A Genome-Wide Characterization of Receptor-like Cytoplasmic Kinase IV Subfamily Members in Populus deltoides Identifies the Potential Role of PdeCRCK6 in Plant Osmotic Stress Responses
Abstract
1. Introduction
2. Results
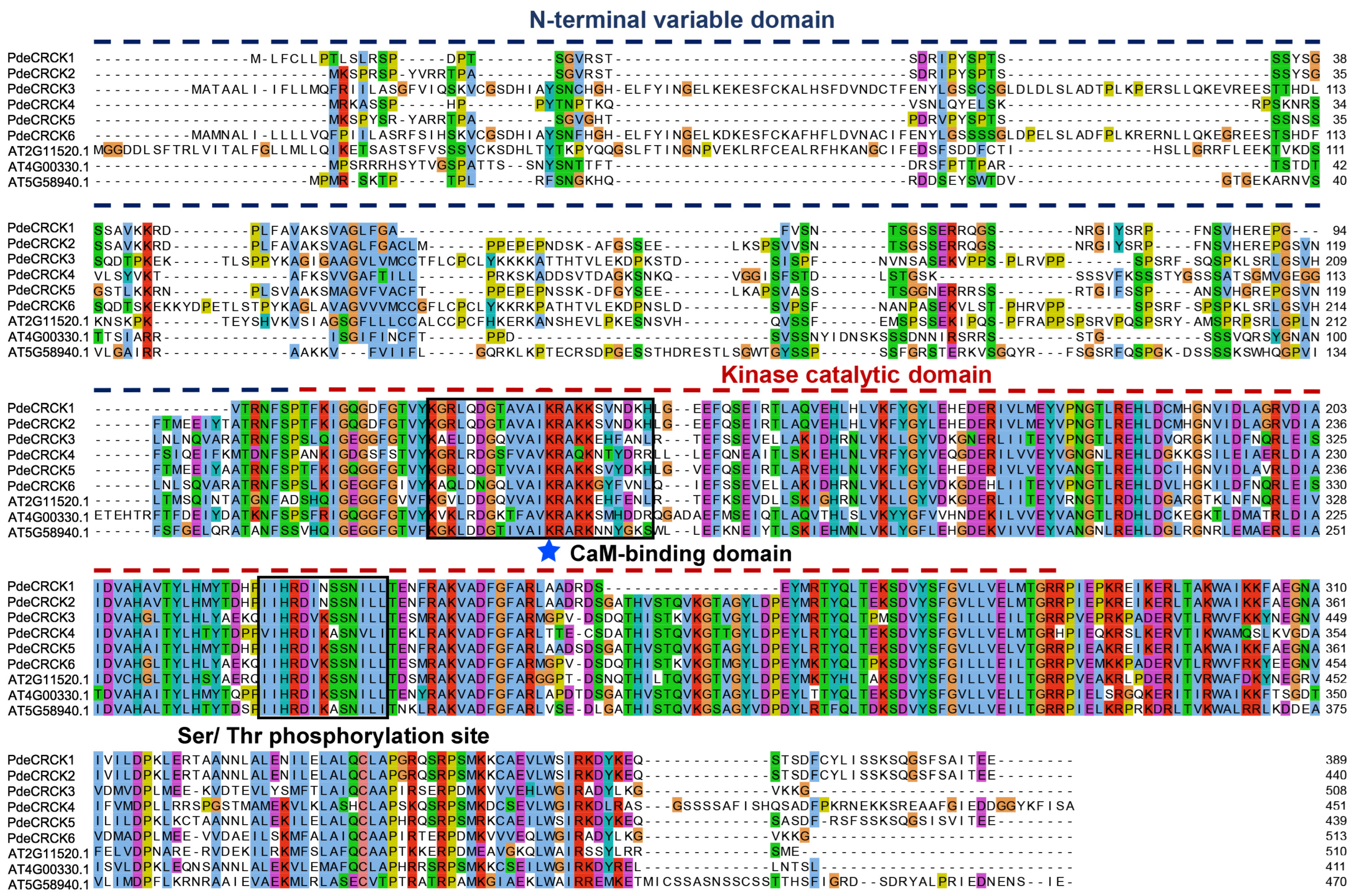
2.1. Identification of RLCK-IV/CRCK Subfamily Members in P. deltoides
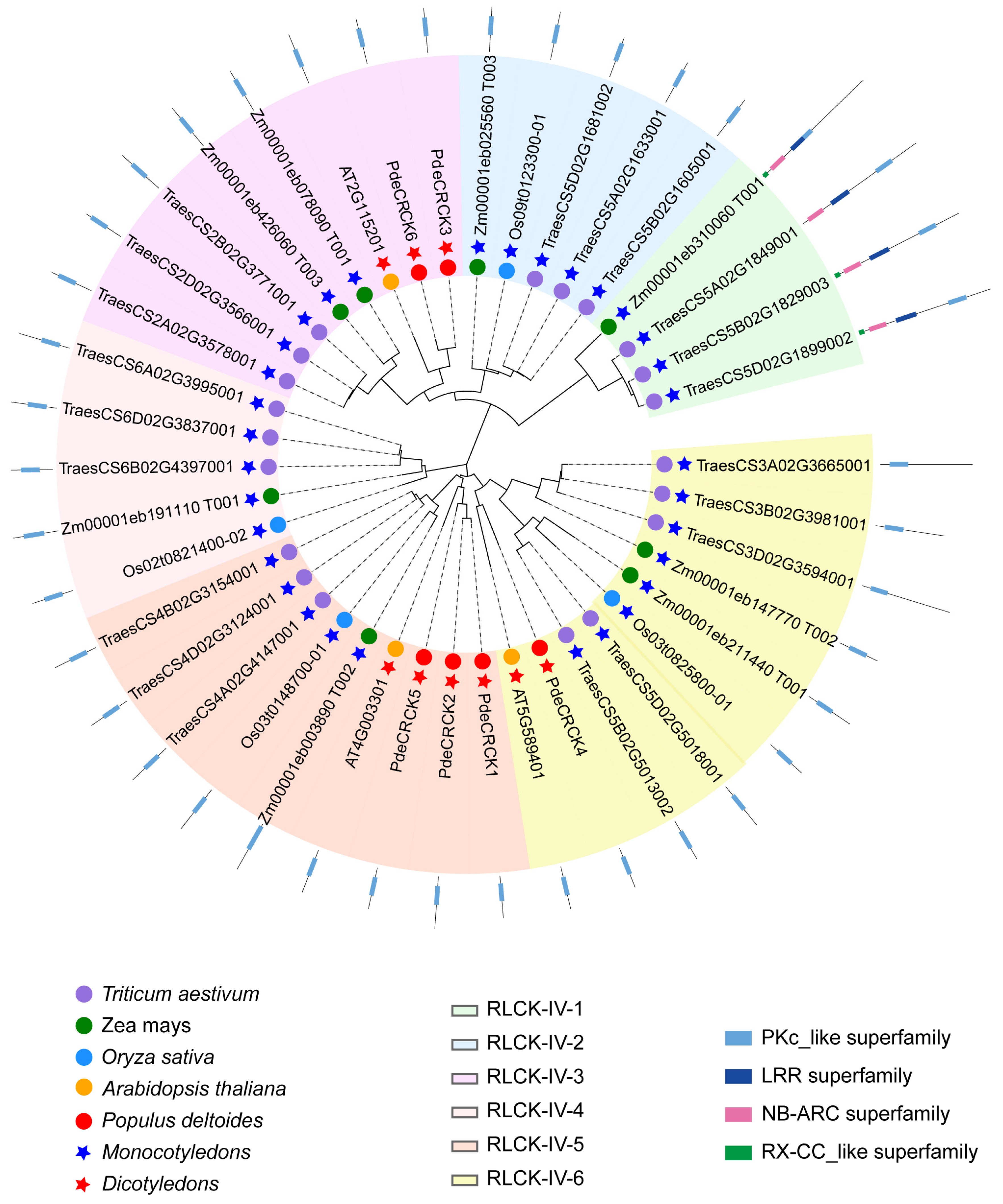
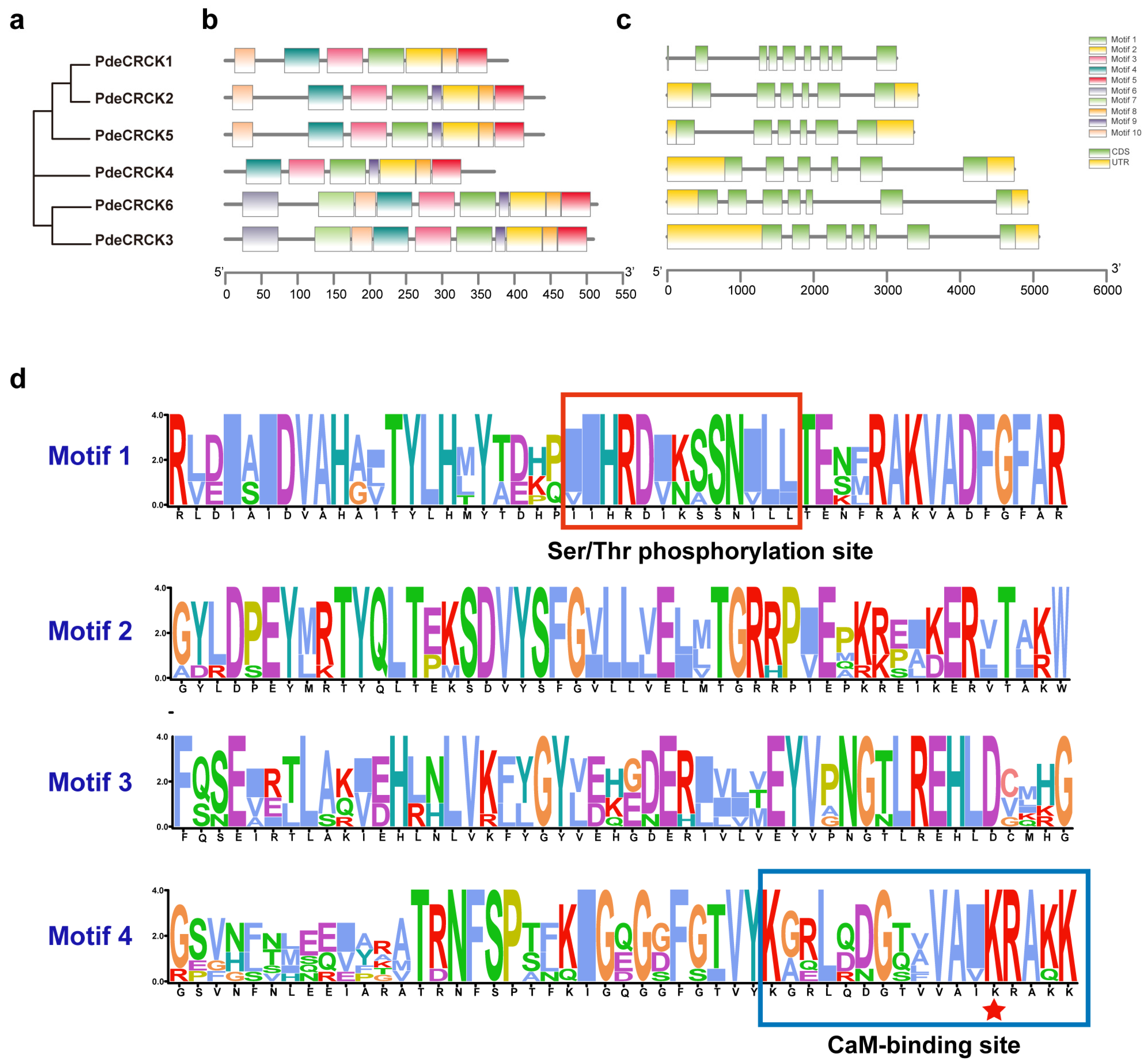
2.2. Phylogenetic Analysis of RLCK-IV/CRCKs
2.3. Gene Structure and Conserved Motifs of PdeCRCKs
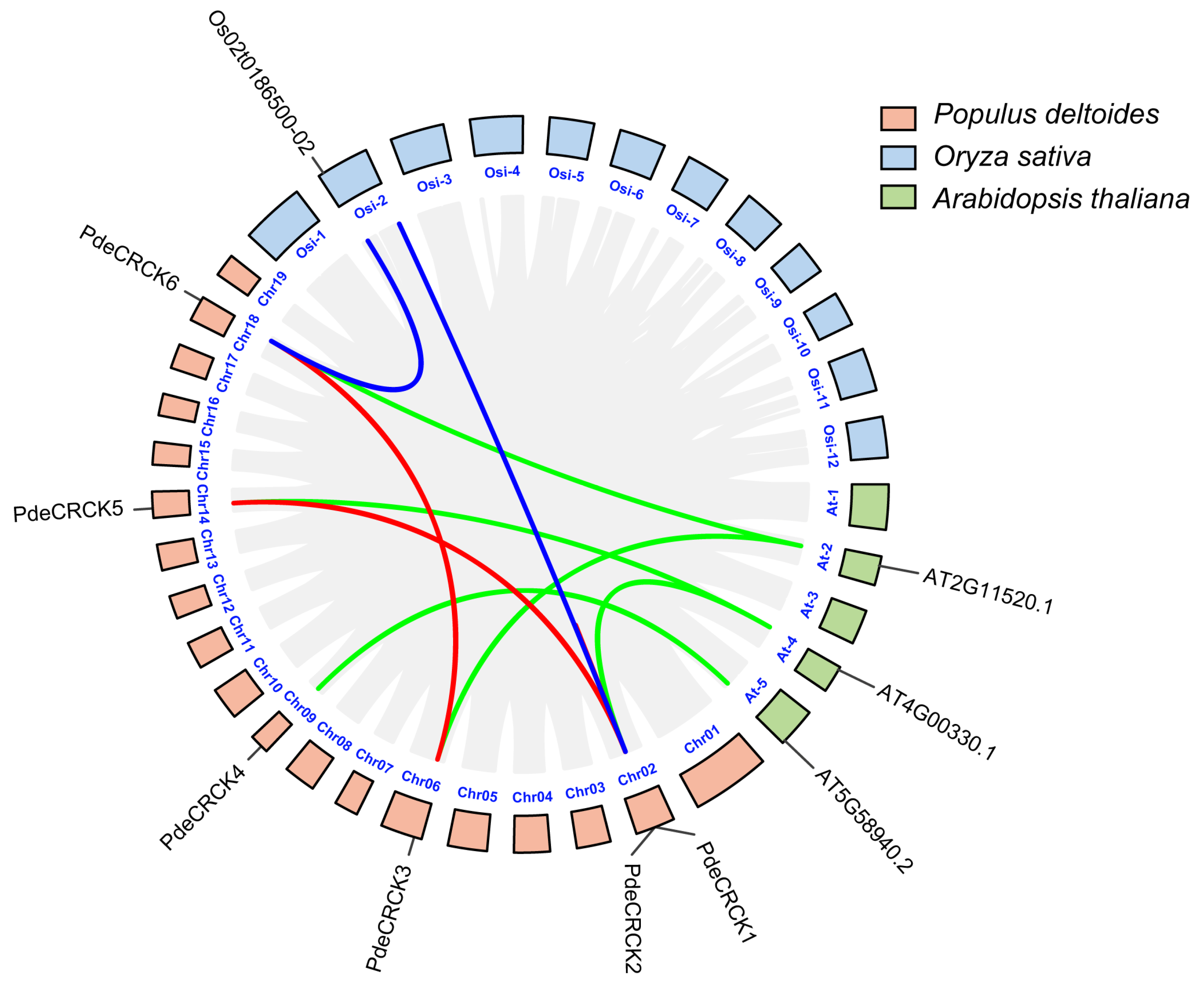
2.4. Chromosome Distribution and Collinearity Analysis of PdeCRCKs
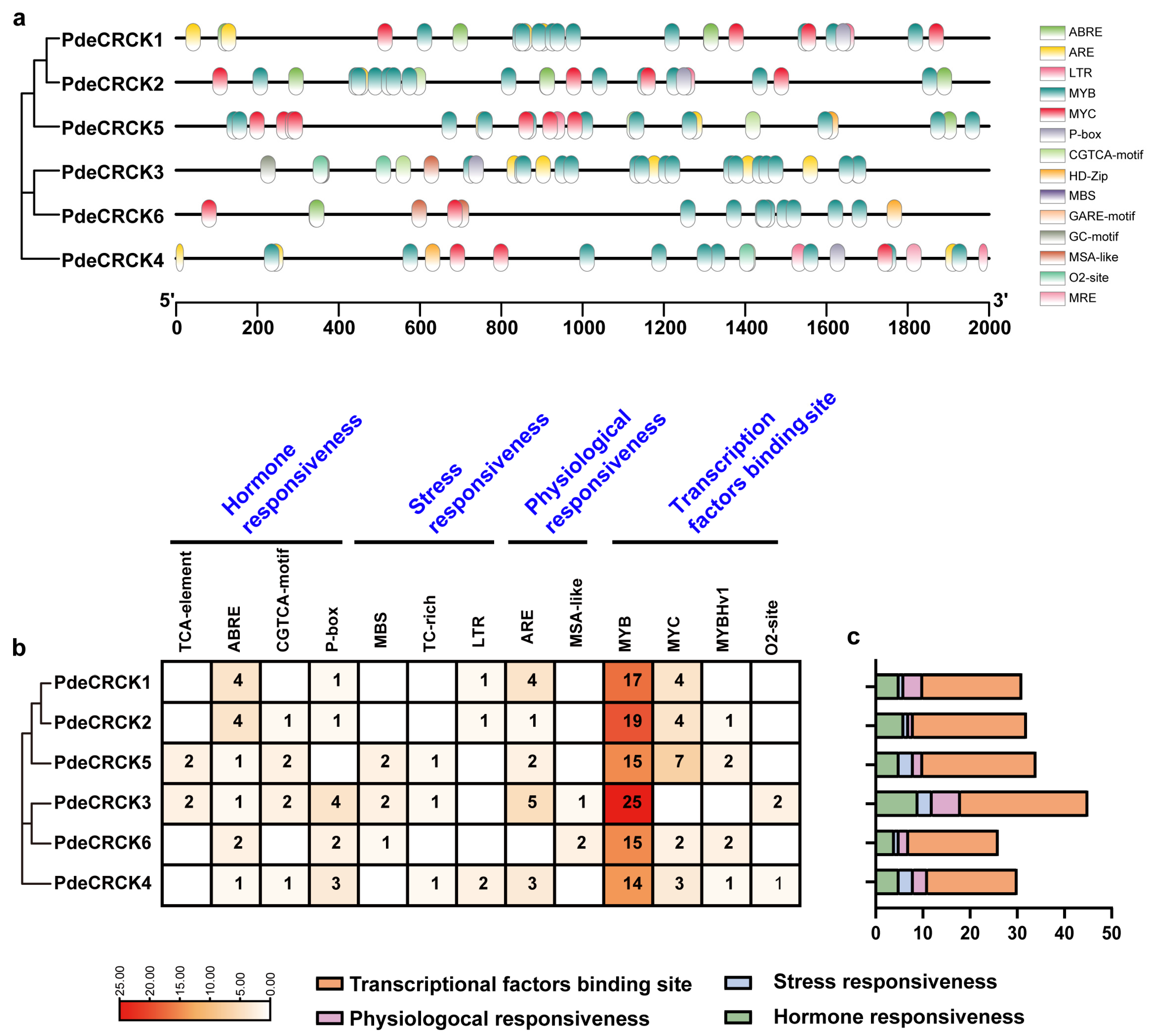
2.5. Cis-Acting Regulatory Element Identification in Promoter of PdeCRCKs
2.6. Expression Profiles of PdeCRCKs Under Polyethylene Glycol (PEG), Mannitol, and ABA Stresses
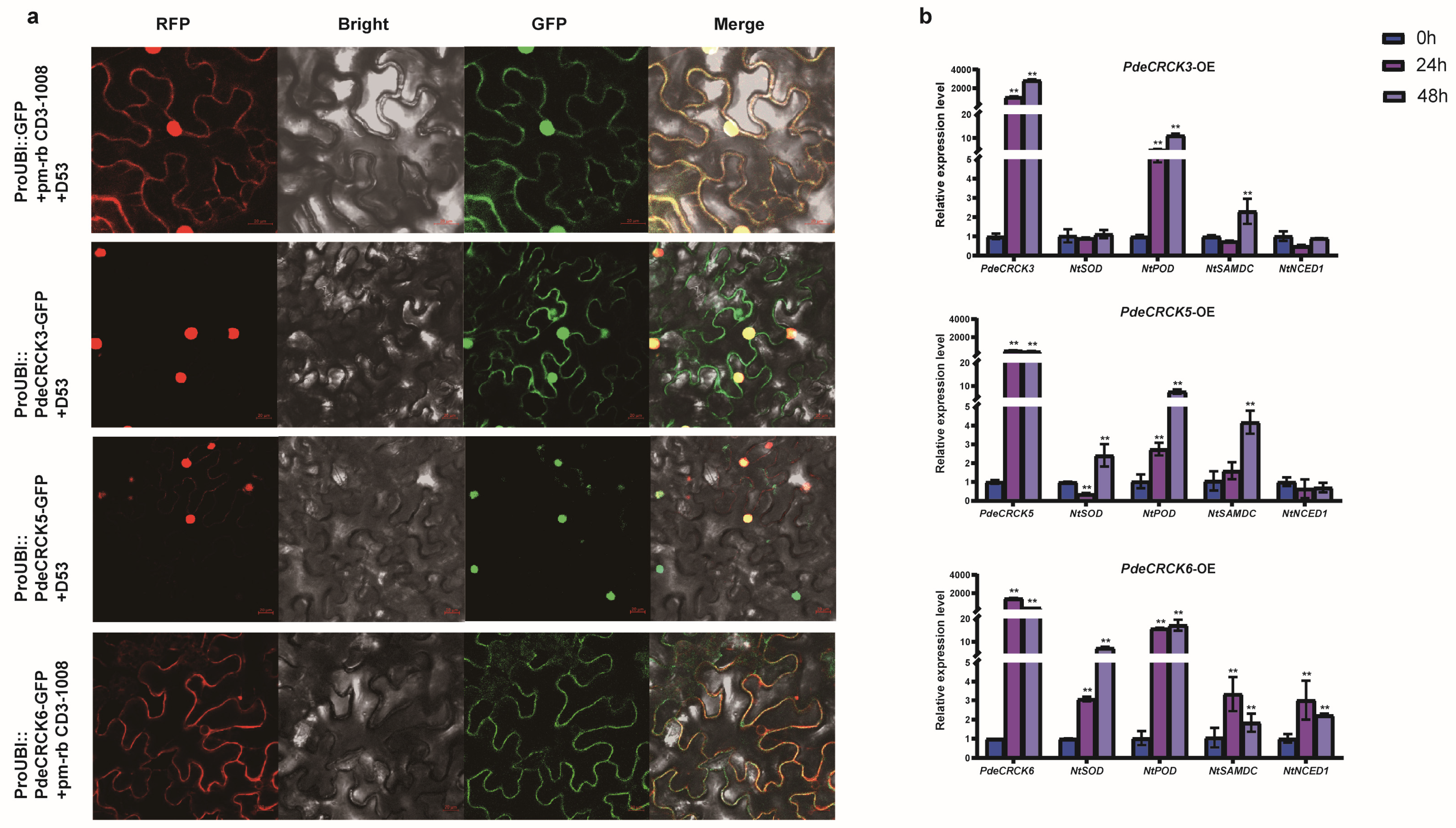
2.7. Subcellular Localization and Potential Functions of PdeCRCK3/5/6
2.8. Proteins That Interact with PdeCRCK6
3. Discussion
4. Materials and Methods
4.1. Identification of RLCK-IV Genes in P. deltoides
4.2. Physicochemical Property Characterization of PdeCRCKs
4.3. Sequence Alignment and Phylogenetic Tree Construction
4.4. Gene Structure, Motif Identification, and Chromosomal Location
4.5. Cis-Acting Regulatory Elements’ Identification in Putative Promoter Sequences
4.6. Synteny Relationships of RLCK-IV/CRCK Subfamily Members
4.7. Plant Materials, Drought Stress, RNA Extraction, and qRT–PCR Analysis
4.8. Subcellular Localization and Transient Expression Analysis of PdeCRCKs in Tobacco
4.9. Quantitative qRT–PCR Analysis
4.10. Immunoprecipitation–Mass Spectrometry (IP–MS)
4.11. Yeast Two-Hybird Assays
4.12. Quantification and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tuskan, G.A. Short-Rotation Woody Crop Supply Systems in the United States: What Do We Know and What Do We Need to Know? Biomass Bioenergy 1998, 14, 307–315. [Google Scholar] [CrossRef]
- Yang, X.; Deng, F.; Ramonell, K.M. Receptor-like Kinases and Receptor-like Proteins: Keys to Pathogen Recognition and Defense Signaling in Plant Innate Immunity. Front. Biol. 2012, 7, 155–166. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Expansion of the Receptor-Like Kinase/Pelle Gene Family and Receptor-Like Proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Plant Receptor-like Kinases; Elsevier: Amsterdam, The Netherlands, 2023; ISBN 978-0-323-90594-7.
