Y.; Golberg, D.; Belausov, E.; Pinto, R.; Swartzberg, D.; Granot, D.; Sela, S. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 2009, 75, 6076–6086. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.V.; Charrier, A.; Schikora, A.; Bigeard, J.; Pateyron, S.; de Tauzia-Moreau, M.L.; Evrard, A.; Mithofer, A.; Martin-Magniette, M.L.; Virlogeux-Payant, I.; et al. Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-triggered immunity in Arabidopsis thaliana. Mol. Plant 2014, 7, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Altier, C.; Martin, G.B. Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ. Microbiol. 2013, 15, 2418–2430. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Ferelli, A.M.C.; Lin, S.S.; Micallef, S.A. Stress response, amino acid biosynthesis and pathogenesis genes expressed in Salmonella enterica colonizing tomato shoot and root surfaces. Heliyon 2020, 6, e04952. [Google Scholar] [CrossRef]
- Iniguez, A.L.; Dong, Y.; Carter, H.D.; Ahmer, B.M.M.; Stone, J.M.; Triplett, E.W. Regulation of Enteric Endophytic Bacterial Colonization by Plant Defenses. Mol. Plant Microbe Interact. 2005, 18, 169–178. [Google Scholar] [CrossRef]
- Zaragoza, W.J.; Noel, J.T.; Teplitski, M. Spontaneous non-rdar mutations increase fitness of Salmonella in plants. Environ. Microbiol. Rep. 2012, 4, 453–458. [Google Scholar] [CrossRef]
- Fratty, I.S.; Shachar, D.; Katsman, M.; Yaron, S. The activity of BcsZ of Salmonella Typhimurium and its role in Salmonella-plants interactions. Front. Cell Infect. Microbiol. 2022, 12, 967796. [Google Scholar] [CrossRef]
- Ferelli, A.M.C.; Bolten, S.; Szczesny, B.; Micallef, S.A. Salmonella enterica Elicits and Is Restricted by Nitric Oxide and Reactive Oxygen Species on Tomato. Front. Microbiol. 2020, 11, 391. [Google Scholar] [CrossRef]
- Schikora, A.; Carreri, A.; Charpentier, E.; Hirt, H. The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS ONE 2008, 3, e2279. [Google Scholar] [CrossRef] [PubMed]
- Oblessuc, P.R.; Matiolli, C.C.; Melotto, M. Novel molecular components involved in callose-mediated Arabidopsis defense against Salmonella enterica and Escherichia coli O157:H7. BMC Plant Biol. 2020, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Litt, P.K.; Kniel, K.E.; Bais, H. Evasion of Plant Innate Defense Response by Salmonella on Lettuce. Front. Microbiol. 2020, 11, 500. [Google Scholar] [CrossRef] [PubMed]
- Schikora, A.; Virlogeux-Payant, I.; Bueso, E.; Garcia, A.V.; Nilau, T.; Charrier, A.; Pelletier, S.; Menanteau, P.; Baccarini, M.; Velge, P.; et al. Conservation of Salmonella infection mechanisms in plants and animals. PLoS ONE 2011, 6, e24112. [Google Scholar] [CrossRef] [PubMed]
- Jacob, C.; Velasquez, A.C.; Josh, N.A.; Settles, M.; He, S.Y.; Melotto, M. Dual transcriptomic analysis reveals metabolic changes associated with differential persistence of human pathogenic bacteria in leaves of Arabidopsis and lettuce. G3 2021, 11, jkab331. [Google Scholar] [CrossRef] [PubMed]
- van Opijnen, T.; Bodi, K.L.; Camilli, A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 2009, 6, 767–772. [Google Scholar] [CrossRef]
- Kroupitski, Y.; Pinto, R.; Belausov, E.; Sela, S. Distribution of Salmonella typhimurium in romaine lettuce leaves. Food Microbiol. 2011, 28, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Bearson, S.M.; Benjamin, W.H.; Swords, W.E.; Foster, J.W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J. Bacteriol. 1996, 178, 2572–2579. [Google Scholar] [CrossRef]
- Sanderson, K.; Stocker, B. Salmonella typhimurium strains used in genetic analysis. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology; ASM Press: Washington, DC, USA, 1987; Volume 2, pp. 1220–1224. [Google Scholar]
- Swords, W.E.; Cannon, B.M.; Benjamin, W.H., Jr. Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect. Immun. 1997, 65, 2451–2453. [Google Scholar] [CrossRef]
- Elviss, N.C.; Little, C.L.; Hucklesby, L.; Sagoo, S.; Surman-Lee, S.; de Pinna, E.; Threlfall, E.J.; Food, W.; Environmental Surveillance Network. Microbiological study of fresh herbs from retail premises uncovers an international outbreak of salmonellosis. Int. J. Food Microbiol. 2009, 134, 83–88. [Google Scholar] [CrossRef]
