Abstract
Cymbidium ensifolium, a prominent orchid species, is both highly valued for its ornamental qualities and commercially cultivated. However, the species has a considerable challenge in its breeding efforts due to the lengthy period of 7–8 years required for it to transition from seed germination to flowering. BBXs are multifunctional proteins that modulate the actions of critical regulators including HY5 and COP1 in response to blue light, ultimately impacting photomorphogenic processes. In this study, BBX proteins, known for their essential roles in regulating developmental processes under various light conditions, were chosen as the main subject of investigation. The outcome reveals the presence of 19 BBX genes in their genome. The genes are classified into four separate clades and dispersed among 12 out of the 20 chromosomes. Located in the nuclear, physicochemical properties of proteins, analysis of the promoter region reveals the existence of almost 800 cis-acting elements, highlighting the complex regulatory mechanisms that control the expression of the CeBBXs in various organs, as well as their response to light and hormone inputs. Moreover, the examination of differential expression under blue light therapy reveals their involvement in photomorphogenic reactions. The expression of CeBBXs exhibits substantial alterations as the duration of exposure to blue light increases. These findings contribute to a deeper understanding of the roles that BBX genes serve in C. ensifolium, providing a basis for future studies on the functions and regulatory mechanisms of BBX members in the context of floral initiation and development within this species.
1. Introduction
Unlike conventional artificial lighting systems, LED technology has evident strengths, such as a long lifespan, compact size, and higher photosynthetic efficiency. Consequently, LED technology has been extensively used in growing different agricultural crops under controlled conditions [1,2]. Recent studies have reported the use of different combinations of LED light spectra to regulate flowering times in horticultural crops, such as strawberries [3], Chrysanthemum spp. [4], Petunia spp. [5], and Dahlia spp. [6]. Research on strawberries has demonstrated that blue LED irradiation significantly promotes flowering [7,8]. Jeong et al. found that chrysanthemums treated with blue light as a supplementary light source formed flower buds 20.5 days after treatment. In contrast, flower buds could not be induced by white light treatment for 35 days [9]. These findings indicate that blue light-induced flowering in short-day plants can override the photoperiodic constraints on plant flowering regulation [8]. The flowering period is considered a critical agricultural characteristic because of the importance of floral features in ornamental horticulture. Furthermore, the utilization of optimized light treatments can enhance the process of plant hybridization and breeding, leading to an expedited flowering period, thereby improving breeding efficiency and the development of high-performing varieties. A comprehensive investigation has been carried out on the photoperiodic flowering pathway in plants. Accumulating data suggest that BBX proteins strongly influence the management of flowering time in Arabidopsis thaliana through photoperiodic mechanisms.
Zinc finger proteins (ZFPs) are abundant protein groups found in eukaryotes and have a vital function in the growth and development of plants [10]. A zinc finger is defined by the presence of cysteine and histidine residues. These residues coordinate around a zinc ion, resulting in the formation of a stable three-dimensional structure [11,12]. The BBX family is distinguished by the presence of the B-box domain. This domain, composed of around 40 amino acids, facilitates the creation of heterodimers with other members of the BBX family or different proteins, hence playing a crucial function in the control of transcription [13]. The B-box domain was initially identified in the Xlxnf7 protein of the African clawed frog (Xenopus laevis) and has since been found in numerous BBX transcription factors in plants [14]. In contrast to animals, certain components from the BBX family in plants exhibit a remarkably preserved CCT (Constans, CO-like, and TOC1) as an area of focus [15], consisting of 42–43 amino acid residues, which has a role in controlling the transcription of BBX proteins and the movement of molecules between the nucleus and cytoplasm [16]. Furthermore, certain BBX proteins possess a Valine–Proline (VP) motif at their C-terminal end, which is involved in interactions with coiled-coil proteins [12,17].