- Singh, P.; Mishra, A.K.; Singh, C.M. Genome-Wide Identification and Characterization of Lectin Receptor-like Kinase (LecRLK) Genes in Mungbean (Vigna Radiata L. Wilczek). J. Appl. Genet. 2021, 62, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Li, J.; Shang, H.; Chen, X.; Hu, X. The RLK Protein TaCRK10 Activates Wheat High-temperature Seedling-plant Resistance to Stripe Rust through Interacting with TaH2A.1. Plant J. 2021, 108, 1241–1255. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Upadhyay, S.K. An Overview of Receptor-like Kinases in Plants. In Plant Receptor-Like Kinases; Elsevier: Amsterdam, The Netherlands, 2023; pp. 1–23. ISBN 978-0-323-90594-7. [Google Scholar]
- Shekhawat, J.; Upadhyay, S.K. DPY1 as an Osmosensor for Drought Signaling. Trends Plant Sci. 2024, 29, 616–619. [Google Scholar] [CrossRef]
- Liu, X.; Liang, C.; Hou, S.; Wang, X.; Chen, D.; Shen, J.; Zhang, W.; Wang, M. The LRR-RLK Protein HSL3 Regulates Stomatal Closure and the Drought Stress Response by Modulating Hydrogen Peroxide Homeostasis. Front. Plant Sci. 2020, 11, 548034. [Google Scholar] [CrossRef]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.X.; Li, W.H. Comparative Analysis of the Receptor-Like Kinase Family in Arabidopsis and Rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Wang, G.; Xiong, L.R.; Sun, J.X.; Chen, Y.; Guo, C.L.; Yu, Y.; He, H.L.; Cai, R.; Pan, J.S. Genome-Wide Identification and Characterization of Lectin Receptor-Like Kinase Gene Family in Cucumber and Expression Profiling Analysis under Different Treatments. Genes 2020, 11, 1032. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, M.-H.; Xiao, D.; Huang, R.L.; Zhan, J.; Wang, A.Q.; He, L.F. Genome-Wide Identification and Evolutionary Analysis of RLKs Involved in the Response to Aluminium Stress in Peanut. BMC Plant Biol. 2021, 21, 281. [Google Scholar] [CrossRef]
- Shumayla; Sharma, S.; Pandey, A.K.; Singh, K.; Upadhyay, S.K. Molecular Characterization and Global Expression Analysis of Lectin Receptor Kinases in Bread Wheat (Triticum Aestivum). PLoS ONE 2016, 11, e0153925. [Google Scholar] [CrossRef] [PubMed]
