Abstract
Kenaf (Hibiscus cannabinus L.), in the Malvaceae family, is an important crop for not only fiber production, but also various other industrial materials. We performed phylogenetic analysis and a genome-wide association study (GWAS) of seven agronomic traits: days to flowering, plant height, fresh weight, dry weight, flower color, stem color, and leaf shape, using 96 kenaf genotypes, including gamma-irradiation-derived mutant lines. Genotypes were determined by genotyping-by-sequencing (GBS) and a total of 49,241 single-nucleotide polymorphisms (SNPs) were used in the analysis. Days to flowering, plant height, fresh weight, and dry weight were positively correlated with each other, and stem color was also correlated with fresh weight and dry weight. The phylogenetic analysis divided the 96 lines into nine related groups within two independent groups, and the GWAS analysis detected a total of 49 SNPs for days to flowering, plant height, fresh weight, dry weight, flower color, stem color, and leaf shape with −log10(P) ≥ 4, of which 22 were located in genic regions. The detected SNPs were located in genes with homology ranging from 45% to 96% to plants of the Malvaceae and Betulaceae, and these genes were found to be involved in plant growth and development via various pathways. Our identification of SNP markers related to agronomic traits is expected to help improve the quality of selective breeding programs for kenaf.
1. Introduction
A member of the Malvaceae family, kenaf (Hibiscus cannabinus L.), is a diploid (2n = 2X = 36) annual herbaceous plant native to Africa [1]. Originating in North Africa, kenaf is now cultivated in many countries, including India, Russia, China, and the United States, and is known to thrive in temperate and tropical environments with abundant sunlight and precipitation [1,2]. Kenaf has a variety of valuable industrial uses, including as a source of edible seed oil, stem fiber, plastic raw materials, and pharmaceuticals. In the past, the main purpose of kenaf production was to produce fiber for the manufacture of carpets, canvas bags, and rope. During World War II, the use of kenaf fiber for rope production came to the forefront, leading to research into its cultivation, production, and processing [3,4]. In Africa, kenaf leaves and stems were used to treat Guinea-worm disease and anemia, while in Ayurvedic medicine, an ancient Indian Hindu tradition, kenaf leaves were used to treat bile, blood, coughing, and diabetes [5,6]. Kenaf is known to have a crude protein content of 6% to 23% in the whole plant, and in particular, the crude protein content in the leaves reaches 14% to 34%, making it suitable as a feed source for livestock [2]. Recently, kenaf fiber has been attracting attention as a material for biodegradable, eco-friendly composites that can replace glass fiber composites [7,8]. Additionally, kenaf is an important medicinal crop as it contains high amounts of phytochemical compounds including polyphenols and anthocyanins. Thus, kenaf has been used in Indian traditional medicine (Ayurvedic medicine) as an aphrodisiac, purgative, and digestive aid including antioxidant, hepatoprotective, and anticancer activities [5].
The yield of kenaf stalks varies from 11 to 18 tons/ha, with cropping season, temperature, and soil moisture acting as major factors [2]. Kenaf plants are mainly known for their light yellow and cream-colored flowers with green stems [2]. Kenaf varieties can be categorized into three maturation groups, determined by their photosensitivity: ultra-early maturing, early-to-medium maturing, and late maturing [3]. Kenaf maturity is closely related to yield: to increase fiber yield, it is advantageous to delay flowering to increase growth rate [9]. However, if flowering is delayed too much, it is disadvantageous for seed maturation, so the photoperiod of the cultivation area according to latitude must be taken into consideration [4]. Mutation breeding involves the use of a mutagen to develop plants, exhibiting novel mutated characteristics that do not disturb elite cultivar traits [10]. Novel kenaf cultivars generated by radiation mutagenesis and showing improved seed and biomass yield characteristics have been developed using radiation breeding techniques. The cultivars of Hibiscus species have been generated via hybridization, but the floriculture and/or pharmaceutical industry relies on a limited number of mutated traits established based on specific flower quality parameters and consumer palatability [11,12,13].
Genetic change, or mutation, is a natural process that creates new genetic variants. Since the frequency of natural mutations is quite low, it is difficult to discover useful genetic variations in a short period of time. Therefore, generating genetic variations by treating crops with physical or chemical mutagens has been used successfully in mutation breeding [14,15]. Among physical mutagens, gamma rays have been used to induce mutations in various crops, such as rice, soybean, and kenaf, and to breed new cultivars [10,14,16]. Anthocyanins play an important role in plant pigmentation, which is strongly associated with the coloration in kenaf. Our research group has also developed novel kenaf cultivars via gamma irradiation on seeds of the introduced cultivars and registered them in the Korea Seed and Variety Service. In our previous work, we explored the potential of a novel flower or stem color mutation cultivar, ‘Jeokbong’ and ‘Bora’, as a functional food [14].
Detection of genomic sequences related to particular traits has the potential not only to study the function of genes, but also to accelerate the speed of future breeding [17,18,19]. Advances in next-generation sequencing (NGS) have made the rearrangement of plant genomes efficient and economical, and have also made it possible to detect the loci, types, and rates of mutations on a genome-wide scale [20,21]. GBS is a sequence-based genotyping method that is characterized by the use of restriction enzymes to sequence multiple samples simultaneously on an NGS platform using a reduced label of the target genome and a DNA barcode adapter [22,23]. GWAS is a method for detecting associations between phenotypes and genes in a population. While linkage mapping methods use bi-parental populations, GWAS is an approach that exploits diverse natural populations [24,25,26]. This approach has been applied to a variety of crops, including rice [27], soybean [28], maize [29], and sorghum [20], and it is being used in breeding programs. To date, no GWAS analyses of kenaf have been reported.
Although the potential industrial value of kenaf is quite high, genetic diversity and genetic studies of the crop are scarce. Therefore, in this study, 96 kenaf genetic resources, including gamma ray mutant lines, were sequenced using GBS, and GWAS analyses were performed for seven agronomic traits: days to flowering, dry weight, fresh weight, plant height, flower color, stem color, and leaf shape.
