Systematic Survey and Analysis Reveal Jasmonate ZIM-Domain Gene Family in Coix lacryma-jobi Under High Temperature
Abstract
1. Introduction
2. Results
2.1. Systematic Identification of Jasmonate ZIM-Domain Family Members in C. lacryma-jobi
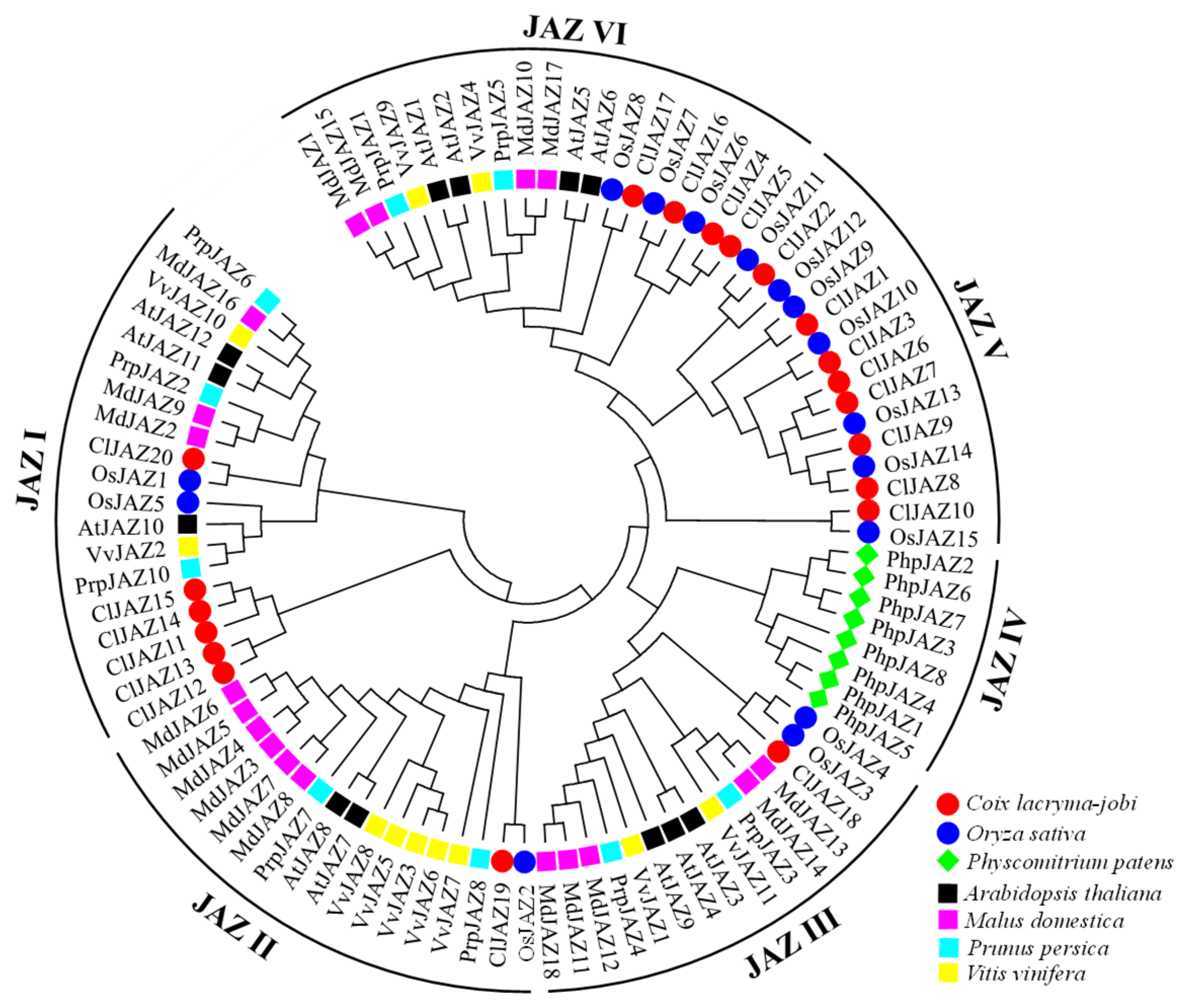
2.2. Phylogenetic Analysis of ClJAZs and Other Plant Jasmonate ZIM-Domain Proteins
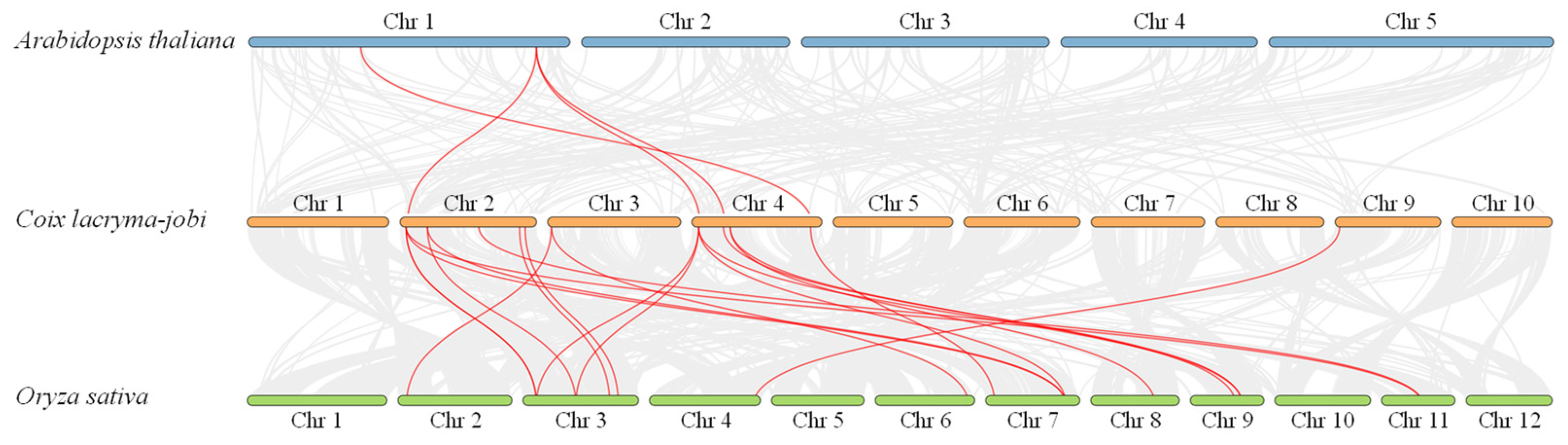
2.3. Chromosome Localization and Collinearity Analysis of ClJAZs Members
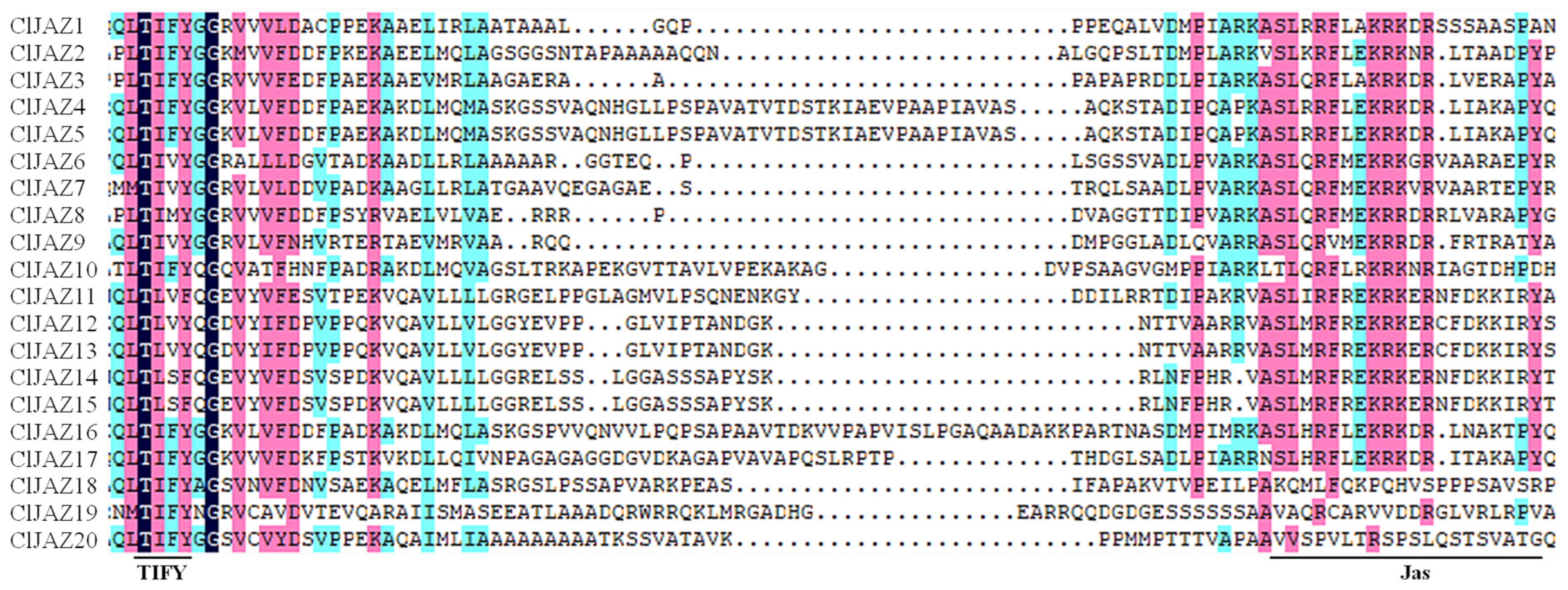
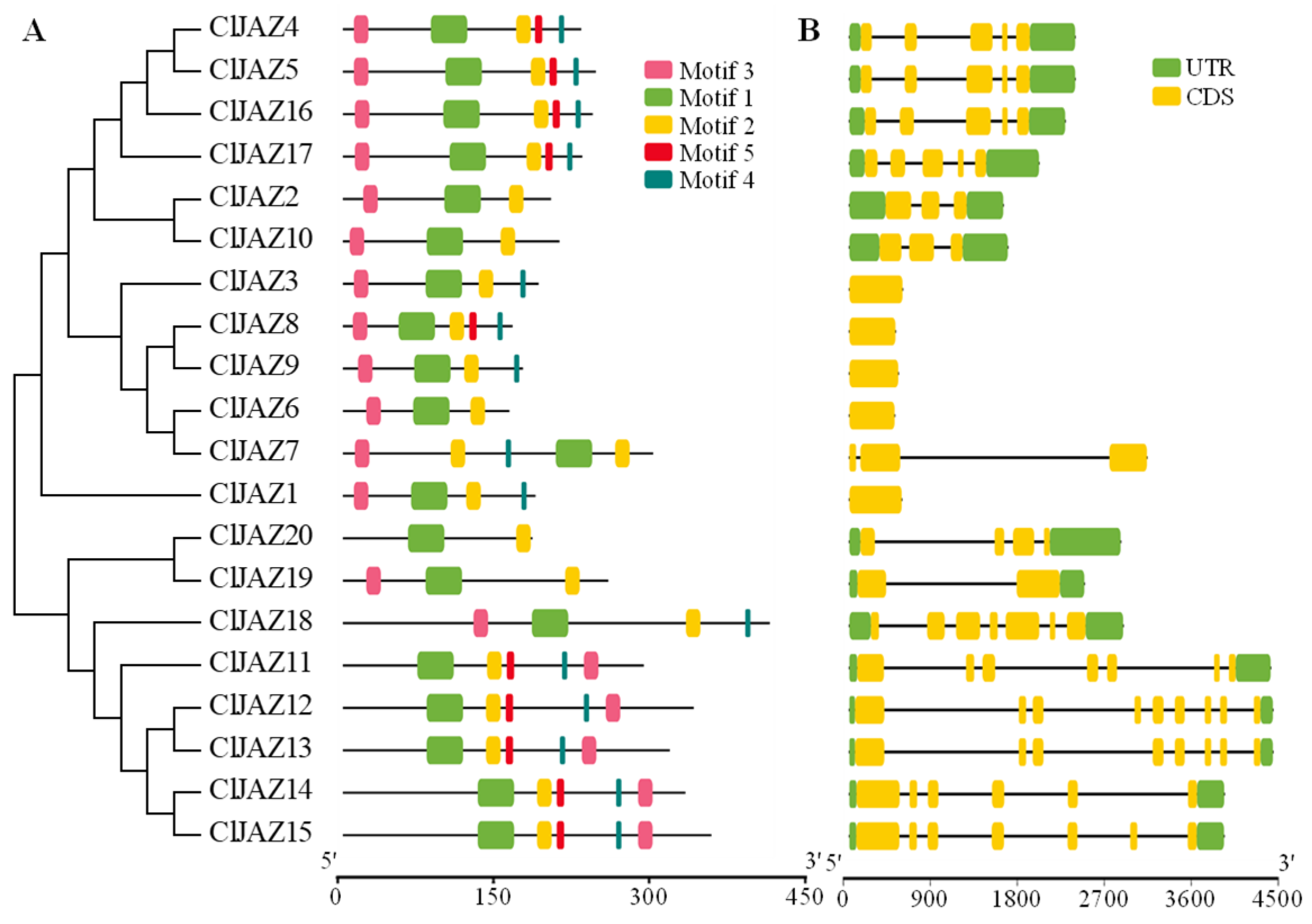
2.4. Conserved Motifs and Gene Structure of ClJAZs Members
2.5. Cis-Regulating Elements Analysis of the Promoter Sequences Before ClJAZs Genes
2.6. Expression Patterns of ClJAZs Genes in Different Tissues
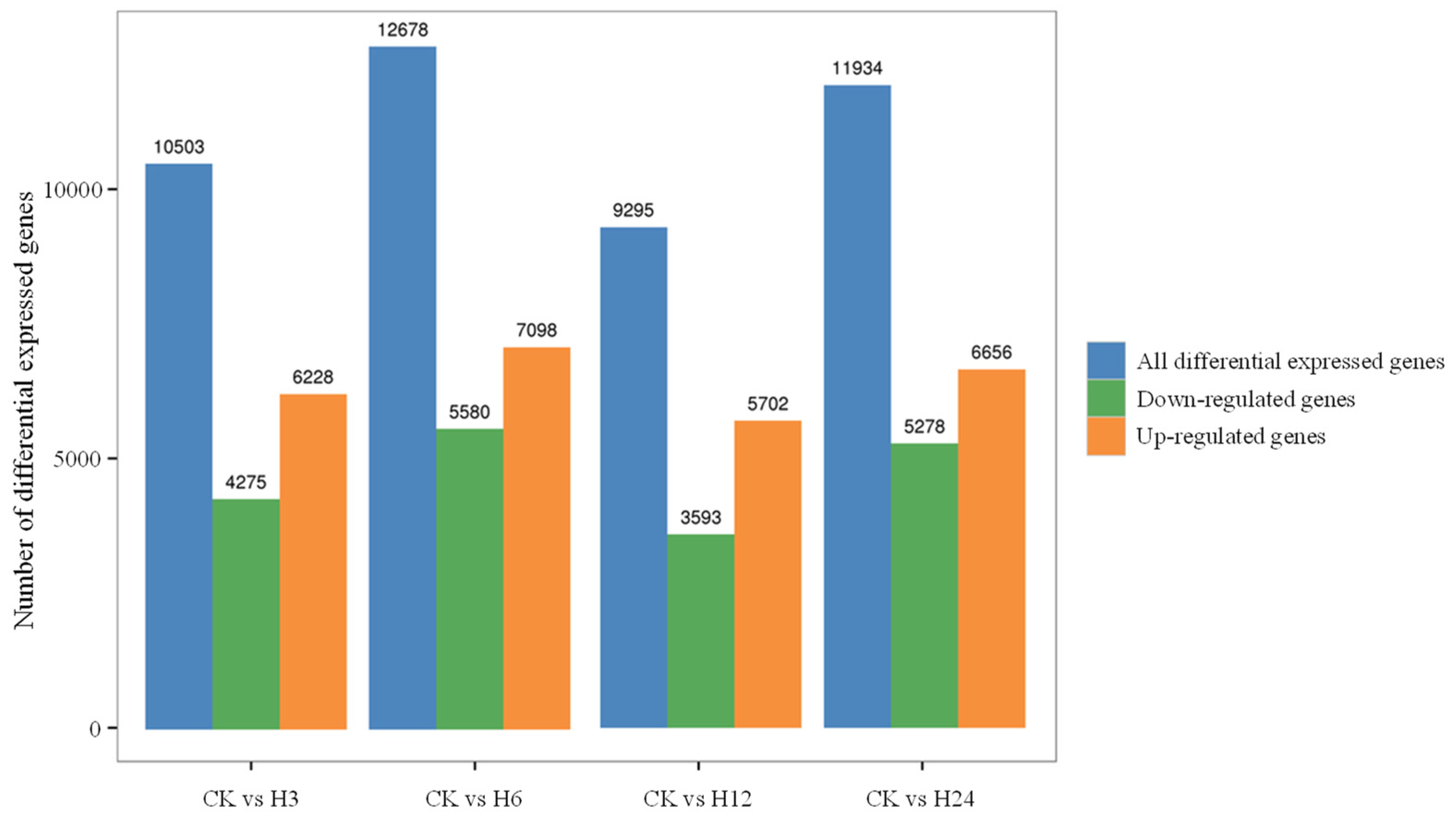
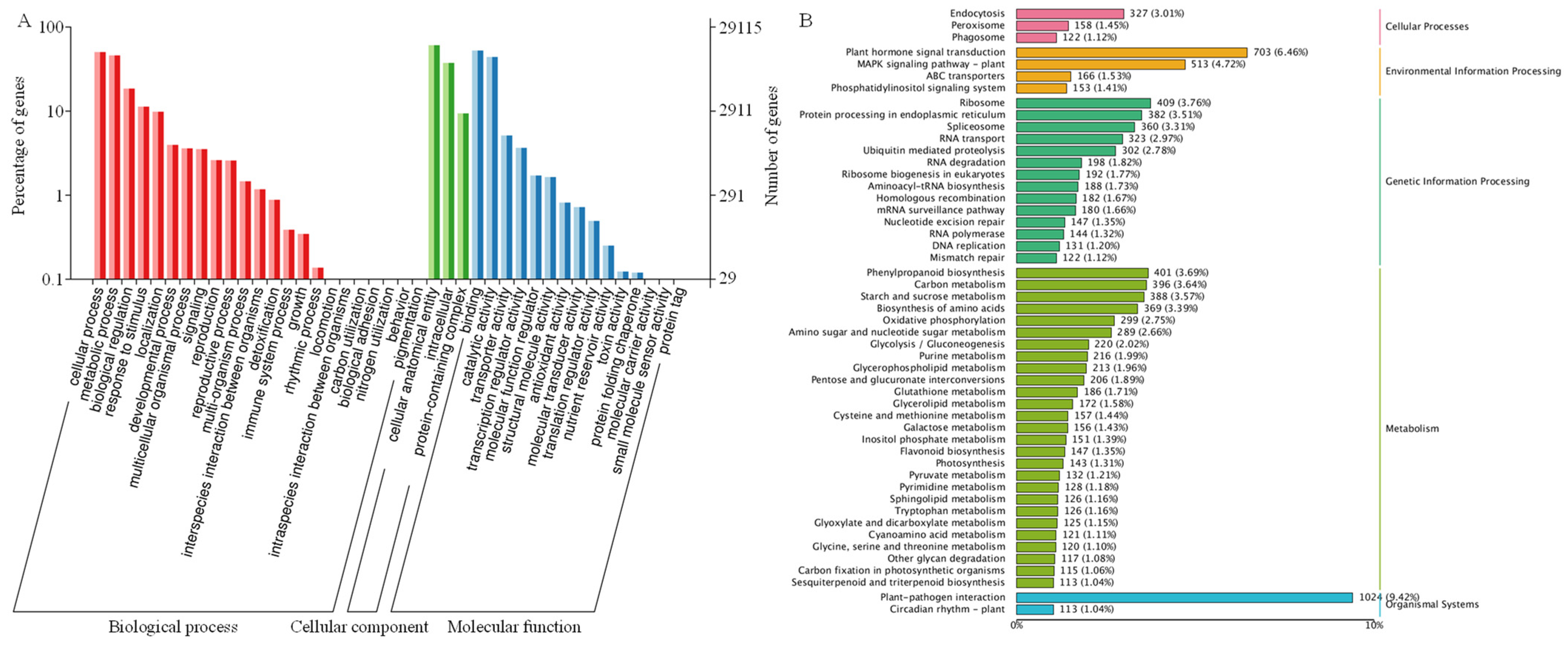
2.7. Transcriptome Analysis of C. lacryma-jobi Under Heat Stress
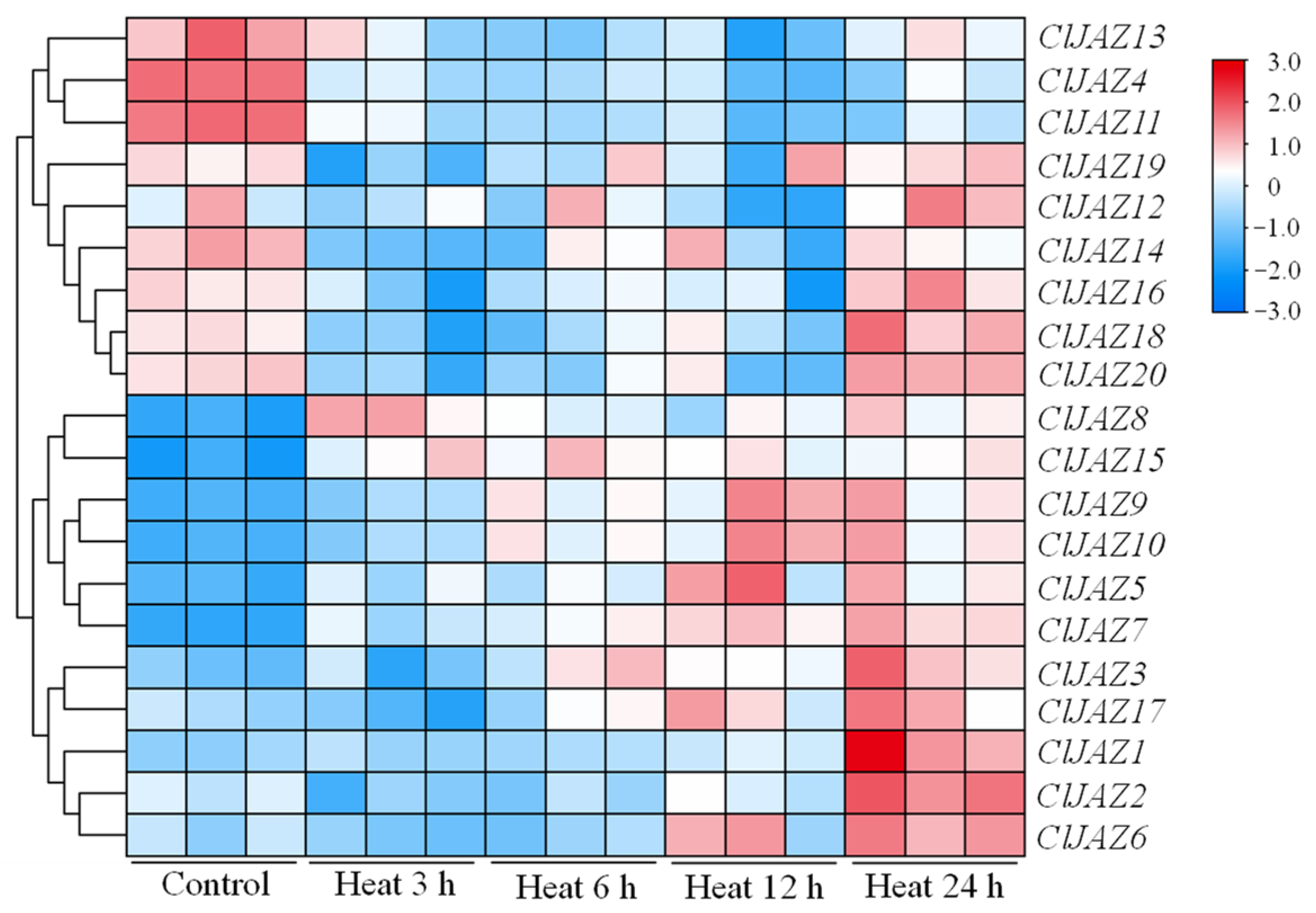
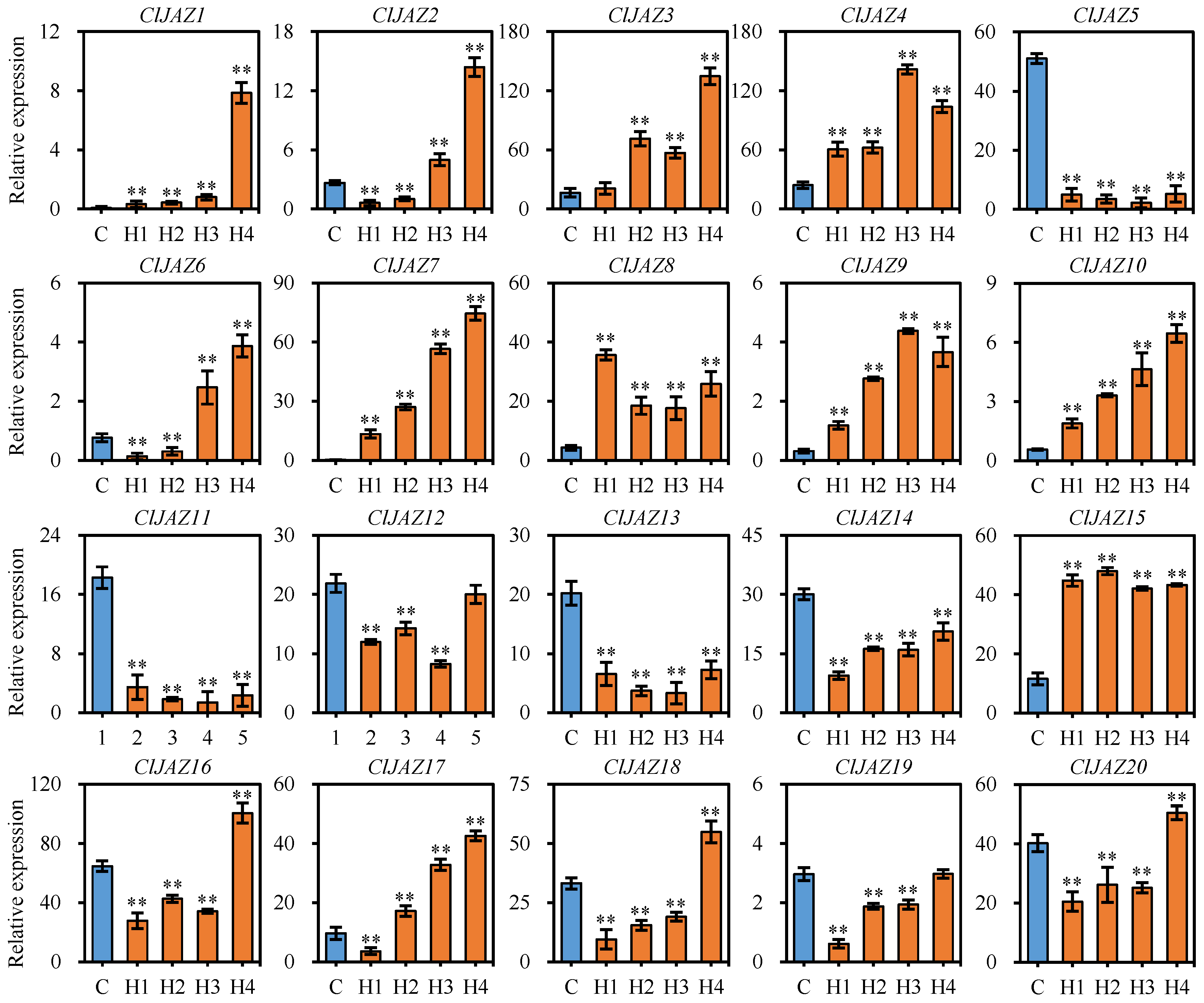
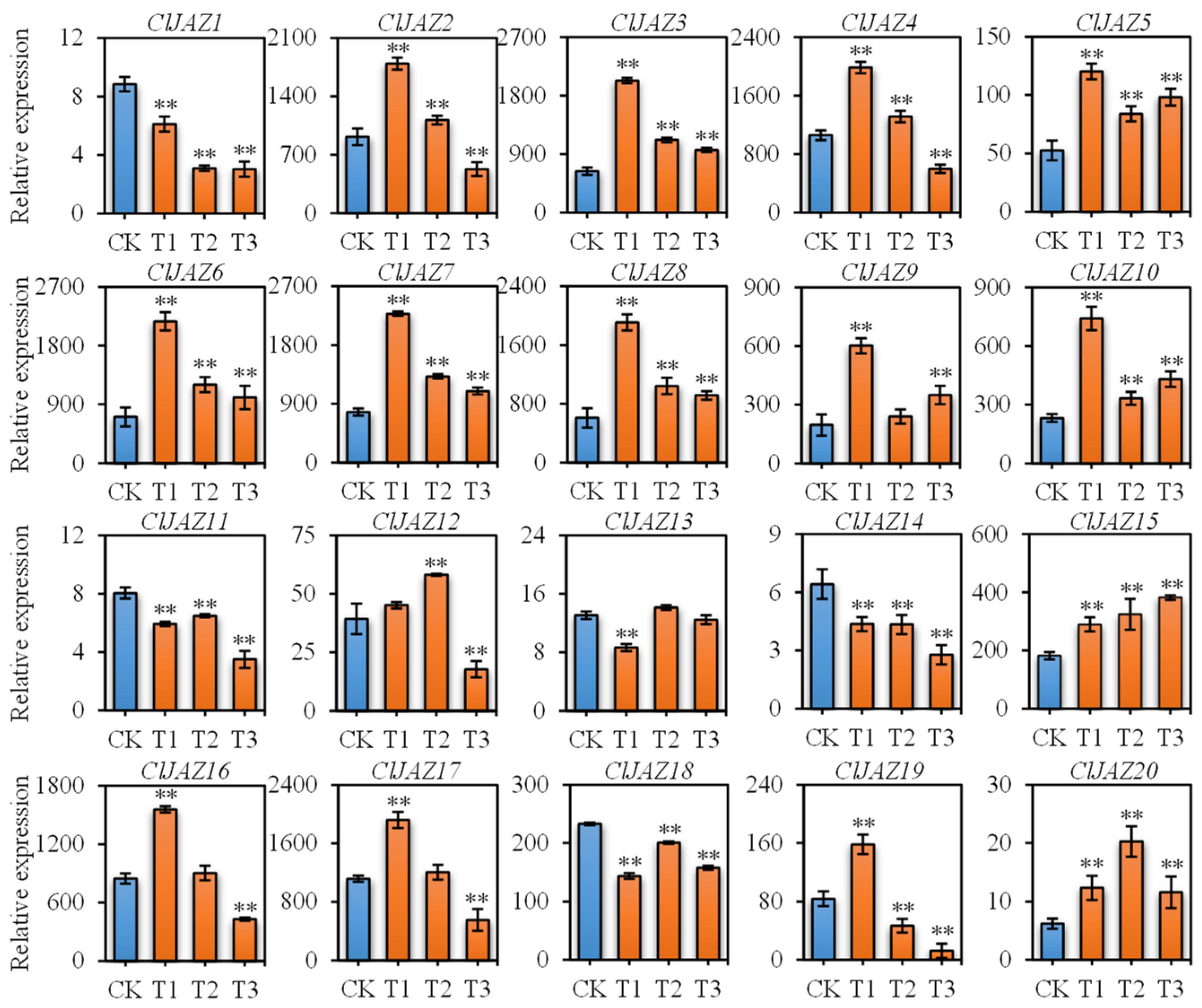
2.8. Expression Patterns of ClJAZs Genes Subjected to Heat Stress
2.9. Expression Patterns of ClJAZs Genes Exposed to ABA and MeJA Treatment
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of Jasmonate ZIM-Domain Family Members in C. lacryma-jobi
4.2. Phylogenetic Analysis of ClJAZ Family Proteins
4.3. Conserved Motifs, Gene Structure and Protein–Protein Interaction of ClJAZ Members
4.4. Chromosomal Location and Collinearity Analysis of ClJAZ Genes