AtCO was the first B-box protein to be cloned and identified. The modulation of AtCO gene expression and the stability of its protein are pivotal for the photoperiodic cues that trigger flowering, which, in turn, initiates the expression of the downstream AtFT gene [18,19]. BBX4, BBX21, BBX22, and BBX23 are recognized for their ability to enhance photomorphogenesis [20,21,22,23,24]. Conversely, BBX24, BBX25, BBX28, BBX30, BBX31, and BBX32 function as inhibitors of light signaling [22,25,26,27,28]. In Cymbidium ensifolium, CRY2 can control cotyledon expansion and flowering time. AtCRY2 has been shown to stimulate the expression of AtFT in response to blue light by inhibiting the degradation of the AtCO protein [29,30]. Under long-day photoperiods, mutants of the AtCO gene exhibit a significant delay in flowering time, whereas AtCO overexpression lines show an early flowering phenotype [31]. In AtBBX7 overexpression lines, the transcription levels of AtCO and AtFT are inhibited under long-day conditions, resulting in delayed flowering, indicating that AtBBX7′s regulation of flowering time in depends on the AtCO gene [32]. Both AtBBX4 and AtBBX32 are negative regulators of flowering in Arabidopsis thaliana; the overexpression of AtBBX4 and AtBBX32 reduces AtFT expression levels and delays flowering under long-day photoperiods. AtBBX4 loss-of-function mutants exhibit an accelerated flowering process under both long-day and short-day photoperiods, but the AtBBX32 mutants do not demonstrate substantial variations in flowering time compared to the wild type under long-day circumstances [20,33]. Simultaneously, a growing body of research revealed the precise roles of BBX family members in different plant species, discovering BBX genes that concern the blooming time. Recently, researchers discovered nine individuals belonging to the BBX family that have a vital function in controlling the timing of blooming in rice [34]. Ye et al. identified FaBBX29 as a significant contributor to the regulation of blooming [35]. Therefore, BBX proteins play a pivotal role in regulating the timing of flowering.
Cymbidium ensifolium is one of the most significant ornamental and commercial orchids in China, renowned for its graceful form, exquisite appearance, and fragrant aroma [36]. However, the growth cycle of this established orchid is long; it takes 7–8 years from seed to flowering under natural conditions, and it requires at least 10–12 years to breed new varieties with superior traits through cross-selection [37]. The extended breeding cycle of C. ensifolium significantly hampers the development and industrialization of new cultivars. While existing research indicates that blue light can be utilized to manipulate flowering time, and the BBX gene family is pivotal in modulating plants’ responses to blue light and their flowering time [35], its role in orchids remains unexplored. Consequently, identifying and elucidating the potential functions of BBX genes in orchids is of considerable importance. The objective of this study is to identify and characterize BBX family genes using genomic data from C. ensifolium. This includes analyzing their physicochemical properties, subcellular localization, gene structure, conserved domains, collinearity, evolutionary relationships, promoter cis-elements, and expression profiles. The results of this study provide new insights into the biological roles of BBX proteins in C. ensifolium and establish a foundation for future investigations into the connection between the blue light signaling pathway and BBX proteins. This could potentially shorten the breeding cycle, thereby enhancing the breeding efficiency and commercial viability of new orchid varieties.
2. Materials and Methods
2.1. Identification and Tertiary Structures of BBX Genes
With the aim of uncovering possible BBX genes in C. ensifolium, we acquired the BBX protein sequences of both C. ensifolium and rice from TAIR (https://www.arabidopsis.org/, accessed on 25 June 2024) [38] and RGAP (http://rice.plantbiology.msu.edu/, accessed on 25 June 2024) [39], respectively. The genomes of C. ensifolium were obtained by downloading the whole-genome sequencing data [40]. Initially, a BLASTp conduct was realized with the help of TBTools (http://cj-chen.github.io/tbtools, accessed on 25 June 2024), a software package that integrates multiple techniques and provides a handy approach for biologists [41]. The BBX domain (PF00643), which is conserved, was obtained according to the Pfam to conduct an HMMER survey with TBtools (http://pfam.xfam.org/, accessed on 25 June 2024) [41]. To further analyze the potential BBX proteins, the results from BLAST and Hmmsearch were subjected to examination (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 25 June 2024). Ultimately, proteins that possessed the entire BBX domain were selected and kept for further examination of their sequence (Table S1). The ExPASy proteomics server (https://www.expasy.org/, accessed on 25 June 2024)) was utilized to forecast the weight, quantization of protein unsteadiness, and hydropathicity index of the CeBBX proteins. Additionally, their position was forecasted [42,43]. In addition, to predict the tertiary protein structure of CeBBX, AlphaFold2.3.2 was utilized (https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb, accessed on 25 June 2024). The SOMPA was employed when the width of output was configured to 70, and conformational status was established at 4 https://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html, accessed on 26 June 2024) [44].