- Zeiner, A.; Colina, F.J.; Citterico, M.; Wrzaczek, M. Cysteine-Rich Receptor-Like Protein Kinases: Their Evolution, Structure, and Roles in Stress Response and Development. J. Exp. Bot. 2023, 74, 4910–4927. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Liang, S.; Wu, Z.; Bi, C.; Yu, Y.T.; Wang, X.F.; Zhang, D.P. Overexpression of an Arabidopsis Cysteine-Rich Receptor-like Protein Kinase, CRK5, Enhances Abscisic Acid Sensitivity and Confers Drought Tolerance. J. Exp. Bot. 2016, 67, 5009–5027. [Google Scholar] [CrossRef] [PubMed]
- Zameer, R.; Alwutayd, K.M.; Alshehri, D.; Mubarik, M.S.; Li, C.; Yu, C.; Li, Z. Identification of Cysteine-Rich Receptor-like Kinase Gene Family in Potato: Revealed StCRLK9 in Response to Heat, Salt and Drought Stresses. Funct. Plant Biol. 2024, 51, FP23320. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Sharma, S.; Kumar, R.; Mendu, V.; Singh, K.; Upadhyay, S.K. Genomic Dissection and Expression Profiling Revealed Functional Divergence in Triticum Aestivum Leucine Rich Repeat Receptor Like Kinases (TaLRRKs). Front. Plant Sci. 2016, 7, 1374. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Madhu; Singh, K.; Upadhyay, S.K. LysM Domain-Containing Proteins Modulate Stress Response and Signalling in Triticum Aestivum L. Environ. Exp. Bot. 2021, 189, 104558. [Google Scholar] [CrossRef]
- Rahim, A.A.; Uzair, M.; Rehman, N.; Rehman, O.U.; Zahra, N.; Khan, M.R. Genome-Wide Identification and Characterization of Receptor-Like Protein Kinase 1 (RPK1) Gene Family in Triticum Aestivum Under Drought Stress. Front. Genet. 2022, 13, 912251. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Tyagi, S.; Sharma, A.; Singh, K.; Upadhyay, S.K. Genomic Dissection and Transcriptional Profiling of Cysteine-Rich Receptor-like Kinases in Five Cereals and Functional Characterization of TaCRK68-A. Int. J. Biol. Macromol. 2019, 134, 316–329. [Google Scholar] [CrossRef]
- Ma, X.L.; Cui, W.N.; Zhao, Q.; Zhao, J.; Hou, X.N.; Li, D.Y.; Chen, Z.L.; Shen, Y.Z.; Huang, Z.J. Functional Study of a Salt-Inducible TaSR Gene in Triticum Aestivum. Physiol. Plant. 2016, 156, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Yang, S.H.; Shin, K.H.; Lee, S.C.; Kim, S.H. The AtLRK10L1.2, Arabidopsis Ortholog of Wheat LRK10, Is Involved in ABA-Mediated Signaling and Drought Resistance. Plant Cell Rep. 2015, 34, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The Receptor-Like Cytoplasmic Kinase (OsRLCK) Gene Family in Rice: Organization, Phylogenetic Relationship, and Expression during Development and Stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef] [PubMed]