2. Results
2.1. Phenotypic Variation and Correlation Analysis
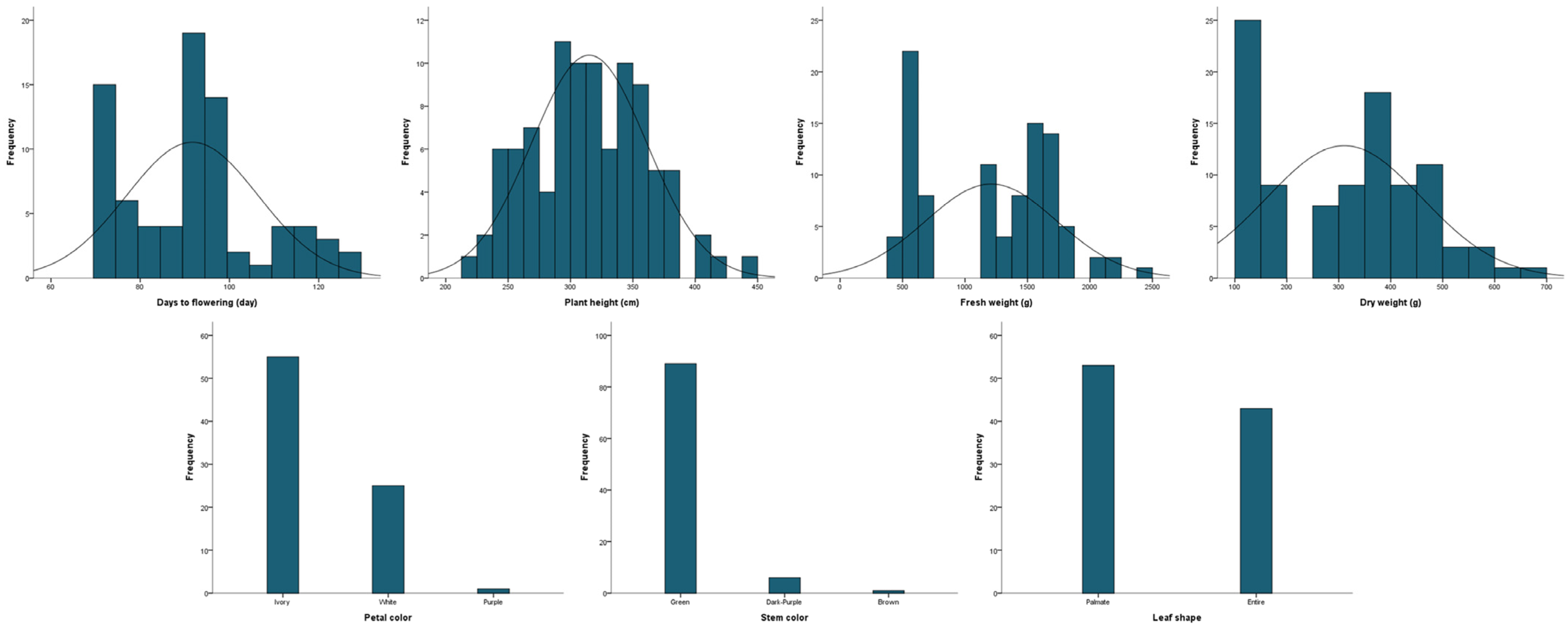
The 96 kenaf lines were determined for four quantitative traits: days to flowering, plant height, fresh weight, and dry weight, and three qualitative traits: flower color, stem color, and leaf shape (Table 1 and Figure 1). The days to flowering ranged from 72 to 125 d (mean 92 d) and plant height was 223–444 cm (mean 315 cm). The fresh and dry weights were 472–2400 g and 110–672 g, respectively (mean 1209 and 312 g). The coefficients of variation for days to flowering and plant height were 16.1% and 14.6%, respectively, which were significantly lower than for fresh and dry weight (43.4% and 47.9%). The skewness of all four traits was close to zero, and the height of the distribution was lower than the normal distribution. Ivory was the most common flower color (55 lines), followed by white (25) and purple (1). The most common stem color was green (89 lines), followed by dark purple (6) and brown (1). For leaf shape, there were 53 palmate types and 43 entire types. The correlations between the seven agricultural traits are shown in Table 2. The four agronomic traits, days to flowering, plant height, fresh weight, and dry weight, each had a positive correlation (p ≤ 0.01). In contrast, stem color showed a negative correlation with fresh and dry weight (p ≤ 0.05).

Table 1.
Descriptive statistics for agronomic traits in 96 kenaf lines.
Figure 1.
Frequency distribution of agronomic traits in 96 kenaf lines.

Table 2.
Correlation coefficients for agronomic traits in 96 kenaf lines.
2.2. Genotyping by Sequencing of 96 Kenaf Lines
The GBS library was constructed from 96 kenaf lines, including gamma-ray-derived mutations, and sequenced using the Illumina Hiseq 2000 platform. A summary of the GBS results is presented in Table 3. Using two biological replicates, a total of 702 million reads comprising 106,096,097,764 nucleotides (106 Gb) were generated, with 7.3 million reads per genotype on average (Table 3). After trimming low quality sequences, 664,405,534 clean reads remained, with 6.6 million reads per genotype on average. The total length of the mapped region was 3,263,929,064 bp, with an average of 33,999,261 bp per sample, which covered approximately 3.17% of the whole genome; the sequences were mapped to the reference genome sequence. Among the 96 lines, the average depth of read mapping ranged from 10.84× to 21.49×.

Table 3.
Summary of GBS sequence data and alignment to the reference genome sequence.
