Characterization of Cell Wall Compositions of Sodium Azide-Induced Brittle Mutant Lines in IR64 Variety and Its Potential Application
Abstract
1. Introduction
2. Results
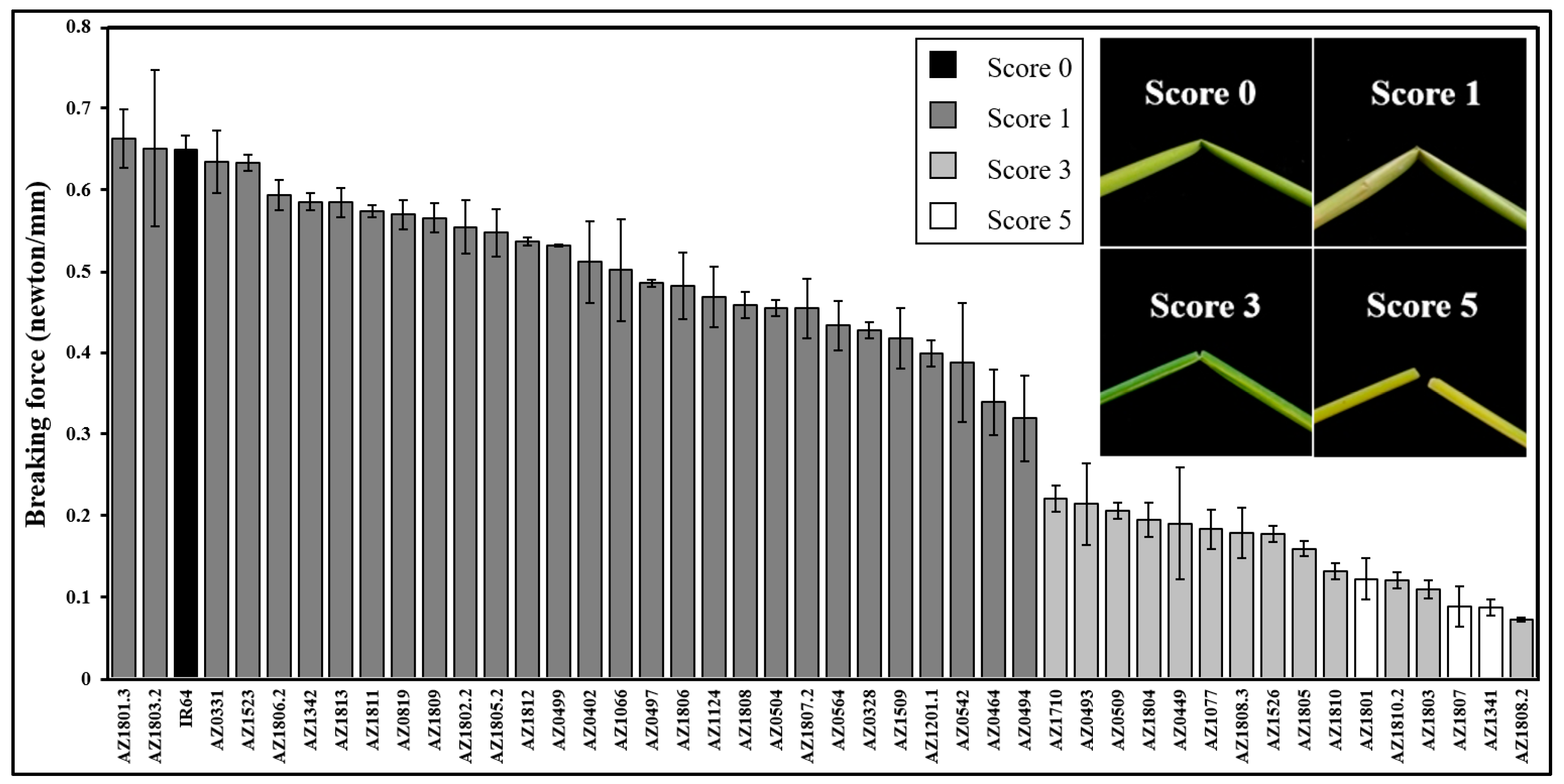
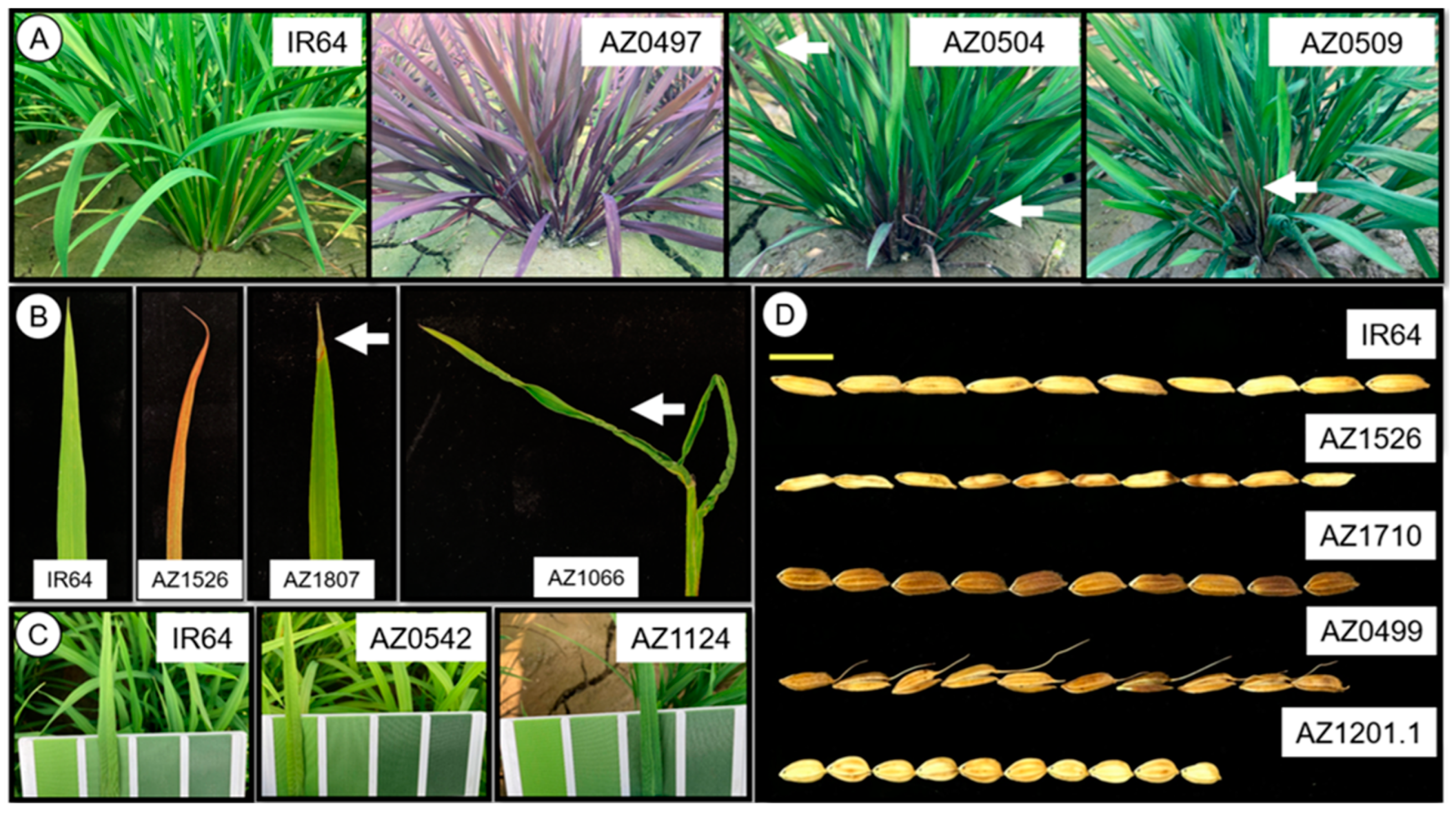
2.1. Qualitative and Quantitative Phenotyping of Brittle Mutant Lines
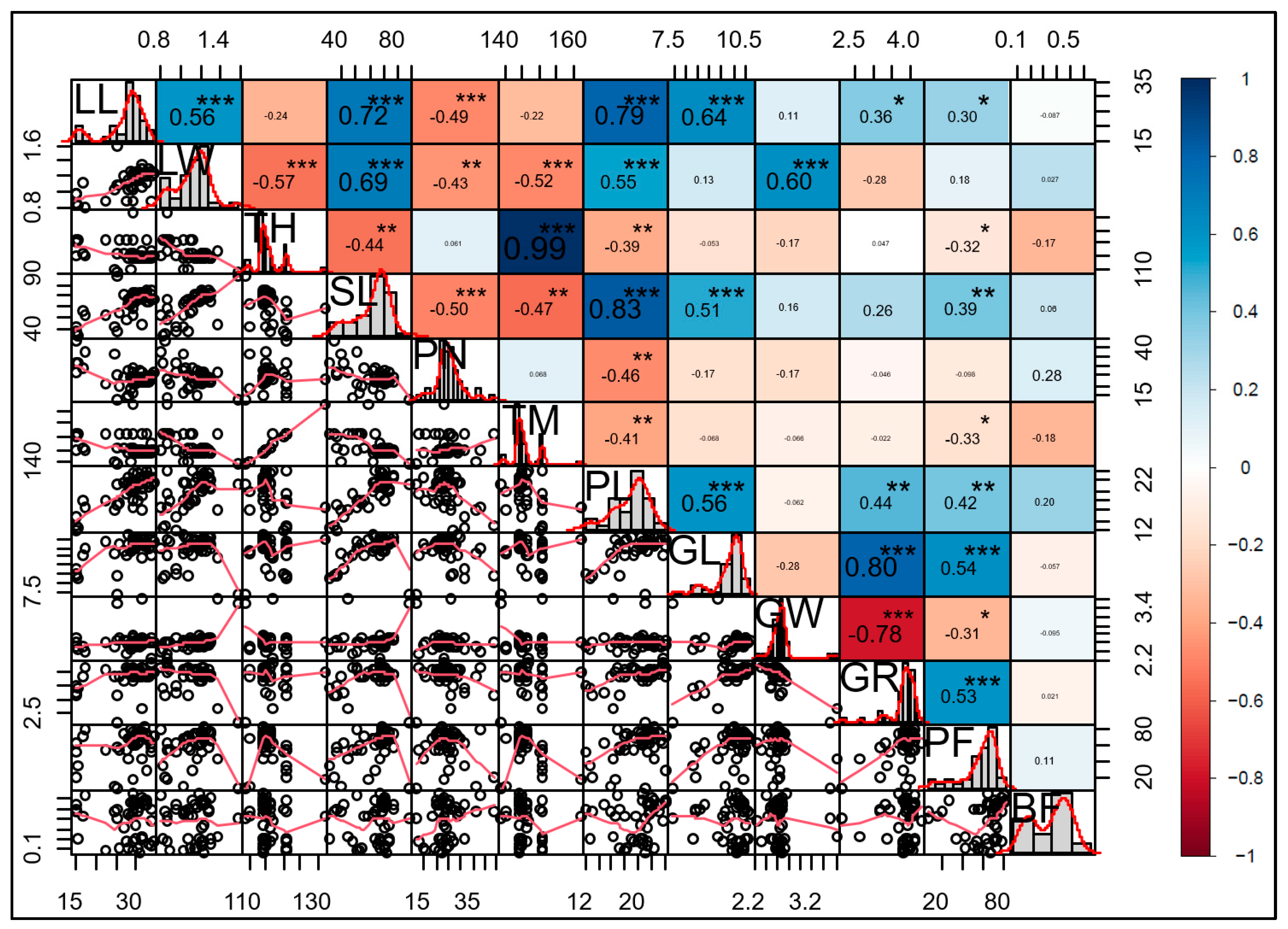
2.2. Correlations Between Morphological Traits
2.3. Association of Brittleness, Breaking Force, and Cell Wall Compositions