4.5. Analysis of Cis-Regulating Elements upon the ClJAZ Promoters
4.6. Transcriptome Analysis of ClJAZ Gene Expression in Different Tissues
4.7. Transcriptome Analysis of ClJAZ Gene Expression Under Heat Stress
4.8. Plant Growth and Experimental Treatments of C. lacryma-jobi
4.9. RNA Extraction and Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Shikha, D.; Jakhar, P.; Satbhai, S.B. Role of jasmonate signaling in the regulation of plant responses to nutrient deficiency. J. Exp. Bot. 2023, 74, 1221–1243. [Google Scholar] [CrossRef] [PubMed]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Duan, G.H.; Li, C.Q.; Liu, L.; Han, G.Y.; Zhang, Y.L.; Wang, C.M. The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bhardwaj, M.; Tran, L.P. JASMONATE ZIM-domain family proteins: Important nodes in jasmonic acid-abscisic acid crosstalk for regulating plant response to drought. Curr. Protein Pept. Sci. 2021, 22, 759–766. [Google Scholar] [CrossRef]
- Zhao, J.; Ying, Y.; Xu, Z.; Chen, J.; Lyu, J.; Yu, Z. Identification and expression profiles of TIFY gene family in Rcbus chingii Hu. J. Nucl. Agri. Sci. 2024, 38, 1823–1835. (In Chinese) [Google Scholar]
- Li, M.Y.; Yu, G.H.; Cao, C.L.; Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef]
- Bisht, N.; Anshu, A.; Singh, P.C.; Chauhan, P.S. Comprehensive analysis of OsJAZ gene family deciphers rhizobacteria-mediated nutrient stress modulation in rice. Int. J. Biol. Macromol. 2023, 253, 126832. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-wide identification, characterization and expression analysis of the JAZ gene family in resistance to gray leaf spots in tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef]