2.3. Construction of Phylogenetic Tree and Genome-Wide Association Study for Agronomic Traits
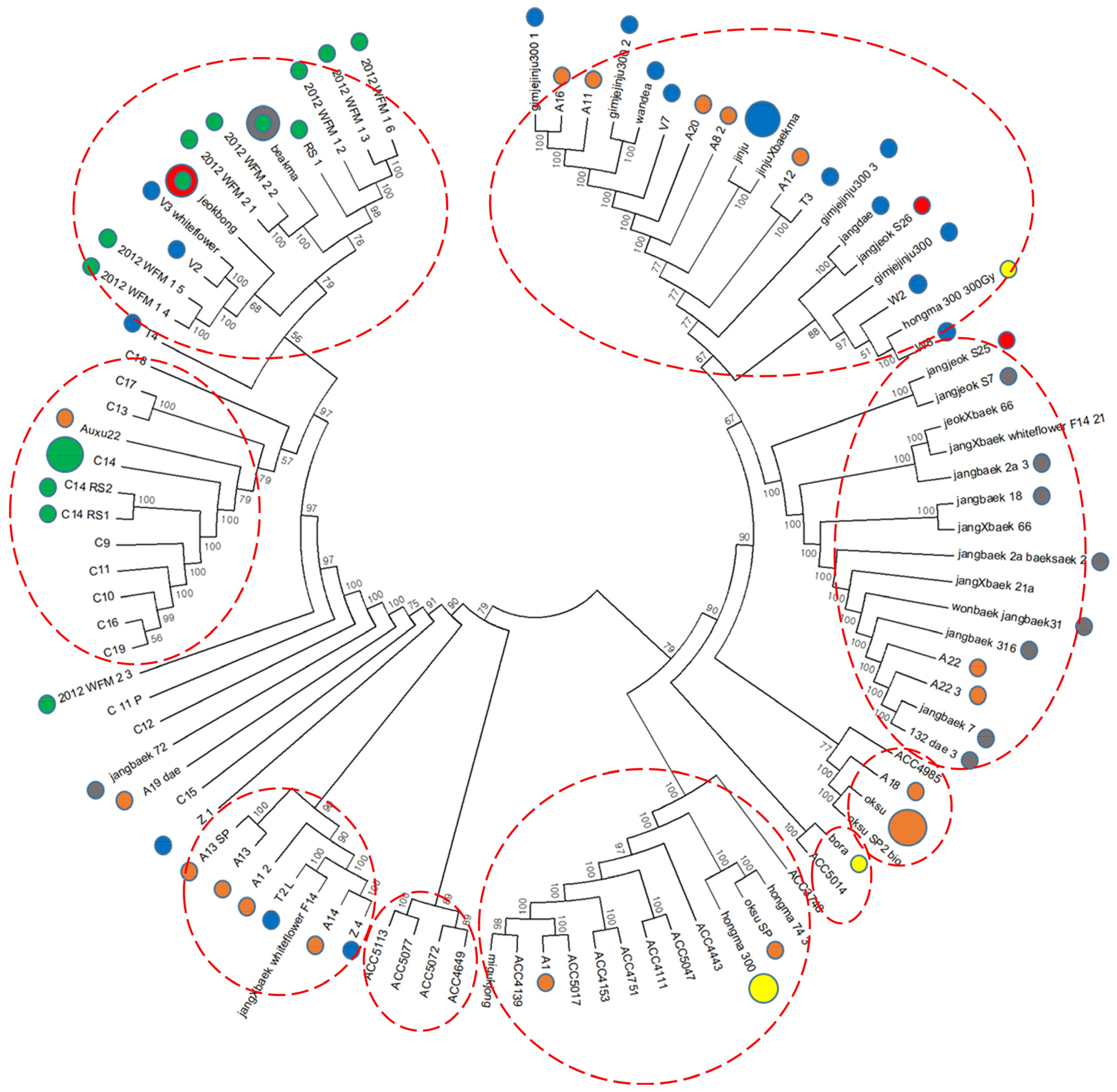
Phylogenetic analysis was performed on 96 kenaf lines, including gamma-ray-derived mutants, based on the UPGMA method, and a dendrogram was generated with 49,241 filtered SNPs using the neighbor-joining method (Figure 2). In the cluster analysis, the 96 kenaf genotypes were divided into nine related groups within two independent groups, with the exception of seven lines (2012_WFM_2_3, C_11_P, C12, jangbaek_72, A19_dae, C15, and Z_1) that could not be grouped.
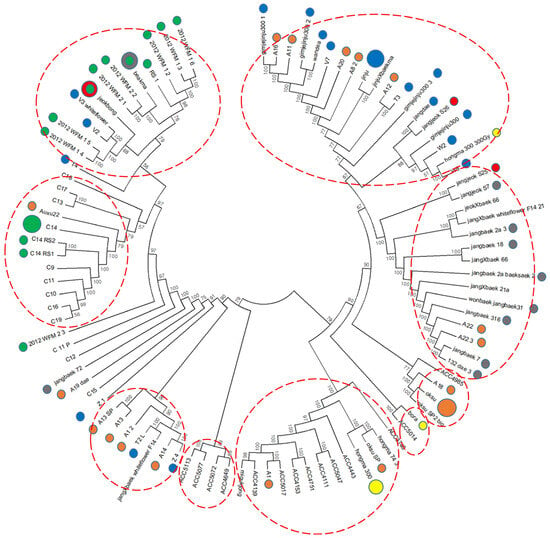
Figure 2.
Phylogenetic tree constructed using 49,241 filtered SNP loci from 96 kenaf genotypes. Colored circles represent mutant lines; the larger colored circles denote the respective original genotypes.
By plotting r2 against the distance (in kb) between a pair of SNPs, it was a very modest decrease in LD decay. When we estimated LD decay for the entire genome, the maximum r2 was halved at around 1433 kb, and as a result, the size of the LD block was considered quite large (Figure S1).
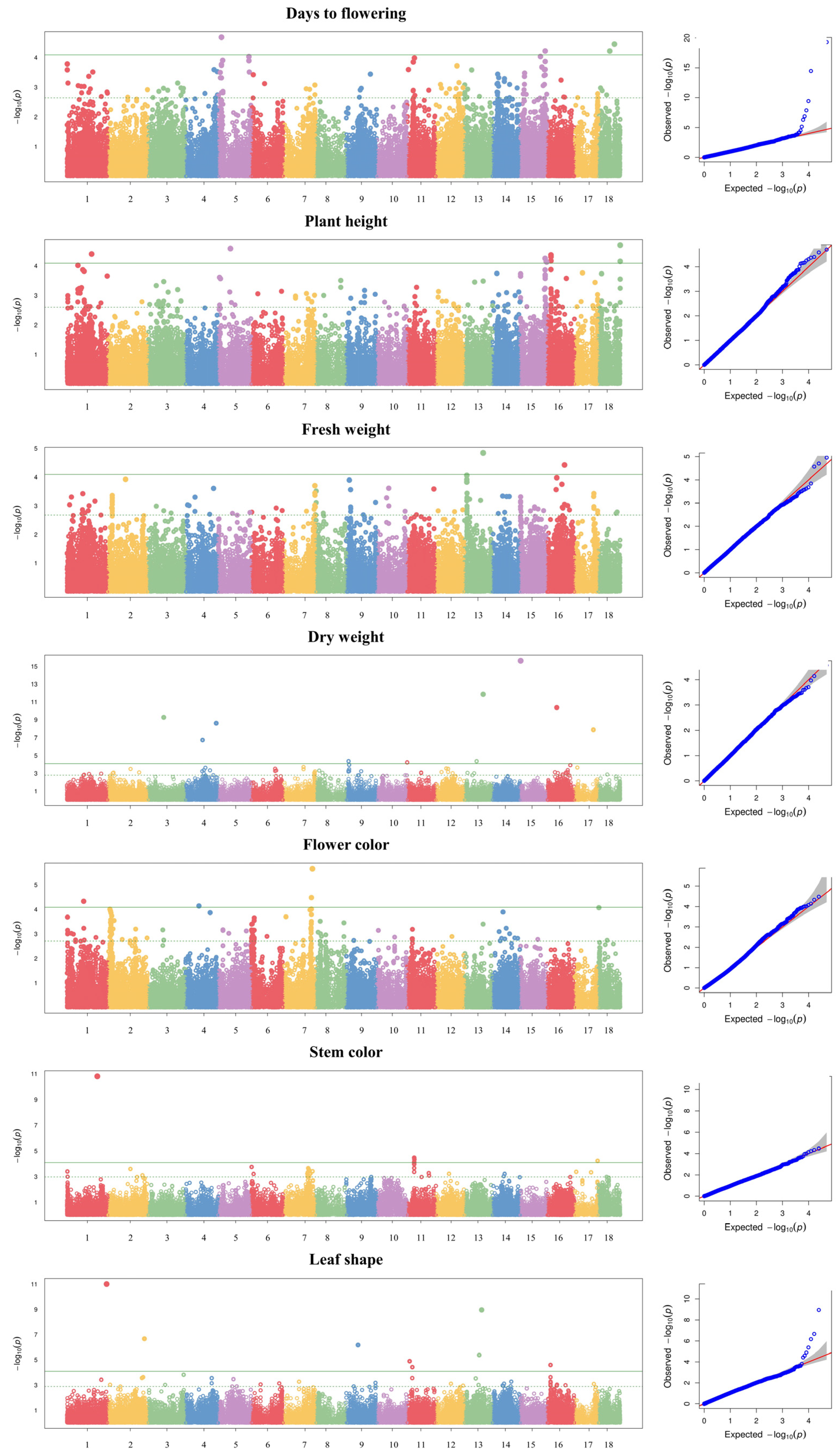
The GWAS was conducted using a multi-locus mixed model for seven agronomic traits: days to flowering, dry weight, fresh weight, plant height, flower color, stem color, and leaf shape. Using a threshold of −log10(P) ≥ 4, a total of 49 SNPs were detected across 13 chromosomes for seven agronomic traits (Table 4 and Figure 3). Of these, 12 SNPs associated with days to flowering were detected on chromosomes 1, 4, 5, 7, 10, 14, 16, and 18; 11 SNPs associated with plant height were detected on chromosomes 1, 5, 15, 16, and 18; three SNPs associated with fresh weight and dry weight were detected on chromosomes 1, 2, and 13; seven SNPs associated with flower color were detected on chromosomes 1, 2, 4, 7, and 18; five SNPs associated with stem color were detected on chromosomes 1, 11, and 17; and eight SNPs associated with leaf shape were detected on chromosomes 1, 2, 9, 11, 13, and 16. Among the 49 SNPs detected, 27 were located in intergenic regions and 22 in genic regions.

Table 4.
SNPs associated with seven agronomic traits detected in GWAS analysis.
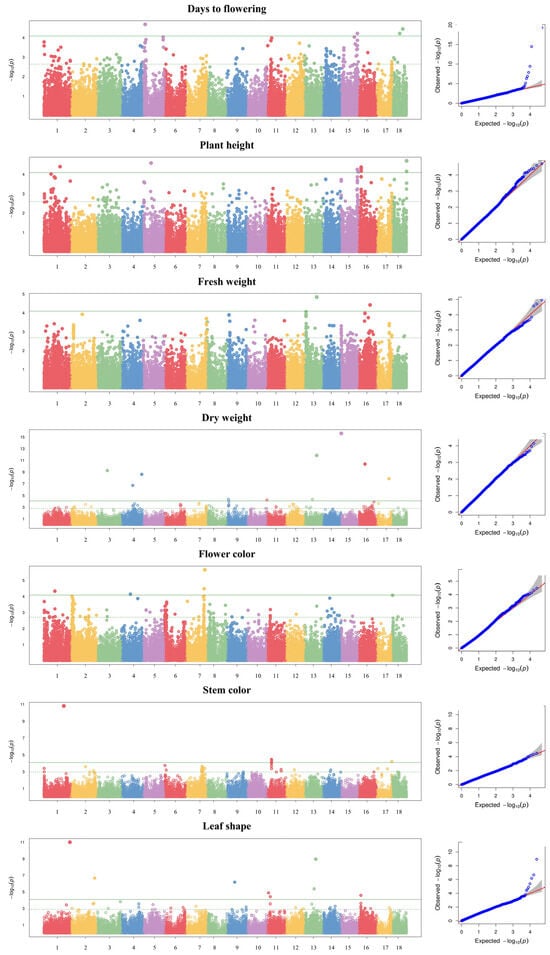
Figure 3.
Manhattan plots and quantile–quantile plots for seven agronomic traits in 96 kenaf genotypes.
2.4. Gene Annotation