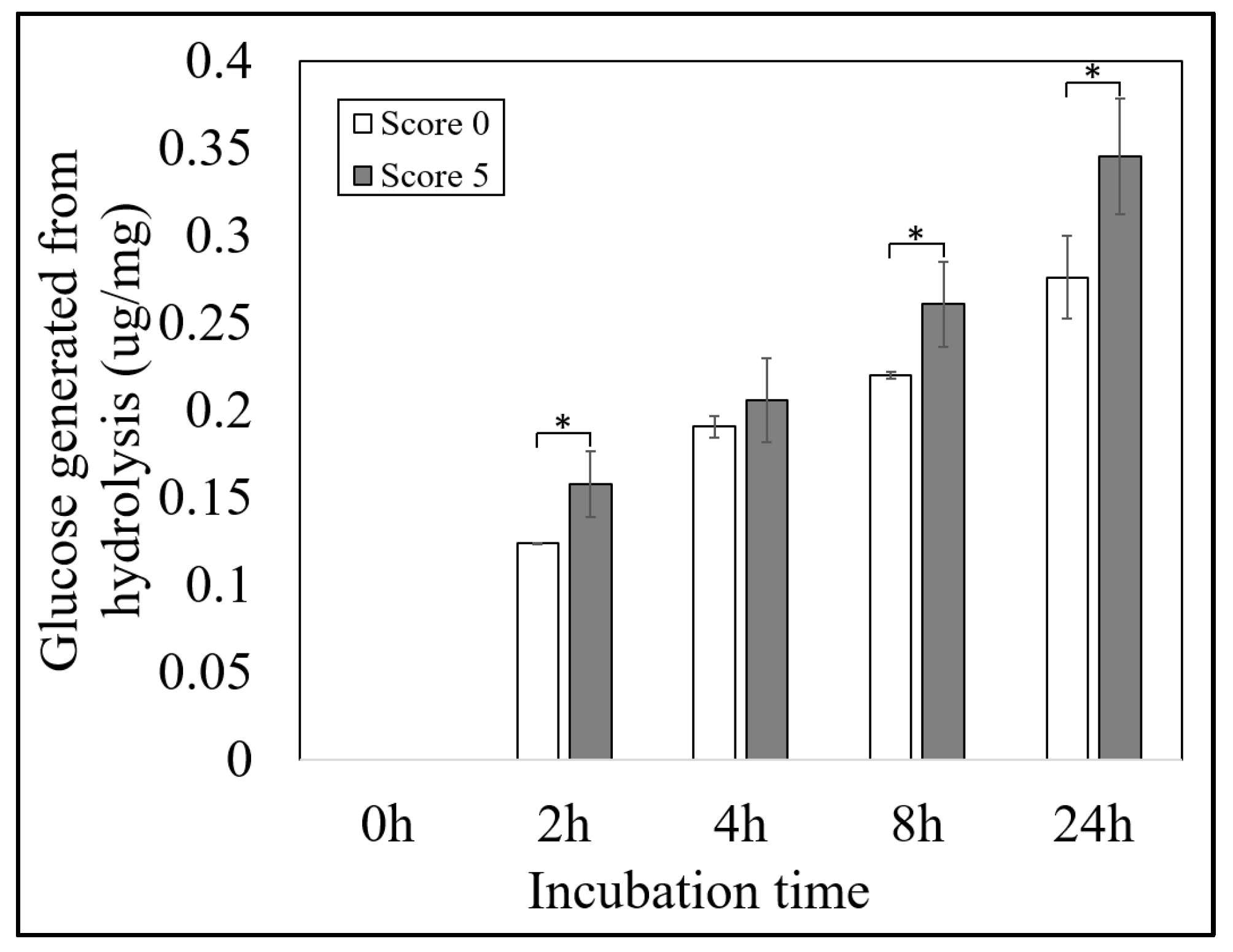
2.4. Evaluation of the Brittle Mutant Lines for Machinery Production and Its Potential for Applications
3. Discussion
3.1. Screening of Diverse BMLs in the NaN3 Mutation Pool
3.2. Development of Methodology for Brittleness Trait Investigation Using BMLs
3.3. Explanation of Mechanical Strength in Rice by Cell Wall Compositions of BMLs
3.4. Potential of the BMLs in Rice Production and Rice Straw Application
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Brittle Culm Mutant Screening and Developed Brittleness Score
4.3. Mechanical Strength Measurements
4.4. Morphological Traits Investigation
4.5. Cell Wall Composition Analysis
4.6. Estimation of Damaged Leaf After Typhoon
4.7. Rice Production Testing of Brittle Mutant Line
4.8. Relative Feed Value (RFV) Calculation
4.9. Hydrolysis and Digestion Analysis
4.10. Data and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- AOSTAT. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 10 September 2021).