- Jurca, M.E.; Bottka, S.; Fehér, A. Characterization of a Family of Arabidopsis Receptor-like Cytoplasmic Kinases (RLCK Class VI). Plant Cell Rep. 2008, 27, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y. Identification of Two Cassava Receptor-like Cytoplasmic Kinase Genes Related to Disease Resistance via Genome-Wide and Functional Analysis. Genomics 2023, 115, 110626. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Osakabe, Y.; Katsura, S.; Mizuno, S.; Maruyama, K.; Kusakabe, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abiotic Stress-inducible Receptor-like Kinases Negatively Control ABA Signaling in Arabidopsis. Plant J. 2012, 70, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Chaudhuri, S.; Yang, L.; Du, L.; Poovaiah, B.W. A Calcium/Calmodulin-Regulated Member of the Receptor-like Kinase Family Confers Cold Tolerance in Plants. J. Biol. Chem. 2010, 285, 7119–7126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP Kinase Cascades Regulate the Cold Response by Modulating ICE1 Protein Stability. Dev. Cell 2017, 43, 618–629.e5. [Google Scholar] [CrossRef] [PubMed]
- Hailemariam, S.; Liao, C.J.; Mengiste, T. Receptor-like Cytoplasmic Kinases: Orchestrating Plant Cellular Communication. Trends Plant Sci. 2024, 29, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Ramegowda, V.; Basu, S.; Krishnan, A.; Pereira, A. Rice GROWTH UNDER DROUGHT KINASE Is Required for Drought Tolerance and Grain Yield under Normal and Drought Stress Conditions. Plant Physiol. 2014, 166, 1634–1645. [Google Scholar] [CrossRef]
- Furio, R.N. Role of Calcium in the Defense Response Induced by Brassinosteroids in Strawberry Plants. Sci. Hortic. 2020, 261, 109010. [Google Scholar] [CrossRef]
- Zhang, L.; Du, L.; Poovaiah, B.W. Calcium Signaling and Biotic Defense Responses in Plants. Plant Signal. Behav. 2014, 9, e973818. [Google Scholar] [CrossRef] [PubMed]
- Raina, M.; Kisku, A.V.; Joon, S.; Kumar, S.; Kumar, D. Calmodulin and Calmodulin-like Ca2+ Binding Proteins as Molecular Players of Abiotic Stress Response in Plants. In Calcium Transport Elements in Plants; Elsevier: Amsterdam, The Netherlands, 2021; pp. 231–248. ISBN 978-0-12-821792-4. [Google Scholar]