The gene annotations of SNPs located in genic regions detected in the GWAS analysis are shown in Table 5. The genic regions had a 45.53% to 96.34% identity with genes from the Malvaceae family (Herrania umbratica, Gossypium mustelinum, Hibiscus syriacus, Durio zibethinus, Gossypium hirsutum, and Gossypium australe) and the Betulaceae family (Carpinus fangiana). Except for fresh weight and dry weight, where no SNPs were detected in the genic region, the majority of SNPs were located in the exon of the respective genic regions: three out of five SNPs were detected for days to flowering, four out of five SNPs for plant height, two out of three SNPs for flower color, one out of two SNPs for stem color, and four SNPs for leaf shape. The highest level of SNPs located in an exon of a gene associated with days to flowering was found on chromosome 5 with −log10(P) = 18.86 and a 90.50% homology to the gene encoding an uncharacterized protein LOC120130459 from Hibiscus syriacus. The next highest −log10(P) values were found for SNPs associated with other traits: plant height showed a 64.05% homology with the gene encoding the abrin-b-like of Durio zibethinus on chromosome 18, flower color showed a 66.89% homology with the gene encoding the hypothetical protein FH972_000750 of Carpinus fangiana on chromosome 7, stem color showed an 88.18% homology with the gene encoding the pentatricopeptide repeat-containing protein of Hibiscus syriacus on chromosome 11, and leaf shape showed an 82.24% homology with the gene encoding the protein DETOXIFICATION 3-like of Hibiscus syriacus on chromosome 1.

Table 5.
SNPs and candidate genes associated with seven agronomic traits in 96 kenaf genotypes.
3. Discussion
Kenaf has wide utility value as it can be used as a source of fiber, edible and pharmaceutical ingredients, as well as for plastic materials, but the crop’s genetics remain underresearched. The main objective of kenaf breeding has been to develop new cultivars that have high yields, are resistant to pests and diseases, are drought tolerant, or can be locally adapted to different environmental and growing conditions [5]. Breeding methods mainly used in kenaf include introduction breeding, exploiting natural variation, hybridizations, and mutation breeding. Recently, cultivars with new characteristics have become required because of changes in various cultivation environments and industrial demands [5,11]. Therefore, remote crossing and modular and mutation breeding are gradually being more widely used. However, the identification of genetic information and relationships using NGS technologies has been limited in kenaf. It is necessary to better understand the genetic basis of important traits in kenaf to improve production and to lay the foundation for molecular breeding efforts [11,12,13]. Our current study was the first to attempt GBS analysis targeting kenaf genetic resources and breeding lines for various agronomic traits and/or useful traits for the pharmaceutical industry.