- Zhang, H.; Li, W.; Niu, D.; Wang, Z.; Yan, X.; Yang, X.; Yang, Y.; Cui, H. Tobacco transcription repressors NtJAZ: Potential involvement in abiotic stress response and glandular trichome induction. Plant Physiol. Biochem. 2019, 141, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.; Joo, H.; Lim, C.W.; Lee, S.C. Roles of the pepper JAZ protein CaJAZ1-03 and its interacting partner RING-type E3 ligase CaASRF1 in regulating ABA signaling and drought responses. Plant Cell Environ. 2023, 46, 3242–3257. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.L.; Zhang, J.X.; Jiang, S.; Lu, Y.; Huang, Y.S.; Dong, X.M.; Hu, Q.; Yao, W.; Zhang, M.Q.; Xiao, S.H. Genome-wide identification of JAZ gene family in sugarcane and function analysis of ScJAZ1/2 in drought stress response and flowering regulation. Plant Physiol. Biochem. 2024, 210, 108577. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Xin, X.F. Regulation and integration of plant jasmonate signaling: A comparative view of monocot and dicot. J. Genet. Genom. 2021, 49, 704–714. [Google Scholar] [CrossRef]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; de Oliveira Ferreira, D.; He, S.Y.; Howe, G.A. Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef]