- Yang, T.; Chaudhuri, S.; Yang, L.; Chen, Y.; Poovaiah, B.W. Calcium/Calmodulin Up-Regulates a Cytoplasmic Receptor-like Kinase in Plants. J. Biol. Chem. 2004, 279, 42552–42559. [Google Scholar] [CrossRef]
- Fahrenkrog, A.M.; Neves, L.G.; Resende, M.F.R.; Dervinis, C.; Davenport, R.; Barbazuk, W.B.; Kirst, M. Population Genomics of the Eastern Cottonwood (Populus Deltoides). Ecol. Evol. 2017, 7, 9426–9440. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Wu, H.; Zhang, J.; Pan, Z.; Zhao, W.; Li, Z.; Tong, C. Genome Assembly of Salicaceae Populus Deltoides (Eastern Cottonwood) I-69 Based on Nanopore Sequencing and Hi-C Technologies. J. Hered. 2021, 112, 303–310. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, D.; Li, S.; Su, Y.; Liang, Q.; Meng, H.; Shen, S.; Fan, Y.; Liu, C.; Zhang, C. FtsHi4 Is Essential for Embryogenesis Due to Its Influence on Chloroplast Development in Arabidopsis. PLoS ONE 2014, 9, e99741. [Google Scholar] [CrossRef]
- Restrepo-Montoya, D.; Brueggeman, R.; McClean, P.E.; Osorno, J.M. Computational Identification of Receptor-like Kinases “RLK” and Receptor-like Proteins “RLP” in Legumes. BMC Genom. 2020, 21, 459. [Google Scholar] [CrossRef] [PubMed]
- Yan, J. Phylogeny of the Plant Receptor-like Kinase (RLK) Gene Family and Expression Analysis of Wheat RLK Genes in Response to Biotic and Abiotic Stresses. BMC Genom. 2023, 24, 224. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Ma, W.; Liu, C.; Zhang, C.; Wu, S.; Chen, M.; Liu, K.; Cai, F.; Lin, F. Evolution and Expression Characteristics of Receptor-Like Cytoplasmic Protein Kinases in Maize, Rice and Arabidopsis. IJMS 2018, 19, 3680. [Google Scholar] [CrossRef] [PubMed]
- Sade, N.; Weng, F.; Tajima, H.; Zeron, Y.; Zhang, L. A Cytoplasmic Receptor-like Kinase Contributes to Salinity Tolerance. Plants 2020, 9, 1383. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Akiyama, Y. Cellular functions, mechanism of action, and regulation of ftsh protease. Annu. Rev. Microbiol. 2005, 59, 211–231. [Google Scholar] [CrossRef] [PubMed]