- Trotereau, J.; Jouan, R.; Naquin, D.; Branger, M.; Schouler, C.; Velge, P.; Mergaert, P.; Virlogeux-Payant, I. Construction and characterization of a saturated Tn-seq library of Salmonella Typhimurium ATCC 14028. Microbiol. Resour. Announc. 2023, 12, e00365-23. [Google Scholar] [CrossRef]
- Linthorst, H.J.M.; Van Loon, L.C. Pathogenesis-related proteins of plants. Crit. Rev. Plant Sci. 1991, 10, 123–150. [Google Scholar] [CrossRef]
- Chalupowicz, L.; Manulis-Sasson, S.; Barash, I.; Elad, Y.; Rav-David, D.; Brandlc, M.T. Effect of Plant Systemic Resistance Elicited by Biological and Chemical Inducers on the Colonization of the Lettuce and Basil Leaf Apoplast by Salmonella enterica. Appl. Environ. Microbiol. 2021, 87, e01151-21. [Google Scholar] [CrossRef] [PubMed]
- Klerks, M.M.; van Gent-Pelzer, M.; Franz, E.; Zijlstra, C.; van Bruggen, A.H. Physiological and molecular responses of Lactuca sativa to colonization by Salmonella enterica serovar Dublin. Appl. Environ. Microbiol. 2007, 73, 4905–4914. [Google Scholar] [CrossRef]
- EFSA. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Patel, J.; Sharma, M. Differences in attachment of Salmonella enterica serovars to cabbage and lettuce leaves. Int. J. Food Microbiol. 2010, 139, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Popoff, M.Y.; Le Minor, L.O. Antigenic Formulas of the Salmonella Serovars; WHO Collaborating Center for Reference and Research on Salmonella: Institute Pasteur: Paris, France, 1997. [Google Scholar]
- Berger, C.N.; Brown, D.J.; Shaw, R.K.; Minuzzi, F.; Feys, B.; Frankel, G. Salmonella enterica strains belonging to O serogroup 1,3,19 induce chlorosis and wilting of Arabidopsis thaliana leaves. Environ. Microbiol. 2011, 13, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, A.L.; Gulig, P.A. The Salmonella typhimurium virulence plasmid encodes a positive regulator of a plasmid-encoded virulence gene. J. Bacteriol. 1991, 173, 7176–7185. [Google Scholar] [CrossRef] [PubMed]
- Bharath, P.; Gahir, S.; Raghavendra, A.S. Abscisic Acid-Induced Stomatal Closure: An Important Component of Plant Defense Against Abiotic and Biotic Stress. Front. Plant Sci. 2021, 12, 615114. [Google Scholar] [CrossRef]
- Roy, D.; Panchal, S.; Rosa, B.A.; Melotto, M. Escherichia coli O157:H7 Induces Stronger Plant Immunity than Salmonella enterica Typhimurium SL1344. Phytopathology 2013, 103, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Montillet, J.L.; Leonhardt, N.; Mondy, S.; Tranchimand, S.; Rumeau, D.; Boudsocq, M.; Garcia, A.V.; Douki, T.; Bigeard, J.; Lauriere, C.; et al. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 2013, 11, e1001513. [Google Scholar] [CrossRef] [PubMed]
- Zarkani, A.A.; Lopez-Pagan, N.; Grimm, M.; Sanchez-Romero, M.A.; Ruiz-Albert, J.; Beuzon, C.R.; Schikora, A. Salmonella Heterogeneously Expresses Flagellin during Colonization of Plants. Microorganisms 2020, 8, 815. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Broekgaarden, C.; Caarls, L.; Vos, I.A.; Pieterse, C.M.; Van Wees, S.C. Ethylene: Traffic Controller on Hormonal Crossroads to Defense. Plant Physiol. 2015, 169, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
- Adie, B.; Chico, J.M.; Rubio-Somoza, I.; Solano, R. Modulation of Plant Defenses by Ethylene. J. Plant Growth Regul. 2007, 26, 160–177. [Google Scholar] [CrossRef]
- Vaghela, B.; Vashi, R.; Rajput, K.; Joshi, R. Plant chitinases and their role in plant defense: A comprehensive review. Enzym. Microb. Technol. 2022, 159, 110055. [Google Scholar] [CrossRef]
- Grover, A. Plant Chitinases: Genetic Diversity and Physiological Roles. Crit. Rev. Plant Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Pandey, V.P.; Awasthi, M.; Singh, S.; Tiwari, S.; Dwivedi, U.N. A Comprehensive Review on Function and Application of Plant Peroxidases. Biochem. Anal. Biochem. 2017, 6, 308. [Google Scholar] [CrossRef]
- Almagro, L.; Gomez Ros, L.V.; Belchi-Navarro, S.; Bru, R.; Ros Barcelo, A.; Pedreno, M.A. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef]