In this study, we performed GWAS to investigate candidate genes for agronomic traits using 96 kenaf genotypes, including gamma-irradiation-derived mutant lines. Mutation induced using gamma radiation has been widely used to create genetic diversity via a variety of gene mutations [27]. In GWAS analyses, progeny lines can be highly correlated between samples and are liable to overestimate the genetic association because of false positives [30,31,32]. In addition, because progeny lines are derived from one or a few parent plants, they do not represent a wide range of genetic diversity, which makes it difficult to capture the many variations in the plant genome and the unusual variants that can occur via any pathway. Therefore, it is common to apply GWAS analyses to natural populations. In our results, among the lines used in the GWAS analysis, we found the phylogeny of the radiation mutant lines was sometimes close to the original line, such as the C14 lines, while in other cases, such as the Auxu and Jinju lines, they diverged into several branches (Table S1 and Figure 2). These results suggest that gamma-irradiation-derived mutant lines can be phylogenetically separated by GWAS, which uses genetic diversity wider than the original lines.
On the basis of their geographic origin, the phylogenetic analysis revealed an unclear pattern of division among the genotypes. Given that the kenaf genotypes evaluated in this study originated from various breeding methods and mixed pedigrees, it is likely that the phylogenetic analysis using many origins and races was not able to differentiate among all the genotypes. Kenaf originated in South Africa and was then introduced into India, China, Russia, and the Americas in the eighteenth century. Nowadays, kenaf is commercially cultivated in more than 20 countries, but the dissemination of genetic information about the crop worldwide is limited. Genotypes collected from Asia and central and North America were found to have close genetic relationships [33].
Correlations between agronomic traits can be used to predict yields or control harvest timing, but they are also important for breeding programs. In various crops, including kenaf, flowering time and biomass traits, such as fresh weight, dry weight, and plant height, are known to be positively correlated [34], and our results in Table 2 show the same pattern. Korean Kenaf cultivars are divided into three maturation groups depending on the flowering date: early maturing, mid–late maturing, and late maturing. Early-maturing groups mature in 70–80 d after sowing, which enables seed harvests, but at the cost of lower biomass. Late-maturing groups grow vegetatively for 130–140 d and yield significantly higher biomass, but late maturation reduces seed quality [14,15]. Therefore, a breeding goal in new Korean kenaf cultivars is to increase both biomass and seed yields per unit area [35,36]. Mutation breeding has the merits of creating new mutant characteristics and adding only few traits without disturbing the other characteristics of a cultivar [37]. The mid–late cultivars ‘Jangdae’ and ‘Wandae’, which afford both high biomass and high seed yield, have been registered with the Korea Seed and Variety Service.
Notably, there is a weak correlation between stem color and biomass. The population used in the analysis had three stem colors: green, dark purple, and brown, with stem colors other than green having a slightly lower biomass content. Although the non-green stem color was associated with lower biomass, such lines are valuable as breeding material because they produce functional substances such as anthocyanins [14]. In general, plant pigments are functional substances with various activities, such as antioxidant, antidiabetic, and anti-inflammatory compounds. Therefore, changes in stem color and flower color are accompanied by changes in the contents of functional compounds that might be valuable in various food and pharmaceutical industries. The Jeokbong, RS1, and RS2 genotypes has distinctive morphological characteristics such as dark purple color. The high levels of phenolic compounds, such as anthocyanin, observed in the previous study for the purple genotypes and their antioxidant activity are approximately 4~5 times higher than other cultivars caused by anthocyanin [14]. In the kenaf plant, the genes of phenylpropanoid biosynthetic enzymes, including HcPAL, Hc4CL, HcC4H, and HcCHS, have been isolated in previous studies [38,39]. Recent progress, including NGS technology, has facilitated various transcriptome analyses in kenaf. Previously, we successfully undertook transcriptome analysis of leaf coloration in ‘Jeokbong’, which exhibited dramatic changes in eight flavonoid structural genes. Identifying genes related to the biosynthesis of anthocyanins and kaempferitrin in kenaf leaves, 29 differentially expressed genes were assigned to eight structural genes, namely 4CL, CHS, CHI, F3H, DFR, ANS, FLS, and 3GT [12,14]. In addition, we performed comparative transcriptome analysis using RNA sequencing and identified putative genes (CHS, F3’H, FLS, DFR, MAT, UFGTs, TT12, GST, and RNS) involved in flower coloration at different flower developmental stages in three kenaf mutants, namely white flower, ivory flower, and purple flower [13]. When the color of petals was changed to purple or white by radiation mutation breeding, the total amount of flavonoids and phenolic compounds was more [12,13,36]. It was assumed that the accumulation of flavonoids and phenolic acids was accelerated. In the reports on the function of inside the plants, flavonoid glycosides were involved in protection against radiation stress and in producing a purple pigment [5,12,13]. We selected the SNPs in the flowers of new kenaf mutants, including white petal, purple petal, and moral types (ivory petal), as well as candidate gene data to better understand the mechanism of flower coloration via radiation.
GWAS analysis confirmed that the gene regions in which the SNPs detected for each trait were located were highly homologous to known gene regions in the Malvaceae and Betulaceae families, but no clearly identified genes for the traits were found. The LRR receptor-like serine/threonine-protein kinase (LRR-RLKs) detected as associated with flowering might be involved in several aspects of plant development, such as cell growth, differentiation, organ formation, and stress responses. Its roles might be similar to those of LRR-RLK BRASSINOSTEROID INSENSITIVE 1 (BRI1), a receptor for brassinosteroids, a class of plant hormones that regulate cell elongation and division [40,41,42]. The E3 ubiquitin-protein ligase BRE1-like 1 (HUB1), detected as associated with plant height, is known to regulate floral development in Arabidopsis by monoubiquitinating H2B [43]. The pentatricopeptide repeat-containing protein (PPR), detected as associated with stem color, is involved in the regulation of genes related to chlorophyll biosynthesis, photosynthesis, and flowering. It is also known to be involved in defense mechanisms against abiotic stresses, such as drought and salinity, and pathogens, such as viruses and bacteria [44]. The Korea Atomic Energy Research Institute discovered a variant with dark purple stems generated using gamma radiation as a mutation source and registered a cultivar ‘Jeokbong’ that is rich in anthocyanins (delphinidin-3-O-sambubioside). This is the same as the anthocyanin reported in roselle (Hibiscus sabdariffa L.). The antioxidant (2,2-diphenyl-1-picrylhydrazyl radical scavenging activities) and angiotensin-converting enzyme inhibitory activity of ‘Jeokbong’ was approximately four times higher than that of three other cultivars [14]. However, ‘Jeokbong’ and other purple stem color mutants (RS1, C14_RS1, jangjeok_S7, jangjeok_S26, and jangjeok_S25) showed poor salt tolerance. These results suggest it should be possible to develop an SNP marker for selection of stem anthocyanin and salt tolerance in kenaf.