- Olinares, P.D.B.; Kim, J.; Van Wijk, K.J. The Clp Protease System; a Central Component of the Chloroplast Protease Network. Biochim. Biophys. Acta (BBA) Bioenerg. 2011, 1807, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Butenko, Y.; Lin, A.; Naveh, L.; Kupervaser, M.; Levin, Y.; Reich, Z.; Adam, Z. Differential Roles of the Thylakoid Lumenal Deg Protease Homologs in Chloroplast Proteostasis. Plant Physiol. 2018, 178, 1065–1080. [Google Scholar] [CrossRef] [PubMed]
- Parcerisa, I.L.; Rosano, G.L.; Ceccarelli, E.A. Biochemical Characterization of ClpB3, a Chloroplastic Disaggregase from Arabidopsis Thaliana. Plant Mol. Biol. 2020, 104, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14–SCFD3-Dependent Degradation of D53 Regulates Strigolactone Signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A Multicolored Set of in Vivo Organelle Markers for Co-localization Studies in Arabidopsis and Other Plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Li, L.; Tang, S.; Yuan, C.; Tian, Q.; Su, Y.; Li, H.G.; Zhao, L.; Yin, W.; Zhao, R.; et al. Evaluation of Appropriate Reference Genes for Reverse Transcription-Quantitative PCR Studies in Different Tissues of a Desert Poplar via Comparision of Different Algorithms. IJMS 2015, 16, 20468–20491. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.S.; Luo, T.; Fu, X.Z.; Fan, Q.J.; Liu, J.H. Cloning and Molecular Characterization of a Mitogen-Activated Protein Kinase Gene from Poncirus Trifoliata Whose Ectopic Expression Confers Dehydration/Drought Tolerance in Transgenic Tobacco. J. Exp. Bot. 2011, 62, 5191–5206. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]


| Gene Name | Gene Locations | CDS (bp) | Protein Length (aa) | MW (kDa) | PI | Instability Index | Aliphatic Index | Sub Location |
|---|---|---|---|---|---|---|---|---|
| PdeCRCK1 | Podel.02G178500.1.v2.1 | 1171 | 389 | 43,623 | 8.78 | 45.49 | 83.5 | Nucleus |
| PdeCRCK2 | Podel.02G180200.1.v2.1 | 1324 | 440 | 49,046 | 8.59 | 50.25 | 76.7 | Nucleus |
| PdeCRCK3 | Podel.06G069100.1.v2.1 | 1527 | 508 | 56,946 | 7.94 | 39.78 | 90.93 | Cell membrane and nucleus |
| PdeCRCK4 | Podel.09G041200.1.v2.1 | 1356 | 451 | 49,881 | 9.49 | 41.82 | 78.69 | Nucleus |
| PdeCRCK5 | Podel.14G089100.1.v2.1 | 1320 | 439 | 48,644 | 9.24 | 49.59 | 81.12 | Nucleus |
| PdeCRCK6 | Podel.18G126600.1.v2.1 | 1542 | 513 | 57,581 | 7.99 | 38.31 | 93.1 | Cell membrane |
| ID | Gene Id | Pfam | GO | Best-Hit-Arabi | Arabi-Defline |
|---|---|---|---|---|---|