- Monte, I.; Caballero, J.; Zamarreño, A.M.; Fernández-Barbero, G.; García-Mina, J.M.; Solano, R. JAZ is essential for ligand specificity of the COI1/JAZ co-receptor. Proc. Natl. Acad. Sci. USA 2022, 119, e2212155119. [Google Scholar] [CrossRef]
- Johnson, L.Y.D.; Major, I.T.; Chen, Y.; Yang, C.; Vanegas-Cano, L.J.; Howe, G.A. Diversification of JAZ-MYC signaling function in immune metabolism. New Phytol. 2023, 239, 2277–2291. [Google Scholar] [CrossRef]
- Liu, H.B.; Shi, J.P.; Cai, Z.X.; Huang, Y.M.; Lv, M.L.; Du, H.L.; Gao, Q.; Zuo, Y.; Dong, Z.B.; Huang, W.; et al. Evolution and domestication footprints uncovered from the genomes of Coix. Mol. Plant 2020, 13, 295–308. [Google Scholar] [CrossRef]
- Chen, J. Essential role of medicine and food homology in health and wellness. Chin. Herb. Med. 2023, 15, 347–348. [Google Scholar] [CrossRef]
- Li, H.; Peng, L.; Yin, F.; Fang, J.; Cai, L.; Zhang, C.; Xiang, Z.; Zhao, Y.; Zhang, S.; Sheng, H.; et al. Research on Coix seed as a food and medicinal resource, it’s chemical components and their pharmacological activities: A review. J. Ethnopharmacol. 2024, 319, 117309. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Hao, D.; Xiao, P. Research progress of Chinese herbal medicine compounds and their bioactivities: Fruitful 2020. Chin. Herb. Med. 2022, 14, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Wang, H.; Ma, H.; Zheng, C. Genome-wide analysis of JAZ family genes expression patterns during fig (Ficus carica L.) fruit development and in response to hormone treatment. BMC Genom. 2022, 23, 170. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.L.; Fang, Y.P.; Jiang, J.M.; Chen, M.Q.; Li, X.Y.; Xie, X. Genome-wide identification and characterization of the JAZ gene family and its expression patterns under various abiotic stresses in Sorghum bicolor. J. Integr. Agric. 2022, 21, 3540–3555. [Google Scholar] [CrossRef]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021, 22, 256. [Google Scholar] [CrossRef]
- Zhang, B.; Zheng, H.; Wu, H.; Wang, C.; Liang, Z. Recent genome-wide replication promoted expansion and functional differentiation of the JAZs in soybeans. Int. J. Biol. Macromol. 2023, 238, 124064. [Google Scholar] [CrossRef]
- Hou, X.; Wang, D.; Cheng, Z.; Wang, Y.; Jiao, Y. A near-complete assembly of an Arabidopsis thaliana genome. Mol. Plant 2022, 15, 1247–1250. [Google Scholar] [CrossRef]
- Shang, L.; He, W.; Wang, T.; Yang, Y.; Xu, Q.; Zhao, X.; Yang, L.; Zhang, H.; Li, X.; Lv, Y.; et al. A complete assembly of the rice Nipponbare reference genome. Mol. Plant 2023, 16, 1232–1236. [Google Scholar] [CrossRef]
- Wei, C.; Gao, L.; Xiao, R.; Wang, Y.; Chen, B.; Zou, W.; Li, J.; Mace, E.; Jordan, D.; Tao, Y. Complete telomere-to-telomere assemblies of two sorghum genomes to guide biological discovery. iMeta 2024, 3, e193. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Tan, K.; Huang, W.; Shi, J.; Li, T.; Hu, J.; Wang, K.; Wang, C.; Xin, B.; et al. A complete telomere-to-telomere assembly of the maize genome. Nat. Genet. 