- Lal, R. World crop residues production and implications of its use as a biofuel. Environ. Int. 2005, 31, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Lo, S.L. Utilization of rice hull and straw. In Rice, 4th ed.; Bao, J.S., Ed.; AACC International Press: St. Paul, MN, USA, 2019; pp. 627–661. [Google Scholar]
- Chhabra, V.; Chandra, M.M. Rice straw management for sustainable agriculture-a review. Plant Arch. 2019, 19, 47–49. [Google Scholar]
- Roy, P.; Kaur, M.; Burman, R.R.; Sharma, J.P.; Roy, T.N. Determinants of paddy straw management decision of farmers in Punjab. JCMSD 2018, 13, 203–210. [Google Scholar]
- Wei, Y.Q.; Wu, D.; Wei, D.; Zhao, Y.; Wu, J.Q.; Xie, X.Y.; Zhang, R.J.; Wei, Z.M. Improved lignocellulose-degrading performance during straw composting from diverse sources with actinomycetes inoculation by regulating the key enzyme activities. Bioresour. Technol. 2019, 271, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Vijayaprabhakar, A.; Durairaj, S.N.; Kalyan, V.S.R.K. Impact of combine harvested rice straw management options on soil microbial population and straw decomposition rate in succeeding rice field. Int. J. Curr. Microbiol. Appl. Sci. 2017, 2, 600–611. [Google Scholar] [CrossRef]
- Béguin, P.; Aubert, J.P. The biological degradation of cellulose. FEMS Microbiol. Rev. 1994, 13, 25–58. [Google Scholar] [CrossRef]
- Sarnklong, C.; Cone, J.; Pellikaan, W.F.; Hendriks, W. Utilization of rice straw and different treatments to improve its feed value for ruminants: A review. Asian-Aust. J. Anim. Sci. 2010, 23, 680–692. [Google Scholar] [CrossRef]
- Aquino, D.; Barrio, A.; Trach, N.; Nguyen Thanh, H.; Khang, D.N.; Nguyen, T.; Nguyen, V.H. Rice straw-based fodder for ruminants. In Sustainable Rice Straw Management; Gummert, M., Hung, N.V., Chivenge, P., Douthwaite, B., Eds.; Springer: Cham, Switzerland, 2020; pp. 111–129. [Google Scholar]
- Dhaliwal, A.K.; Mohan, A.; Sidhu, G.; Maqbool, R.; Gill, K.S. An ethylmethane sulfonate mutant resource in pre-green revolution hexaploid wheat. PLoS ONE 2015, 10, e0145227. [Google Scholar] [CrossRef]
- Li, Y.H.; Qian, Q.; Zhou, Y.H.; Yan, M.X.; Sun, L.; Zhang, M.; Fu, Z.M.; Wang, Y.H.; Han, B.; Pang, X.M.; et al. BRITTLE CULM1, which encodes a COBRA-Like protein, affects the mechanical properties of rice plants. Plant Cell 2003, 15, 2020–2031. [Google Scholar] [CrossRef]
- Xu, J.D.; Zhang, Q.F.; Zhang, T.; Zhang, H.Y.; Xu, P.Z.; Wang, X.D.; Wu, X.J. Phenotypic characterization, genetic analysis and gene-mapping for a brittle mutant in rice. J. Integr. Plant Biol. 2008, 50, 319–328. [Google Scholar] [CrossRef]
- Zhang, M.L.; Wei, F.; Guo, K.; Hu, Z.; Li, Y.Y.; Xie, G.S.; Wang, Y.T.; Cai, X.W.; Peng, L.C.; Wang, L.Q. A novel FC116/BC10 mutation distinctively causes alteration in the expression of the genes for cell wall polymer synthesis in rice. Front. Plant Sci. 2016, 7, 1366–1381. [Google Scholar] [CrossRef] [PubMed]
- Aohara, T.; Kotake, T.; Kaneko, Y.; Takatsuji, H.; Tsumuraya, Y.; Kawasaki, S. Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes. Plant Cell Physiol. 2009, 50, 1886–1897. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, Y.L.; Chen, S.H.; Xu, Y.; Zhou, K.N.; Zhang, L.; Ming, M.; Wu, F.Q.; Lin, Q.B.; Wang, J.L.; et al. BRITTLE CULM16 (BRITTLE NODE) is required for the formation of secondary cell walls in rice nodes. J. Integr. Agric. 2017, 16, 1286–1293. [Google Scholar] [CrossRef]
- Wang, X.L.; Cheng, Z.J.; Zhao, Z.C.; Gan, L.; Qin, R.Z.; Zhou, K.N.; Ma, W.W.; Zhang, B.C.; Wang, J.L.; Zhai, H.Q.; et al. BRITTLE SHEATH1 encoding OsCYP96B4 is involved in secondary cell wall formation in rice. Plant Cell Rep. 2016, 35, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.F.; Qin, Y.L.; Fang, J.J.; Yuan, S.J.; Peng, L.X.; Zhao, J.F.; Li, X.Y. A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PLoS ONE 2016, 11, e0153993. [Google Scholar] [CrossRef]
- Zhang, B.C.; Deng, L.W.; Qian, Q.; Xiong, G.Y.; Zeng, D.L.; Li, R.; Guo, L.B.; Li, J.Y.; Zhou, Y.H. A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol. Biol. 2009, 71, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Yang, Y.; Yao, J.L.; Chen, G.X.; Li, X.H.; Zhang, Q.F.; Wu, C.Y. FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol. Biol. 2009, 69, 685–697. [Google Scholar] [CrossRef]
- Li, M.; Xiong, G.Y.; Li, R.; Cui, J.J.; Tang, D.; Zhang, B.C.; Pauly, M.; Cheng, Z.K.; Zhou, Y.H. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009, 60, 1055–1069. [Google Scholar] [CrossRef]
- Sawasdee, A.; Tsai, T.H.; Liao, W.C.; Wang, C.S. Identification of the CesA7 gene encodes brittleness mutation derived from IR64 variety and breeding for ruminant feeding. Agriculture 2024, 14, 706. [Google Scholar] [CrossRef]
- Kotake, T.; Aohara, T.; Hirano, K.; Sato, A.; Kaneko, Y.; Tsumuraya, Y.; Takatsuji, H.; Kawasaki, S. Rice Brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J. Exp. Bot. 2011, 62, 2053–2062. [Google Scholar] [CrossRef]
- Jiang, Y.D.; He, P.L.; Liao, H.X.; Zhang, X.B.; Wu, G.C.; He, G.H.; Lin, T.T.; Sang, X.C. Identification and gene mapping of a fragile and leaf-tip dead mutant fld1 in Oryza sativa. Chin. Bull. Bot. 2014, 49, 663–671. [Google Scholar] [CrossRef]
- Sang, X.C.; Du, C.; Wang, X.W.; Yang, Z.L.; Ling, Y.H.; Zhao, F.M.; Li, Y.F.; He, G.H. Identification and gene mapping of dwarf and brittle culm mutant dbc1 in Oryza sativa. Acta Agron. Sin. 2013, 39, 626–631. [Google Scholar] [CrossRef]