| 1 | Podel.02G006800.1 | PF02672, PF00044, and PF02800 | GO:0055114 and GO:0016620 | AT1G42970.1 | Glyceraldehyde-3-phosphate dehydrogenase B subunit |
| 2 | Podel.04G229700.1 | ||||
| 3 | Podel.05G065100.1 | PF05496 | GO:0009378, GO:0006310, and GO:0006281 | AT5G64580.1 | AAA-type ATPase family proteins |
| 4 | Podel.10G049300.1 | PF00044 and PF02800 | GO:0055114 and GO:0016620 | AT3G04120.1 | Glyceraldehyde-3-phosphate dehydrogenase C subunit 1 |
| 5 | Podel.12G142900.1 | PF05199 and PF00732 | GO:0055114, GO:0016614, and GO:0050660 | AT5G51950.1 | Glucose–methanol–choline (GMC) oxidoreductase family proteins |
| 6 | Podel.14G119200.1 | PF00240 and PF01599 | GO:0005515 and GO:0006412, GO:0005840, and GO:0003735 | AT2G47110.2 | Ubiquitin 6 |
| 7 | Podel.T155200.1 | PF00657 | GO:0016788 | AT1G29670.1 | GDSL-like Lipase/Acylhydrolase superfamily proteins |
| Gene ID | Primer Sequence F (5′-3′) | Primer Sequence R (5′-3′) |
|---|---|---|
| PdeRG5-RT | CCCAGAGCCGCACCAACT | TGGGTTTCTTGATGCCATTTTG |
| Podel.06G209700.1-RT | AAGCCTCCGGAGCAAATGAA | CCTGCACTTGGTGTCCTCTT |
| Podel.02G016400.1-RT | ATGCATCGGCACAGACTTGA | TCATGCTCGCAAACTCCTCA |
| Podel.16G071000.1-RT | TGGGTCCTTGACTAAAGTGC | GAGGCTTGTTTGGTCTTGCG |
| Podel.01G118600.1-RT | GGGCATGATGGGAAGTGGAA | GGTCTTGCCTTTCATGGGGA |
| Podel.05G261000.1-RT | ACCAACCAACACCACCTATACC | TTCCCGACGCCTTCTCTGTA |
| PdeCRCK1-RT | TTTTGTCTTTTGCCCACGCT | GCCTCGATTTGAACCTTGCC |
| PdeCRCK2-RT | TTCTGGGAGCAGTGAAAGAAGG | CACCTTGTCCAATCTTGAATGTG |
| PdeCRCK3-RT | ACCCCCATGAGTGACGTGTA | ACCTCCGTGTCCACCTTTTC |
| PdeCRCK4-RT | TGCGAGATTGACCACCGAAT | TCAATCGGGTGTCTTCCTGT |
| PdeCRCK5-RT | GTAGGCGCCCTATTGAAGCA | GCCAAGTTATTTGCTGCGGT |
| PdeCRCK6-RT | GCGCCCTGTGGAGATGAAG | TACACCTTTCAGGTAGTCTGCT |
| NtUBQ-RT | TCCAGGACAAGGAGGGTAT | CATCAACAACAGGCAACCTAG |
| NtSOD-RT | AGCTACATGACGCCATTTCC | CCCTGTAAAGCAGCACCTTC |
| NtPOD-RT | AAATGGTGGCGCTAGCCGGTG | GCATTGAAGACGTGCCGCTGG |
| NtSAMDC-RT | CATTCACATTACCCCGGAAG | AGCAACATCAGCATGCAAAG |
| NtNCED1-RT | AAGAATGGCTCCGCAAGTTA | GCCTAGCAATTCCAGAGTGG |
| PdeCRCK3-GFP | gtgttacttctgcaggagctcATGGCCACGGCTGCATT | catggatccggtaccgagctcCCCTTTCTTTACACCTTTCAG |
| PdeCRCK5-GFP | gtgttacttctgcaggagctcATGAAGAGCCCATATTC | catggatccggtaccgagctcTTCTTCTGTTATTACTGAAAT |
| PdeCRCK6-GFP | gtgttacttctgcaggagctcATGGCCACGGCTGCATT | catggatccggtaccgagctcCCCTTTCTTTACACCTTTCAG |