2023, 55, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhang, Q.; Huang, J.; Zhang, S.; Yao, W.; Yu, Z.; Deng, Z.; Yu, J.; Kong, W.; Yu, X.; et al. A chromosomal-scale genome assembly of modern cultivated hybrid sugarcane provides insights into origination and evolution. Nat. Commun. 2024, 15, 3041. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Chu, Y.; Chen, Y.; Zhao, Q.; Liao, B.; Zhang, J.; Gao, Y.; Xu, J.; Chen, S. Genome-wide identification and transcriptional profiling analysis of PIN/PILS auxin transporter gene families in Panax ginseng. Chin. Herb. Med. 2022, 14, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, G.; Chen, J.; Ying, Y.; Yao, L.; Li, X.; Teixeira da Silva, J.A.; Yu, Z. Role of Rubus chingii BBX gene family in anthocyanin accumulation during fruit ripening. Front. Plant Sci. 2024, 15, 1427359. [Google Scholar] [CrossRef]
- Sun, Y.; Oh, D.H.; Duan, L.; Ramachandran, P.; Ramirez, A.; Bartlett, A.; Tran, K.N.; Wang, G.; Dassanayake, M.; Dinneny, J.R. Divergence in the ABA gene regulatory network underlies differential growth control. Nat. Plants 2022, 8, 549–560. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; He, Y.; Lang, Z.; Zhao, Y.; Tao, H.; Li, Q.; Hong, G. The CsHSFA-CsJAZ6 module-mediated high temperature regulates flavonoid metabolism in Camellia sinensis. Plant Cell Environ. 2023, 46, 2401–2418. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, G.; Teixeira da Silva, J.A.; Li, M.; Zhao, C.; He, C.; Si, C.; Zhang, M.; Duan, J. Genome-wide identification and analysis of DNA methyltransferase and demethylase gene families in Dendrobium officinale reveal their potential functions in polysaccharide accumulation. BMC Plant Biol. 2021, 21, 21. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, X.; Fu, Y.; Wang, H.; Yu, C.; Chu, J.; Jiang, B.; Zhu, J. Genome-wide identification and expression analysis of VQ gene family under abiotic stress in Coix lacryma-jobi L. BMC Plant Biol. 2023, 23, 327. [Google Scholar] [CrossRef]




| Name | Size/aa | Mw/kD 1 | pI 2 | II 3 | AI 4 | GRAVY 5 | Localization 6 |
|---|---|---|---|---|---|---|---|
| ClJAZ1 | 184 | 19.17 | 9.36 | 84.60 | 70.87 | −0.234 | Nucleus |
| ClJAZ2 | 199 | 21.18 | 9.77 | 66.41 | 68.44 | −0.424 | Nucleus |
| ClJAZ3 | 187 | 19.41 | 7.83 | 61.91 | 74.06 | −0.160 | Nucleus |
| ClJAZ4 | 228 | 24.00 | 6.62 | 38.33 | 69.61 | −0.368 | Nucleus |
| ClJAZ5 | 242 | 25.51 | 6.62 | 41.85 | 75.66 | −0.287 | Nucleus |
| ClJAZ6 | 159 | 16.55 | 10.09 | 45.17 | 80.13 | −0.272 | Nucleus |
| ClJAZ7 | 297 | 31.27 | 11.18 | 52.52 | 68.96 | −0.468 | Nucleus |
| ClJAZ8 | 162 | 17.39 | 9.84 | 71.82 | 76.48 | −0.244 | Nucleus |
| ClJAZ9 | 172 | 18.53 | 10.47 | 67.92 | 75.00 | −0.257 | Nucleus |
| ClJAZ10 | 207 | 22.28 | 10.05 | 46.08 | 68.02 | −0.416 | Nucleus |
| ClJAZ11 | 288 | 30.92 | 4.64 | 62.42 | 64.83 | −0.679 | Nucleus |
| ClJAZ12 | 336 | 35.90 | 8.99 | 51.68 | 66.31 | −0.494 | Nucleus |
| ClJAZ13 | 313 | 33.45 | 8.94 | 54.62 | 65.88 | −0.489 | Nucleus |