- Wu, G.C.; Sang, X.C.; Ma, J.; Zhu, X.Y.; Ren, D.Y.; Guo, S.; Jiang, Y.D.; Yang, Z.L.; Ling, Y.H.; He, G.H. Genetic analysis and fine mapping of a dwarf and fragile mutant dwf1 in rice. J. Plant Genet. Res. 2014, 15, 795–801. [Google Scholar]
- Wang, X.W.; Tang, Y.Q.; Liao, H.X.; Jiang, Y.D.; Yang, Z.L.; Sang, X.C. Identification and gene fine mapping of fragile-plant and brown-panicle mutant fb1 in rice. J. Nucl. Agric. Sci. 2017, 31, 2298–2305. [Google Scholar]
- Wang, D.F.; Yuan, S.J.; Yin, L.; Zhao, J.F.; Guo, B.T.; Lan, J.H.; Li, X.Y. A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice. Plant Sci. 2012, 196, 117–124. [Google Scholar] [CrossRef]
- Shu, Y.; Zeng, D.; Qin, R.; Jin, X.; Zheng, X.; Shi, C. Identification and gene fine mapping of a brittle culm 16 (bc16) Mutant in Rice. Chin. J. Rice Sci. 2016, 30, 345–355. [Google Scholar] [CrossRef]
- Ma, X.Z.; Li, C.M.; Huang, R.; Zhang, K.; Wang, Q.; Fu, C.Y.; Liu, W.G.; Sun, C.H.; Wang, P.R.; Wang, F.; et al. Rice Brittle Culm19 encoding cellulose synthase subunit CESA4 causes dominant brittle phenotype but has no distinct influence on growth and grain yield. Rice 2021, 14, 95. [Google Scholar] [CrossRef]
- Xu, S.L.; Zhang, M.C.; Ye, J.H.; Hu, D.X.; Zhang, Y.Y.; Li, Z.; Liu, J.R.; Sun, Y.F.; Wang, S.; Yuan, X.P.; et al. Brittle culm 25, which encodes an UDP-xylose synthase, affects cell wall properties in rice. Crop J. 2023, 11, 733–743. [Google Scholar] [CrossRef]
- Rao, Y.C.; Yang, Y.L.; Xin, D.D.; Li, X.J.; Zhai, K.E.; Ma, B.J.; Pan, J.W.; Qian, Q.; Zeng, D.L. Characterization and cloning of a brittle culm mutant (bc88) in rice (Oryza sativa L.). Chin. Sci. Bull. 2013, 58, 3000–3006. [Google Scholar] [CrossRef][Green Version]
- Li, G.Z.; Zeng, X.F.; Li, Y.; Li, J.R.; Huang, X.Z.; Zhao, D.G. BRITTLE CULM17, a novel allele of TAC4, affects the mechanical properties of rice plants. Int. J. Mol. Sci. 2022, 23, 5305. [Google Scholar] [CrossRef]
- Cao, X.Y.; Zhou, T.; Sun, Y.; Zhang, Y.H.; Xu, H.; Liu, W.; Zou, Y.; Chen, Q.Q.; Ma, H.; Gu, D.F.; et al. Identification and gene cloning of a brittle culm mutant (bc22) in rice. Agriculture 2024, 14, 235. [Google Scholar] [CrossRef]
- Yan, C.J.; Yan, S.; Zeng, X.H.; Zhang, Z.Q.; Gu, M.H. Fine mapping and isolation of bc7(t), allelic to OsCesA4. Acta Genet. Sin. 2007, 34, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.Q.; Wang, J.M.; Bai, L.; Zhao, Z.G.; Chen, K.M. Anatomical and chemical alterations but not photosynthetic dynamics and apoplastic transport changes are involved in the brittleness culm mutation of rice. J. Integr. Plant Biol. 2008, 50, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.P.; Tong, C.; Wang, Y.; Ren, S.J.; Shen, S.Q. Character identification, genetic analysis and gene mapping of a low cellulose mutant LCM527-1 in rice. J. Zhejiang Univ.-Agric. Life Sci. 2015, 41, 261–268. [Google Scholar] [CrossRef]
- Jin, Z.M.; Ping, B.Z.; Shen, H.J.; Du, H.Q.; Li, R.Q.; Zhu, L.; Zhang, D.B.; Yuan, Z. Characterisation and gene mapping of a brittle culm mutant bc-s1 in rice. Chin. Bull. Bot. 2016, 51, 167–174. [Google Scholar] [CrossRef]
- Xu, Z.P.; Zhong, C.Y.; Zhang, L.J.; Liu, Q.Q. Identification and gene cloning of the brittle culm mutant bc1-wu3 in rice. Chin. J. Rice Sci. 2017, 31, 157–165. [Google Scholar] [CrossRef]
- Hirano, K.; Kotake, T.; Kamihara, K.; Tsuna, K.; Aohara, T.; Kaneko, Y.; Takatsuji, H.; Tsumuraya, Y.; Kawasaki, S. Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta 2010, 232, 95–108. [Google Scholar] [CrossRef]
- Song, X.Q.; Liu, L.F.; Jiang, Y.J.; Zhang, B.C.; Gao, Y.P.; Liu, X.L.; Lin, Q.S.; Ling, H.Q.; Zhou, Y.H. Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol. Plant 2013, 6, 768–780. [Google Scholar] [CrossRef]
- Tanaka, K.; Murata, K.; Yamazaki, M.; Onosato, K.; Miyao, A.; Hirochika, H. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 2003, 133, 73–83. [Google Scholar] [CrossRef]
- Li, Y. Genetic Mapping of Rice Brittle Culm Gene C8. Master’s Thesis, Huazhong Agriculture University, Wuhan, China, 2011. [Google Scholar]
- Fanata, W.I.D.; Lee, K.H.; Son, B.H.; Yoo, J.Y.; Harmoko, R.; Ko, K.S.; Ramasamy, N.K.; Kim, K.H.; Oh, D.B.; Jung, H.S.; et al. N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J. 2013, 73, 966–979. [Google Scholar] [CrossRef]
- Sun, H.Z.; Sun, J.J.; Yuan, Z.K.; Li, F.H.; Li, X.R.; Li, J.Z.; Du, Y.X.; Wang, F.Q. A Tos17 transposon insertion in OsCesA9 causes brittle culm in rice. Gene 2024, 890, 147818. [Google Scholar] [CrossRef] [PubMed]
- Chern, C.G.; Fan, M.J.; Yu, S.M.; Hour, A.L.; Lu, P.C.; Lin, Y.C.; Wei, F.J.; Huang, S.C.; Chen, S.; Lai, M.H.; et al. A rice phenomics study—Phenotype scoring and seed propagation of a T-DNA insertion-induced rice mutant population. Plant Mol. Biol. 2007, 65, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.X.; You, C.J.; Chen, G.X.; Li, X.H.; Zhang, Q.F.; Wu, C.Y. OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol. Biol. 2011, 75, 333–345. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Li, S.B.; Qian, Q.; Zeng, D.L.; Zhang, M.; Guo, L.B.; Liu, X.L.; Zhang, B.C.; Deng, L.W.; Liu, X.F.; et al. BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J. 2009, 57, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, B.C.; Qian, Q.; Yu, Y.C.; Li, R.; Zhang, J.W.; Liu, X.L.; Zeng, D.L.; Li, J.Y.; Zhou, Y.H. Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 2010, 63, 312–328. [Google Scholar] [CrossRef]