| PdeCRCK6-2300 | cggggatcctctagagtcgacATGGCTATGAATGCATT | tccggtaccgagctcgtcgacCCCTTTCTTTACACC |
| PdeCRCK6-BD | tggccatggaggccgaattcATGGCTATGAATGCATT | cgctgcaggtcgacggatccTTACCCTTTCTTTACACC |
| Podel.02G006800.1-AD | gccatggaggccagtgaattcATGGCCACCCACGCAG | atgcccacccgggtggaattcCTAAGCTTCATAGACTTTGC |
| Podel.04G229700.1-AD | gccatggaggccagtgaattcATGAACATTGATAGAC | atgcccacccgggtggaattcTCAAGGACGTACGATAGCA |
| Podel.05G065100.1-AD | gccatggaggccagtgaattcATGAAATCCCTCGTTTC | atgcccacccgggtggaattcTCAAAGGAAATGGCTGGCC |
| Podel.10G049300.1-AD | gccatggaggccagtgaattcATGGCATGTGATAAGA | atgcccacccgggtggaattcTCAAGCTTGAGTCTTGGCC |
| Podel.14G119200.1-AD | gccatggaggccagtgaattcATGGCTAAATCCGTACT | atgcccacccgggtggaattcTTAATCACCACCAGCCGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, H.; He, Z.; Liu, L.; Cai, R.; Huang, H.; Xie, X.; Cao, X.; Li, Y.; Qiu, W.; Lu, Z.; et al. A Genome-Wide Characterization of Receptor-like Cytoplasmic Kinase IV Subfamily Members in Populus deltoides Identifies the Potential Role of PdeCRCK6 in Plant Osmotic Stress Responses. Plants 2024, 13, 3371. https://doi.org/10.3390/plants13233371
Pan H, He Z, Liu L, Cai R, Huang H, Xie X, Cao X, Li Y, Qiu W, Lu Z, et al. A Genome-Wide Characterization of Receptor-like Cytoplasmic Kinase IV Subfamily Members in Populus deltoides Identifies the Potential Role of PdeCRCK6 in Plant Osmotic Stress Responses. Plants. 2024; 13(23):3371. https://doi.org/10.3390/plants13233371
Chicago/Turabian StylePan, Huanhuan, Zhengquan He, Linxiu Liu, Renyue Cai, Hu Huang, Xinru Xie, Xun Cao, Yanan Li, Wenmin Qiu, Zhuchou Lu, and et al. 2024. "A Genome-Wide Characterization of Receptor-like Cytoplasmic Kinase IV Subfamily Members in Populus deltoides Identifies the Potential Role of PdeCRCK6 in Plant Osmotic Stress Responses" Plants 13, no. 23: 3371. https://doi.org/10.3390/plants13233371
APA StylePan, H., He, Z., Liu, L., Cai, R., Huang, H., Xie, X., Cao, X., Li, Y., Qiu, W., Lu, Z., Han, X., Qiao, G., Zhuo, R., Hu, J., & Xu, J. (2024). A Genome-Wide Characterization of Receptor-like Cytoplasmic Kinase IV Subfamily Members in Populus deltoides Identifies the Potential Role of PdeCRCK6 in Plant Osmotic Stress Responses. Plants, 13(23), 3371. https://doi.org/10.3390/plants13233371