2.2. Examination of the Exon–Intron Architecture and Identification
The conserved motifs of the CeBBX proteins were validated through the utilization of the MEME program (http://meme-suite.org/tools/meme, accessed on 25 June 2024) [45], while the conserved domain information was collected from NCBI. The exon–intron structure was examined using the genome-wide GFF annotation file, and visualization was performed using TBtools software [41].
2.3. Phylogenetic Analysis and Cis-Element Identification
The BBX proteins found in C. ensifolium and rice were utilized to categorize the BBX proteins specifically found in C. ensifolium. The Clustalx program was utilized for conducting diverse sequence correspondence of the entire set of BBX proteins [46]. The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with a thousand bootstrap replications, utilizing MEGA6.0 software [47]. Afterward, the tree underwent additional modifications using iTOL (https://itol.embl.de, accessed on 25 June 2024) [48]. The sequences located 2000 base pairs upstream of the transcriptional start sites and were designated as the proximal promoter region sequences. The promoter sequences of the CeBBX genes were obtained from the C. ensifolium Genomics Database. The promoter sequences were analyzed for cis-elements by submitting them to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 25 June 2024) [49].
2.4. Analyzing the Chromosome Position, Duplication Events, and Synteny of the CeBBX Gene in C. ensifolium
The genomes of C. goeringii were obtained by downloading its whole-genome sequencing data [50]. TBTools software was utilized to determine the positions of the CeBBX members on the chromosomes of C. ensifolium. Additionally, it was employed to visually represent the gene duplications of CeBBX genes and highlight regions that exhibit homology [41].
2.5. Plant Materials and Treatments
The plant materials utilized in this investigation were somaclones of C. ensifolium acquired from the Fujian Agriculture and Forestry University. Seedlings with three leaves were inoculated in a culture medium and subjected to different light treatments. The experimental plants were exposed to blue light, whereas the control group was subjected to white light. Leaf samples were collected at four specific time intervals: 0, 1, 3, 5, 7, and 15 days following the treatment. Three separate biological replicates were included in each sample collection to confirm the data’s trustworthiness. The specimens were swiftly frozen and preserved at a temperature of −80 °C for later extraction of RNA.
2.6. Extraction of RNA and Analysis Using Real-Time Quantitative PCR (RT-qPCR)
The FastPure® Plant Total RNA Isolation Kit, designed for samples high in polysaccharides and polyphenolics, was sourced from Vazyme in Nanjing, China, for the purpose of extracting RNA. The concentration of RNA was measured with a Nanodrop 2000 spectrophotometer, and it was evaluated through agarose gel electrophoresis. For the conversion of RNA to complementary DNA (cDNA), the HiScript III 1st Strand cDNA Synthesis Kit, which includes a gDNA removal option, was also procured from Vazyme. The primers necessary for RT-qPCR were designed using the Primer3Plus online tool, with the actin gene serving as a control (Table S1). The RT-qPCR reactions were conducted with the Taq Pro Universal SYBR qPCR Master Mix Kit manufactured by Vazyme, located in Nanjing, China. The expression levels were determined using the 2−∆∆Ct method [51].
2.7. Subcellular Localization of BBX Proteins
In order to ascertain the subcellular distribution of CeBBX in plant cells, the complete sequences of CeBBX18 were integrated into the pCAMBIA1302-GFP vector. The recombinant vector was subsequently inserted into the Agrobacterium tumefaciens strain GV3101 (ANGYUBio, Fuzhou, China). Undamaged tobacco leaves (Nicotiana benthamiana) were chosen and infused with A. tumefaciens carrying the modified vector. The tobacco plants were placed in a dark environment and kept at a temperature of 25 °C for a duration of 12 h. Fluorescence images of GFP were acquired using a confocal scanning microscope called Axio-Imager_LSM-800, manufactured by Zeiss in Oberkochen, Germany.