- Wang, C.L.; Wang, L.Q.; Mou, T.M. Characterization and gene mapping of a brittle culm mutant nbc (t) in rice. J. Huazhong Agric. Univ. 2012, 31, 159–164. [Google Scholar]
- Li, F.C.; Liu, S.T.; Xu, H.; Xu, Q. A novel FC17/CESA4 mutation causes increased biomass saccharification and lodging resistance by remodeling cell wall in rice. Biotechnol. Biofuels 2018, 11, 298–310. [Google Scholar] [CrossRef]
- Peng, Y.C.; Liu, W.Z.; Fu, Y.P.; Wang, H.T.; Hu, G.C.; Chen, W.F.; Xu, Z.J. Characterization and gene mapping of a dominant Brittle culm mutant Bc18 in rice (Oryza sativa L.). Chin. J. Rice Sci. 2016, 30, 127–135. [Google Scholar] [CrossRef]
- Zhang, B.C.; Zhang, L.J.; Li, F.; Zhang, D.M.; Liu, X.L.; Wang, H.; Xu, Z.P.; Chu, C.C.; Zhou, Y.H. Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat. Plants 2017, 3, 17017. [Google Scholar] [CrossRef]
- Wu, B.; Zhang, B.C.; Dai, Y.; Zhang, L.; Shangguan, K.k.; Peng, Y.G.; Zhou, Y.H.; Zhu, Z. Brittle culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol. 2012, 159, 1440–1452. [Google Scholar] [CrossRef]
- Li, F.C.; Zhang, M.L.; Guo, K.; Hu, Z.; Zhang, R.; Feng, Y.Q.; Yi, X.Y.; Zou, W.H.; Wang, L.Q.; Wu, C.Y.; et al. High-level hemicellulosic arabinose predominately affects lignocellulose crystallinity for genetically enhancing both plant lodging resistance and biomass enzymatic digestibility in rice mutants. Plant Biotechnol. J. 2015, 13, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.C.; Liu, X.L.; Qian, Q.; Liu, L.F.; Dong, G.J.; Xiong, G.Y.; Zeng, D.L.; Zhou, Y.H. Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc. Natl. Acad. Sci. USA 2011, 108, 5110–5115. [Google Scholar] [CrossRef] [PubMed]
- Ahloowalia, B.S.; Maluszynski, M.; Nichterlein, K. Global impact of mutation-derived varieties. Euphytica 2004, 135, 187–204. [Google Scholar] [CrossRef]
- Tseng, H.Y.; Lin, D.G.; Hsieh, H.Y.; Tseng, Y.J.; Tseng, W.B.; Chen, C.W.; Wang, C.S. Genetic analysis and molecular mapping of QTLs associated with resistance to bacterial blight in a rice mutant, SA0423. Euphytica 2015, 205, 231–241. [Google Scholar] [CrossRef]
- Viana, V.E.; Pegoraro, C.; Busanello, C.; Costa de Oliveira, A. Mutagenesis in rice: The basis for breeding a new super plant. Front. Plant Sci. 2019, 10, 1326–1353. [Google Scholar] [CrossRef]
- Wang, C.S.; Lo, K.L.; Wang, A.Z. Sodium azide mutagenesis generated diverse and broad spectrum blast resistance mutants in rice. Euphytica 2019, 215, 145–155. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nakamura, A.; Iwai, H.; Ishii, T.; Ma, J.F.; Yokoyama, R.; Nishitani, K.; Satoh, S.; Furukawa, J. Effect of silicon deficiency on secondary cell wall synthesis in rice leaf. J. Plant Res. 2012, 125, 771–779. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Cui, D.T.; Ye, Z.H. Secondary cell wall biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef]
- Ye, Y.F.; Liu, B.M.; Zhao, M.; Wu, K.; Cheng, W.M.; Chen, X.B.; Liu, Q.; Liu, Z.; Fu, X.D.; Wu, Y.J. CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice. Plant Mol. Biol. 2015, 89, 385–401. [Google Scholar] [CrossRef]
- Jahn, C.E.; McKay, J.K.; Mauleon, R.; Stephens, J.; McNally, K.L.; Bush, D.R.; Leung, H.; Leach, J.E. Genetic variation in biomass traits among 20 diverse rice varieties. Plant Physiol. 2011, 155, 157–168. [Google Scholar] [CrossRef]
- Soebarinoto, S.; Chuzaemi, S.; van Bruchem, J.; Hartutik, H.; Mashudi, M. The nutritive value of rice straw in relation to variety, urea treatment, location of growth and season, and its prediction from in Sacco degradability. Asian-Australas. J. Anim. Sci. 1997, 10, 215–222. [Google Scholar] [CrossRef]
- Wei, C.X.; Xie, P.S.; Chen, Y.F.; Yu, H.G.; Su, Y.J.; Gu, M.H.; Yan, C.J. Anatomical and chemical characteristics of culm of rice brittle mutant bc7(t). Funct. Plant Biol. 2011, 38, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Fog, K. The effect of added nitrogen on the rate of decomposition of organic matter. Biol. Rev. 1988, 63, 433–462. [Google Scholar] [CrossRef]
- Su, Y.J.; Zhao, G.Q.; Wei, Z.W.; Yan, C.J.; Liu, S.J. Mutation of cellulose synthase gene improves the nutritive value of rice straw. Asian-Australas. J. Anim. Sci. 2012, 25, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhang, W.; Ren, S.F.; Liu, F.; Zhao, C.Q.; Liao, H.F.; Xu, Z.D.; Huang, J.F.; Li, Q.; Tu, Y.Y.; et al. Hemicelluloses negatively affect lignocellulose crystallinity for high biomass digestibility under NaOH and H2SO4 pretreatments in Miscanthus. Biotechnol. Biofuels 2012, 5, 58. [Google Scholar] [CrossRef]
- Jeng, T.L.; Lin, Y.W.; Wang, C.S.; Sung, J.M. Comparisons and selection of rice mutants with high iron and zinc contents in their polished grains that were mutated from the indica type cultivar IR64. J. Food Compos. Anal. 2012, 28, 149–154. [Google Scholar] [CrossRef]
- Wang, C.S.; Tseng, T.H.; Lin, C.Y. Rice biotech research at the taiwan agricultural research institute. APBN 2002, 06, 950–956. [Google Scholar] [CrossRef]
- Sawasdee, A. Screening and Characterization of Brittle Mutants from the Mutation Pools of TNG67 and IR64 Rice Varieties and Breeding to Improve Bacterial Blight Resistance. Master’s Thesis, National Chung Hsing University, Taichung, Taiwan, 2017. [Google Scholar]
- UPOV. Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability (Rice). 2020; pp. 1–35. Available online: https://www.upov.int/edocs/tgdocs/en/tg016.pdf (accessed on 1 January 2015).