The protein FAR1-RELATED SEQUENCE 11 (FRS11), detected as associated with leaf shape, is a member of the FRS family of transcription factors that play important roles in plant growth and development, light signaling, plant hormone responses, and stress resistance. Using a mutant with knocked out FRS11, it was confirmed that FRS11 regulates plant growth and development in response to light via its response to far-infrared light. It delayed flowering in potatoes [45] and is involved in leaf senescence in Arabidopsis [46]. It is clear that most of the genes identified in our study might play important roles in growth and development. However, it is likely that they influence each trait via a complex mechanism rather than a direct causal link, so further research is needed. Our findings will provide useful information for a better understanding of kenaf genetics and demonstrate the utility of mutations, either directly in breeding programs, or indirectly as a research tool.
4. Materials and Methods
4.1. Plant Materials and DNA Isolation
Ninety-six genotypes were studied (Table S1). These included 55 mutant genotypes derived from gamma irradiation (300 Gy) of seed. Thirty genotypes were obtained from the Genebank of Rural Development Administration (RDA) in Korea and the Bangladesh Jute Mills Corporation (BJC) (Dhaka, Bangladesh). These kenaf lines were collected from South Korea (1), United States of America (1), Iran (1), India (2), Russia (2), China (7), Italy (2), and Bangladesh (14). Eight white flower mutant genotypes were generated from selection breeding (natural variants). Four genotypes were developed from hybridization between ‘Jangdae’ and ‘Baekma’ (JangXbaek21, jangXbaek21a), ‘Jeokbong’ and ‘Baekma’ (jeokXbaek66), and ‘Jinju’ and ‘Baekma’ (jinjuXbaekma). The agronomic traits investigated were days to flowering, plant height, fresh weight, dry weight, flower color, stem color, and leaf shape. Genomic DNA was isolated from 20 mg of lyophilized leaf tissue using a DNeasy Plant Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol.
4.2. Sequence Pre-Processing and Alignment to Reference Genome
For demultiplexing, we used barcode sequences, adapter sequence trimming with cutadapt (version 1.8.3) [47], and sequence quality trimming with DynamicTrim and LengthSort programs from the SolexaQA (v.1.13) package [48]. DynamicTrim trims bad quality bases at both ends of short reads on the basis of their phred score and refines them into good-quality cleaned reads, while LengthSort removes reads with too many bases trimmed by DynamicTrim. DynamicTrim uses a phred score ≥ 20 and the LengthSort process uses a short read length ≥ 25 bp. The cleaned reads that passed the pre-processing were mapped to the reference genome using the BWA (0.7.17-r1188) program [49].
4.3. Raw SNP Detection and Generation of SNP Matrix
BAM-formatted files generated by mapping clean reads to the reference genome were used to perform SNP validation and raw SNP (In/Del) detection using the SAMtools (0.1.16) program [50] and SEEDERS own scripts [51], as well as to extract matching sequences. Default values were used except for the following options: minimum mapping quality for SNPs (−Q) = 30, minimum mapping quality for gaps (−q) = 15, minimum read depth (−d) = 3, maximum read depth (−D) = 398, minimum indel score for filtering nearby SNPs (−G) = 30, SNPs within INT bp around gaps to filter (−w) = 15, and window size for filtering high density SNPs (−W) = 15. We created a unified SNP matrix between samples to perform SNP comparison analysis between analytes. In this method, the raw SNP positions obtained by comparing each sample with the reference genome are used as candidate SNP positions to build a unified list, and the empty regions (non-SNP loci) are filled with the consensus sequence of the sample to create a matrix. The final SNP matrix is then created by filtering out mis-called SNP (In/Del) loci via SNP comparisons between samples. Among these loci, SNPs (In/Del) are classified according to the following type classification criteria: Homozygous, SNP read depth ≥ 90%; Heterozygous, 40% ≤ SNP read depth ≤ 60%; etc.: 20% ≤ read rate < 40% and 60% < read rate < 90%.
4.4. Linkage Disequilibrium Estimation
LD decay analysis with PopLDdecay program (https://github.com/BGI-shenzhen/PopLDdecay accessed on 5 December 2023) using a VCF file of 49,241 SNP loci from 96 kenaf samples. PopLDdecay Option was set as follows: Min minor allele frequency filter: 0.05, Max ratio of het allele filter: 0.6, and Max ratio of miss allele filter: 0.3. Using the calculated LD values, the LD decay plot was plotted using the HW method (the Hill and Weir method) [52] in R package.
4.5. Genome-Wide Association Study and Phylogenetic Analysis
To perform association analysis, we used an SNP filter process with the following conditions: SNP loci of biallelic, minor allele frequency > 5%, and missing data < 30%. The representative SNP loci and trait information selected via the filter process were used for association analysis using GAPIT [53]. The GWAS analysis was performed using a multiple mixed linear model. A total of 49,241 positions were used in the final dataset and the phylogenetic analysis was performed in MEGA6 [54]. Bootstrap consensus trees inferred from 1000 replicates were used to represent the evolutionary history of the analyzed taxa using the UPGMA method.
5. Conclusions
In this study, we performed phylogenetic and GWAS analyses using GBS data to investigate candidate genes for agronomic traits in 96 kenaf genotypes, including mutant lines derived from gamma irradiation. Gamma-irradiation-derived mutant lines were phylogenetically separated using wider genetic diversity compared with the original varieties. As a result of our GWAS analysis of agronomic traits, kenaf genes showing homology to known genes related to growth were discovered. The results of our study identified mutant genotypes and 19 candidate genes significantly associated with five traits of kenaf. This is the first study describing the SNPs generated using GBS and an association analysis to identify genes related to the large variability in morphological characteristics and yield-related traits among kenaf mutants derived from gamma-ray irradiation. The mutants obtained in this study will be useful genetic resources for the development of novel kenaf cultivars with improved ornamental value and chemical compound profiles. Our results may help breeders select kenaf mutants with suitable SNPs as optimal genotypes for the bioplastics and pharmaceutical industry. These findings highlight the value of mutant lines in genetic analyses and should prove useful for kenaf breeding programs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13020249/s1, Table S1: Country of origin for the 96 kenaf assessed in this study. Figure S1: Distribution of the marker density and decay of the linkage disequilibrium (LD) over distance presented as an accumulative distribution.