| ClJAZ14 | 328 | 35.01 | 5.11 | 53.43 | 58.78 | −0.628 | Nucleus |
| ClJAZ15 | 353 | 37.57 | 5.12 | 55.84 | 64.28 | −0.565 | Nucleus |
| ClJAZ16 | 239 | 25.35 | 9.25 | 47.20 | 74.85 | −0.426 | Nucleus |
| ClJAZ17 | 229 | 23.84 | 9.28 | 43.83 | 74.76 | −0.403 | Nucleus |
| ClJAZ18 | 409 | 43.00 | 9.83 | 70.23 | 67.97 | −0.281 | Nucleus |
| ClJAZ19 | 254 | 25.55 | 9.78 | 64.91 | 69.76 | −0.350 | Nucleus |
| ClJAZ20 | 181 | 19.22 | 9.26 | 77.91 | 77.79 | −0.367 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Shen, Y.; Sun, Y.; Xu, Z.; Zheng, F.; Shen, X. Systematic Survey and Analysis Reveal Jasmonate ZIM-Domain Gene Family in Coix lacryma-jobi Under High Temperature. Plants 2024, 13, 3230. https://doi.org/10.3390/plants13223230
Yu Z, Shen Y, Sun Y, Xu Z, Zheng F, Shen X. Systematic Survey and Analysis Reveal Jasmonate ZIM-Domain Gene Family in Coix lacryma-jobi Under High Temperature. Plants. 2024; 13(22):3230. https://doi.org/10.3390/plants13223230
Chicago/Turabian StyleYu, Zhenming, Yufeng Shen, Yiming Sun, Zhangting Xu, Feixiong Zheng, and Xiaoxia Shen. 2024. "Systematic Survey and Analysis Reveal Jasmonate ZIM-Domain Gene Family in Coix lacryma-jobi Under High Temperature" Plants 13, no. 22: 3230. https://doi.org/10.3390/plants13223230
APA StyleYu, Z., Shen, Y., Sun, Y., Xu, Z., Zheng, F., & Shen, X. (2024). Systematic Survey and Analysis Reveal Jasmonate ZIM-Domain Gene Family in Coix lacryma-jobi Under High Temperature. Plants, 13(22), 3230. https://doi.org/10.3390/plants13223230