- IRRI. Standard Evaluation System for Rice, 5th ed.; IRRI: Manila, Philippines, 2013. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Wang, J.W.; Du, M.H.; Zhang, J.P. Method for Producing Silicon Supplementation Preparation by Silicon Enriched Plants. CN1879666A, 20 December 2006. [Google Scholar]
- Central Weather Bureau. 2016. Available online: https://www.cwa.gov.tw/eng/ (accessed on 27 September 2016).
- Kao, M.R.; Kuo, H.W.; Lee, C.C.; Huang, K.Y.; Huang, T.Y.; Li, C.W.; Chen, C.W.; Wang, A.H.J.; Yu, S.M.; Ho, T.H.D. Chaetomella raphigera β-glucosidase D2-BGL has intriguing structural features and a high substrate affinity that renders it an efficient cellulase supplement for lignocellulosic biomass hydrolysis. Biotechnol. Biofuels 2019, 12, 258. [Google Scholar] [CrossRef]


| Type of Mutagen | Source of Mutation | Mutant Name | Wild Type | Investigation of Brittleness | Analysis State of Compositions | References | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Growth Stage/ Tissue | TPA a | Growth Stage/ Tissue | C | H | L | S | |||||
| Chemical | NaN3 | (45) b BMLs | IR64 | M/L c | + | M/L | + | + | + | + | This study |
| EMS | fp2 | E-you 532 | 1st IN | + | 2nd IN | + | + | + | + | [13] | |
| Bc6 | IR68 | C, L | − | 2WH/C, L | + | + | + | − | [23] | ||
| fld1 | Jinhui10 | - | + | - | + | − | + | − | [24] | ||
| dbc1 | Jinhui10 | - | + | - | + | − | + | − | [25] | ||
| dwf1 | Jinhui10 | - | + | - | + | + | + | + | [26] | ||
| fb1 | Jinhui10 | - | + | - | + | − | + | − | [27] | ||
| bc11 | Nipponbare | 2nd IN, FL | + | 2nd IN | + | + | + | − | [19] | ||
| S1-60 | Nipponbare | Hd/1st, 2nd IN, L | + | 2nd IN | + | + | − | − | [28] | ||
| bc16 | Nipponbare | M/C | − | M, C | + | + | + | − | [29] | ||
| S1-24 | Nipponbare | Hd/1st, 2nd IN, L | + | 2nd IN | + | + | − | − | [18] | ||
| Bc19 | Nipponbare | 2WH/2nd IN, FL | + | 2WH, 2nd IN, FL | + | + | + | − | [30] | ||
| bc25 | Nipponbare | 2nd IN, FL | + | C | + | + | − | − | [31] | ||
| bc88 | Wuyunjing7 | All | − | - | + | − | − | − | [32] | ||
| fc116 | Zhonghua11 | M/2nd IN, FL | + | C | + | + | + | − | [14] | ||
| bc17 | Pingtangheinuo | Hd/FL, C | + | Hd/FL, SH, C | + | − | + | − | [33] | ||
| bc22 | LR005 | Hd/2nd IN | + | Hd/IN | + | + | + | − | [34] | ||
| Physical | 60Co-γ rays | bc1 | Shuang Ke Zao | 1st IN, FL | + | 1st IN | + | + | + | − | [12] |
| bc7(t) | Zhonghua11 | C, L | − | LGF/C | + | + | + | − | [35] | ||
| bcm | Xiushui110 | C, L | − | C, L | + | − | + | − | [36] | ||
| lcm527-1 | 527 | - | + | - | + | + | + | − | [37] | ||
| bc-s1 | 9522 | - | + | - | + | + | + | − | [38] | ||
| bc1-wu3 | Wuyujing 3 | - | + | - | + | + | − | − | [39] | ||
| bc16(node) | 93-11 | Hd/UMN | + | Hd/UMN | + | + | + | − | [16] | ||
| γ-rays | bc3 | Nourin8 | Hd/C | − | 2WH/C | + | + | − | − | [40] | |
| Microwave | bc13 | Yinhuazhan | 2nd IN, FL | + | R, S | + | + | − | − | [41] | |
| Biological | Tos17 | (5) mutants | Nipponbare | 2nd IN, L | − | 2nd IN | + | − | − | − | [42] |
| C8 | Nipponbare | - | + | - | + | + | + | − | [43] | ||
| Gnt1 | Nipponbare | - | + | - | + | + | + | − | [44] | ||
| bc26 | ZH15 | - | − | M/C, L | + | + | − | − | [45] | ||
| T-DNA | (14) lines | Tainung67 | C or L | − | - | − | − | − | − | [46] | |
| bc1l4 | Zhonghua11 | - | − | M/IN | + | + | + | − | [47] | ||
| Other | Nature | bc10 | Huang Jin Qin | C, L | + | C | + | + | + | − | [48] |
| bc12 | C418 | 2nd IN, FL | + | 2nd IN | + | + | + | − | [49] | ||
| nbc(t) | 93-11/IRBB21 | - | + | - | + | − | + | − | [50] | ||