Author Contributions
Writing—original draft preparation, W.J.K.; methodology, B.Y., Y.-j.L. and S.-H.K.; writing—review and editing, J.H.K.; resources, S.H.K., J.-W.A. and S.-Y.K.; supervision, J.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the research program of KAERI, Republic of Korea (Project No. 523410-24) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (RS-2022-00156231).
Data Availability Statement
The original contribution presented in the study are publicly available.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Afzal, M.Z.; Ibrahim, A.K.; Xu, Y.; Niyitanga, S.; Li, Y.; Li, D.; Yang, X.; Zhang, L. Kenaf (Hibiscus cannabinus L.) breeding. J. Nat. Fibers 2022, 19, 4063–4081. [Google Scholar] [CrossRef]
- Webber, C.L., III; Bhardwaj, H.L.; Bledsoe, V.K. Kenaf production: Fiber, feed, and seed. Trends New Crops New Uses 2002, 13, 327–339. [Google Scholar]
- Dempsey, J.M. Fiber Crops; Univ. Presses of Florida: Gainesville, FL, USA, 1975. [Google Scholar]
- Scott, A. Kenaf Seed Production: 1981–82; Biennial Report for 1980; Rio Farms, Inc.: Monte Alto, TX, USA, 1981; pp. 60–63. [Google Scholar]
- Alexopoulou, E.; Papatheohari, Y.; Christou, M.; Monti, A. Origin, description, importance, and cultivation area of kenaf. In Kenaf: A Multi-Purpose Crop for Several Industrial Applications: New Insights from the Biokenaf Project; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–15. [Google Scholar]
- Khare, C.P. Indian Medicinal Plants: An Illustrated Dictionary; Springer Science & Business Media: Berlin, Germany, 2008. [Google Scholar]
- Taufiq, M.; Mansor, M.R.; Mustafa, Z. Characterisation of wood plastic composite manufactured from kenaf fibre reinforced recycled-unused plastic blend. Compos. Struct. 2018, 189, 510–515. [Google Scholar] [CrossRef]
- Ramesh, P.; Durga Prasad, B.; Narayana, K. Characterization of kenaf fiber and its composites: A review. J. Reinf. Plast. Compos. 2018, 37, 731–737. [Google Scholar] [CrossRef]
- Dryer, J. In Kenaf seed cultivars. In Proceedings of the First Conference on Kenaf for Pulp, Gainesville, FL, USA, 31 October–1 November 1967; pp. 44–46. [Google Scholar]
- Ha, B.-K.; Lee, K.J.; Velusamy, V.; Kim, J.-B.; Kim, S.H.; Ahn, J.-W.; Kang, S.-Y.; Kim, D.S. Improvement of soybean through radiation-induced mutation breeding techniques in Korea. Plant Genet. Resour. 2014, 12 (Suppl. S1), S54–S57. [Google Scholar] [CrossRef]
- Li, D.; Huang, S. The Breeding of Kenaf. In Kenaf: A Multi-Purpose Crop for Several Industrial Applications: New Insights from the Biokenaf Project; Springer: Berlin/Heidelberg, Germany, 2013; pp. 45–58. [Google Scholar]
- Lyu, J.I.; Choi, H.-I.; Ryu, J.; Kwon, S.-J.; Jo, Y.D.; Hong, M.J.; Kim, J.-B.; Ahn, J.-W.; Kang, S.-Y. Transcriptome analysis and identification of genes related to biosynthesis of anthocyanins and kaempferitrin in kenaf (Hibiscus cannabinus L.). J. Plant Biol. 2020, 63, 51–62. [Google Scholar] [CrossRef]
- Lyu, J.I.; Ryu, J.; Kim, D.-G.; Kim, J.M.; Ahn, J.-W.; Kwon, S.-J.; Kim, S.H.; Kang, S.-Y. Comparative Transcriptome Analysis Identified Potential Genes and Transcription Factors for Flower Coloration in Kenaf (Hibiscus cannabinus L.). Agronomy 2023, 13, 715. [Google Scholar] [CrossRef]
- Ryu, J.; Kwon, S.-J.; Ahn, J.; Kim, S.H.; Lee, S.Y.; Kim, J.-B.; Jo, Y.; Ha, B.-K.; Kang, S.-Y. Development of a stem-color mutant kenaf (Hibiscus cannabinus L.) cultivar, ‘Jeokbong’, and analysis of its functional compounds. Hortic. Sci. Technol. 2018, 36, 77–81. [Google Scholar] [CrossRef]
- Kim, D.-G.; Ryu, J.; Lee, M.-K.; Kim, J.M.; Ahn, J.-W.; Kim, J.-B.; Kang, S.-Y.; Bae, C.-H.; Kwon, S.-J. Nutritional properties of various tissues from new kenaf cultivars. J. Crop Sci. Biotechnol. 2018, 21, 229–239. [Google Scholar] [CrossRef]
- Kang, S.; Shin, I.; Kim, D.; Lee, G.; Kim, J.; Lee, D.; Lee, S.; Lee, D. A new green-kerneled glutinous rice mutant variety, “Nogwonchalbyeo” developed by gamma ray irradiation. Korean J. Breed. Sci. 2008, 40, 303–307. [Google Scholar]
- Lu, L.; Zhai, X.; Li, X.; Wang, S.; Zhang, L.; Wang, L.; Jin, X.; Liang, L.; Deng, Z.; Li, Z. Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains. Nat. Commun. 2022, 13, 4672. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, S.; Lu, X.; Guo, H.; Chen, S.; Yin, X.; Li, H.; Dai, G.; Liu, L. Integrating Genomics and Metabolomics for the Targeted Discovery of New Cyclopeptides with Antifungal Activity from a Marine-Derived Fungus Beauveria felina. J. Agric. Food Chem. 2023, 71, 9782–9795. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zaman, W.; Lu, J.; Niu, Q.; Zhang, X.; Ayaz, A.; Saqib, S.; Yang, B.; Zhang, J.; Zhao, H. Natural lupeol level variation among castor accessions and the upregulation of lupeol synthesis in response to light. Ind. Crops Prod. 2023, 192, 116090. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Yang, B.; Kim, W.J.; Kim, J.; Kwon, S.-J.; Kim, J.H.; Ahn, J.-W.; Kim, S.H.; Rha, E.-S.; Ha, B.-K. Genome-Wide Association Study (GWAS) of the Agronomic Traits and Phenolic Content in Sorghum (Sorghum bicolor L.) Genotypes. Agronomy 2023, 13, 1449. [Google Scholar] [CrossRef]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child. -Educ. Pract. 2013, 98, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Poland, J.A.; Rife, T.W. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 2012, 5, 92–101. [Google Scholar] [CrossRef]