- Robert, N.; Roche, K.; Lebeau, Y.; Breda, C.; Boulay, M.; Esnault, R.; Buffard, D. Expression of grapevine chitinase genes in berries and leaves infected by fungal or bacterial pathogens. Plant Sci. 2002, 162, 389–400. [Google Scholar] [CrossRef]
- Rakwal, R.; Yang, G.; Komatsu, S. Chitinase induced by jasmonic acid, methyl jasmonate, ethylene and protein phosphatase inhibitors in rice. Mol. Biol. Rep. 2004, 31, 113–119. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yue, X.; Wang, R.; Zhang, Y. Ethylene mediates UV-B-induced stomatal closure via peroxidase-dependent hydrogen peroxide synthesis in Vicia faba L. J. Exp. Bot. 2011, 62, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Schierstaedt, J.; Duan, Y.; Trotereau, J.; Virlogeux-Payant, I.; Schikora, A. Novel method to recover Salmonella enterica cells for Tn-Seq approaches from lettuce leaves and agricultural environments using combination of sonication, filtration, and dialysis membrane. J. Microbiol. Methods 2023, 208, 106724. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, M.H.; Desai, P.; Porwollik, S.; Canals, R.; Perez, D.R.; Chu, W.; McClelland, M.; Teplitski, M. Salmonella Persistence in Tomatoes Requires a Distinct Set of Metabolic Functions Identified by Transposon Insertion Sequencing. Appl. Environ. Microbiol. 2017, 83, e03028-16. [Google Scholar] [CrossRef] [PubMed]
- Elpers, L.; Deiwick, J.; Hensel, M. Effect of Environmental Temperatures on Proteome Composition of Salmonella enterica Serovar Typhimurium. Mol. Cell. Proteom. 2022, 21, 100265. [Google Scholar] [CrossRef] [PubMed]
- Prusky, D.B.; Wilson, R.A. Does increased nutritional carbon availability in fruit and foliar hosts contribute to modulation of pathogen colonization? Postharvest Biol. Technol. 2018, 145, 27–32. [Google Scholar] [CrossRef]
- Han, M.; Schierstaedt, J.; Duan, Y.; Nietschke, M.; Jechalke, S.; Wolf, J.; Hensel, M.; Neumann-Schaal, M.; Schikora, A. Salmonella enterica relies on carbon metabolism to adapt to agricultural environments. Front. Microbiol. 2023, 14, 1213016. [Google Scholar] [CrossRef]
- Noel, J.T.; Arrach, N.; Alagely, A.; McClelland, M.; Teplitski, M. Specific responses of Salmonella enterica to tomato varieties and fruit ripeness identified by in vivo expression technology. PLoS ONE 2010, 5, e12406. [Google Scholar] [CrossRef]
- Liu, B.; Hou, W.; Li, K.; Chen, Q.; Liu, Y.; Yue, T. Specific gene SEN1393 contributes to higher survivability of Salmonella Enteritidis in egg white by regulating sulfate assimilation pathway. Int. J. Food Microbiol. 2021, 337, 108927. [Google Scholar] [CrossRef]
- Fornefeld, E.; Schierstaedt, J.; Jechalke, S.; Grosch, R.; Schikora, A.; Smalla, K. Persistence of Salmonella Typhimurium LT2 in Soil Enhanced after Growth in Lettuce Medium. Front. Microbiol. 2017, 8, 757. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Su, X.; Wang, B.; Geng, X.; Du, Y.; Yang, Q.; Liang, B.; Meng, G.; Gao, Q.; Yang, W.; Zhu, Y.; et al. A high-continuity and annotated tomato reference genome. BMC Genom. 2021, 22, 898. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, M.; Zarkani, A.A.; Duan, Y.; Grimm, M.; Trotereau, J.; Virlogeux-Payant, I.; Schikora, A. Bidirectional Comparisons Revealed Functional Patterns in Interaction between Salmonella enterica and Plants. Plants 2024, 13, 414. https://doi.org/10.3390/plants13030414
Han M, Zarkani AA, Duan Y, Grimm M, Trotereau J, Virlogeux-Payant I, Schikora A. Bidirectional Comparisons Revealed Functional Patterns in Interaction between Salmonella enterica and Plants. Plants. 2024; 13(3):414. https://doi.org/10.3390/plants13030414
Chicago/Turabian StyleHan, Min, Azhar A. Zarkani, Yongming Duan, Maja Grimm, Jérôme Trotereau, Isabelle Virlogeux-Payant, and Adam Schikora. 2024. "Bidirectional Comparisons Revealed Functional Patterns in Interaction between Salmonella enterica and Plants" Plants 13, no. 3: 414. https://doi.org/10.3390/plants13030414
APA StyleHan, M., Zarkani, A. A., Duan, Y., Grimm, M., Trotereau, J., Virlogeux-Payant, I., & Schikora, A. (2024). Bidirectional Comparisons Revealed Functional Patterns in Interaction between Salmonella enterica and Plants. Plants, 13(3), 414. https://doi.org/10.3390/plants13030414