| fc17 | ShenNong265 | C, L | + | M/C | + | + | + | − | [51] | ||
| Bc18 | II-32B//Xqz B/Dular | - | + | - | + | + | + | − | [52] | ||
| bs1 | Nipponbare | - | − | M/SH | + | + | − | − | [53] | ||
| Tissue culture | bsh1 | H3774 | 6W/SH | − | 6W/SH | + | + | + | − | [17] | |
| bc15 | Zhonghua8 | 2nd IN, L | + | 2nd IN | + | + | + | − | [54] | ||
| Collection | T-DNA and EMS | (36) lines | Nipponbare | M/4th IN | + | M/S | + | + | + | − | [55] |
| - | - | bc14 | NE17 | 2nd IN, L | + | 2nd IN | + | + | − | − | [56] |
| Traits | IR64 (Mean ± SD) | 45 Brittle Mutant Lines | |||
|---|---|---|---|---|---|
| Min | Max | Mean | CV (%) | ||
| Leaf length (cm) | 31.68 ± 3.48 | 14.53 | 34.49 | 27.92 | 19.96 |
| Leaf width (cm) | 1.30 ± 0.06 | 0.80 | 1.58 | 1.13 | 14.11 |
| Day to heading | 114 ± 0.00 | 109 | 132 | 115.96 | 3.29 |
| Culm length (cm) | 70.33 ± 3.20 | 32.67 | 88.00 | 62.15 | 20.60 |
| Day to maturity | 144 ± 0.00 | 139 | 162 | 146.04 | 2.66 |
| Panicle length (cm) | 22.01 ± 2.40 | 12.22 | 24.06 | 19.84 | 15.13 |
| Panicle no./plant | 23.00 ± 1.15 | 11.67 | 44.00 | 24.31 | 24.49 |
| Grain length (cm) | 10.23 ± 0.52 | 7.35 | 10.76 | 9.92 | 7.52 |
| Grain width (cm) | 2.67 ± 0.20 | 2.24 | 3.60 | 2.63 | 8.54 |
| Grain length/grain width | 3.83 ± 0.32 | 2.11 | 4.29 | 3.80 | 10.65 |
| Fertility (%) | 77.89 ± 4.65 | 6.98 | 82.35 | 60.25 | 31.74 |
| Breaking force (N) | 0.61 ± 0.02 | 0.07 | 0.66 | 0.39 | 49.20 |
| Cellulose content (%) | 28.85 ± 1.69 | 18.39 | 28.85 | 23.58 | 8.61 |
| Hemicellulose content (%) | 33.63 ± 1.28 | 24.85 | 39.08 | 35.28 | 9.09 |
| Lignin content (%) | 3.20 ± 0.73 | 1.26 | 7.00 | 3.40 | 43.41 |
| Silica content (%) | 7.81 ± 1.41 | 3.31 | 18.67 | 9.45 | 30.50 |
| Brittleness Score | NDF | ADF | DDM | DMI | RFV |
|---|---|---|---|---|---|
| 0 | 65.84 ± 0.98 | 33.72 ± 2.39 | 62.63 ± 1.86 | 1.78 ± 0.07 | 86.65 ± 5.75 |
| 1 | 65.57 ± 2.93 | 31.00 ± 2.71 | 64.75 ± 2.11 | 1.83 ± 0.08 | 92.02 ± 4.87 |
| 3 | 65.29 ± 2.22 | 28.65 ± 1.28 | 66.59 ± 0.99 | 1.84 ± 0.06 | 95.00 ± 4.31 |
| 5 | 60.51 ± 2.38 | 25.63 ± 1.60 | 68.93 ± 1.24 | 1.99 ± 0.08 | 106.14 ± 5.99 |
| Anova | p = 0.0175 | p = 0.0002 | p = 0.0002 | p = 0.0135 | p = 0.0002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawasdee, A.; Tsai, T.-H.; Chang, Y.-H.; Shrestha, J.K.; Lin, M.-C.; Chiang, H.-I.; Wang, C.-S. Characterization of Cell Wall Compositions of Sodium Azide-Induced Brittle Mutant Lines in IR64 Variety and Its Potential Application. Plants 2024, 13, 3303. https://doi.org/10.3390/plants13233303
Sawasdee A, Tsai T-H, Chang Y-H, Shrestha JK, Lin M-C, Chiang H-I, Wang C-S. Characterization of Cell Wall Compositions of Sodium Azide-Induced Brittle Mutant Lines in IR64 Variety and Its Potential Application. Plants. 2024; 13(23):3303. https://doi.org/10.3390/plants13233303
Chicago/Turabian StyleSawasdee, Anuchart, Tsung-Han Tsai, Yi-Hsin Chang, Jeevan Kumar Shrestha, Meng-Chun Lin, Hsin-I Chiang, and Chang-Sheng Wang. 2024. "Characterization of Cell Wall Compositions of Sodium Azide-Induced Brittle Mutant Lines in IR64 Variety and Its Potential Application" Plants 13, no. 23: 3303. https://doi.org/10.3390/plants13233303
APA StyleSawasdee, A., Tsai, T.-H., Chang, Y.-H., Shrestha, J. K., Lin, M.-C., Chiang, H.-I., & Wang, C.-S. (2024). Characterization of Cell Wall Compositions of Sodium Azide-Induced Brittle Mutant Lines in IR64 Variety and Its Potential Application. Plants, 13(23), 3303. https://doi.org/10.3390/plants13233303