- Deschamps, S.; Llaca, V.; May, G.D. Genotyping-by-sequencing in plants. Biology 2012, 1, 460–483. [Google Scholar] [CrossRef]
- Josephs, E.B.; Stinchcombe, J.R.; Wright, S.I. What can genome-wide association studies tell us about the evolutionary forces maintaining genetic variation for quantitative traits? New Phytol. 2017, 214, 21–33. [Google Scholar] [CrossRef]
- Ayaz, A.; Huang, H.; Zheng, M.; Zaman, W.; Li, D.; Saqib, S.; Zhao, H.; Lü, S. Molecular cloning and functional analysis of GmLACS2-3 reveals its involvement in cutin and suberin biosynthesis along with abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 9175. [Google Scholar] [CrossRef]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef]
- Hwang, S.-G.; Lee, S.C.; Lee, J.; Lee, J.W.; Kim, J.-H.; Choi, S.Y.; Kim, J.-B.; Choi, H.-I.; Jang, C.S. Resequencing of a core rice mutant population induced by gamma-ray irradiation and its application in a genome-wide association study. J. Plant Biol. 2020, 63, 463–472. [Google Scholar] [CrossRef]
- Kim, W.J.; Kang, B.H.; Kang, S.; Shin, S.; Chowdhury, S.; Jeong, S.-C.; Choi, M.-S.; Park, S.-K.; Moon, J.-K.; Ryu, J. A Genome-Wide Association Study of Protein, Oil, and Amino Acid Content in Wild Soybean (Glycine soja). Plants 2023, 12, 1665. [Google Scholar] [CrossRef]
- Tian, F.; Bradbury, P.J.; Brown, P.J.; Hung, H.; Sun, Q.; Flint-Garcia, S.; Rocheford, T.R.; McMullen, M.D.; Holland, J.B.; Buckler, E.S. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011, 43, 159–162. [Google Scholar] [CrossRef]
- Clayton, D.G.; Walker, N.M.; Smyth, D.J.; Pask, R.; Cooper, J.D.; Maier, L.M.; Smink, L.J.; Lam, A.C.; Ovington, N.R.; Stevens, H.E. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 2005, 37, 1243–1246. [Google Scholar] [CrossRef]
- Korte, A.; Vilhjálmsson, B.J.; Segura, V.; Platt, A.; Long, Q.; Nordborg, M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 2012, 44, 1066–1071. [Google Scholar] [CrossRef]
- Brachi, B.; Morris, G.P.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, L.; Wu, J.; Qi, J.; Zhou, R. Genetic effect analysis of some field and quality traits of kenaf hybrid and parents. Plant Fibers Prod. 2004, 26, 261–266. [Google Scholar]
- Cheng, Z.; Lu, B.-R.; Sameshima, K.; Fu, D.-X.; Chen, J.-K. Identification and genetic relationships of kenaf (Hibiscus cannabinus L.) germplasm revealed by AFLP analysis. Genet. Resour. Crop Evol. 2004, 51, 393–401. [Google Scholar] [CrossRef]
- Kang, S.; Kwon, S.; Jeong, S.; Kim, J.; Kim, S.; Ryu, J. An improved kenaf cultivar ‘Jangdae’ with seed harvesting in Korea. Korean J. Breed. Sci. 2016, 48, 349–354. [Google Scholar] [CrossRef]
- Ryu, J.; Kwon, S.-J.; Kim, D.-G.; Lee, M.-K.; Kim, J.M.; Jo, Y.D.; Kim, S.H.; Jeong, S.W.; Kang, K.-Y.; Kim, S.W. Morphological characteristics, chemical and genetic diversity of kenaf (Hibiscus cannabinus L.) genotypes. J. Plant Biotechnol. 2017, 44, 416–430. [Google Scholar] [CrossRef]
- Spencer-Lopes, M.; Forster, B.P.; Jankuloski, L. Manual on Mutation Breeding; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2018. [Google Scholar]
- Lyu, J.I.; Ramekar, R.; Kim, D.-G.; Kim, J.M.; Lee, M.-K.; Hung, N.N.; Kim, J.-B.; Ahn, J.-W.; Kang, S.-Y.; Choi, I.-Y. Characterization of gene isoforms related to cellulose and lignin biosynthesis in Kenaf (Hibiscus cannabinus L.) mutant. Plants 2020, 9, 631. [Google Scholar] [CrossRef]
- Ryu, J.; Kwon, S.-J.; Sung, S.Y.; Kim, W.-J.; Kim, D.S.; Ahn, J.-W.; Kim, J.-B.; Kim, S.H.; Ha, B.-K.; Kang, S.-Y. Molecular cloning, characterization, and expression analysis of lignin biosynthesis genes from kenaf (Hibiscus cannabinus L.). Genes Genom. 2016, 38, 59–67. [Google Scholar] [CrossRef]
- Lease, K.A.; Lau, N.Y.; Schuster, R.A.; Torii, K.U.; Walker, J.C. Receptor serine/threonine protein kinases in signalling: Analysis of the erecta receptor-like kinase of Arabidopsis thaliana. New Phytol. 2001, 151, 133–143. [Google Scholar] [CrossRef]
- Lin, F.; Li, S.; Wang, K.; Tian, H.; Gao, J.; Zhao, Q.; Du, C. A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 2020, 296, 110465. [Google Scholar] [CrossRef]
- Liu, P.-L.; Du, L.; Huang, Y.; Gao, S.-M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef]
- Cao, Y.; Dai, Y.; Cui, S.; Ma, L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 2008, 20, 2586–2602. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Chen, Q.; Song, Y.; Liu, K.; Su, C.; Yu, R.; Li, Y.; Yang, Y.; Zhou, B.; Wang, J.; Hu, G. Genome-Wide Identification and Functional Characterization of FAR1-RELATED SEQUENCE (FRS) Family Members in Potato (Solanum tuberosum). Plants 2023, 12, 2575. [Google Scholar] [CrossRef]
- Ma, L.; Li, G. FAR1-related sequence (FRS) and FRS-related factor (FRF) family proteins in Arabidopsis growth and development. Front. Plant Sci. 2018, 9, 692. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-E.; Oh, S.-K.; Lee, J.-H.; Lee, B.-M.; Jo, S.-H. Genome-wide SNP calling using next generation sequencing data in tomato. Mol. Cells 2014, 37, 36. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.; Weir, B. Variances and covariances of squared linkage disequilibria in finite populations. Theor. Popul. Biol. 1988, 33, 54–78. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).