3. Results
3.1. Identification and Characterization of BBX Proteins in C. ensifolium
A BLASTP search was conducted on the C. ensifolium genome database using BBX protein sequences from the same species to identify BBX family members. After eliminating duplicate sequences, 19 BBX genes were identified and designated as CeBBX1 to CeBBX19 based on their gene IDs in ascending order (Figure 1 and Table S1). The result shows detailed information for each CeBBX. The characteristics assessed included the count of amino acids, theoretical isoelectric point, and the average of GRAVY. The CeBBX proteins exhibit significant variation in size, with amino acid counts ranging from 189 (CeBBX10) to 587 (CeBBX15). Correspondingly, their molecular weights range from 20,281.83 Da (CeBBX10) to 64,770.75 Da (CeBBX15). The theoretical pI values span from 4.87 (CeBBX1) to 8.01 (CeBBX13), with most CeBBX proteins having a pI below 7, indicating they are predominantly acidic. Proteins with higher pI values, such as CeBBX10 and CeBBX13, are more basic. The instability index varies from 40.06 (CeBBX5) to 73.86 (CeBBX11), suggesting that many CeBBX proteins may be prone to rapid degradation or require stabilization through interactions with other cellular components. The aliphatic index ranges from 59.42 (CeBBX13) to 86.79 (CeBBX4), with higher values suggesting greater thermostability. The GRAVY values range from −0.755 (CeBBX13) to −0.11 (CeBBX4), emphasizing that the proteins are generally hydrophilic. Subcellular localization analysis reveals that all CeBBX members are localized in the nucleus. Additionally, transient expression of a recombinant CeBBX-GFP protein in tobacco epidermal cells shows that CeBBX18 is confined to the nucleus, suggesting that CeBBX proteins are involved in the regulation of nuclear transcription, consistent with their predicted subcellular localization.
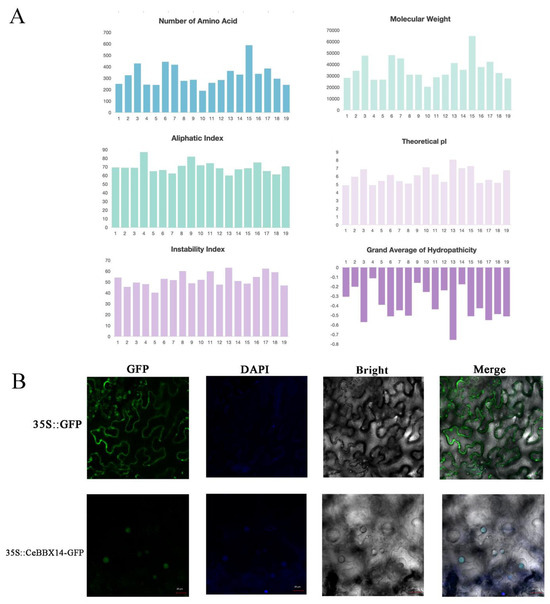
Figure 1.
Identification and characterization of BBX proteins in C. ensifolium. (A) Physicochemical properties of 19 CeBBX proteins; (B) Subcellular localization of CeBBX14. The images, arranged from left to right, depict the green fluorescent protein (GFP), the DAPI field (representing nuclear staining), the bright field, and an overlay of GFP, DAPI, and brilliant field from the same sample. Scale bar = 20 μm.
3.2. Gene Structures, Conserved Domains, and Motif Analysis
Our findings display the conserved patterns found in 19 CeBBX proteins (Figure 2A). BBX genes with comparable gene architectures and identical conserved motifs often group in the evolutionary tree. Among these 19 BBX genes, 15 conserved motifs have been identified. Notably, CeBBX6 and CeBBX3 each contain the highest number of conserved motifs, with seven. Motifs 1, 2, and 4 are notably recurrent across various CeBBX proteins, suggesting their pivotal roles in the proteins’ functions. CeBBX15, in particular, displays a diverse array of motifs, including Motif 5 and Motif 10, potentially endowing this protein with unique functional properties. Additionally, a total of 10 BBX genes possess a CCT domain. Comprehensive gene structure analysis reveals that all 19 BBX genes contain between one and four introns. Of these, CeBBX18 has the longest intron, followed by CeBBX6 and CeBBX16. The gene CeBBX15 is marked by a complex arrangement of multiple exons. Conversely, CeBBX14 has a simplified gene structure with fewer introns. CeBBX6 and CeBBX7 feature extended untranslated regions (UTRs), which might influence their mRNA stability and translation efficiency.
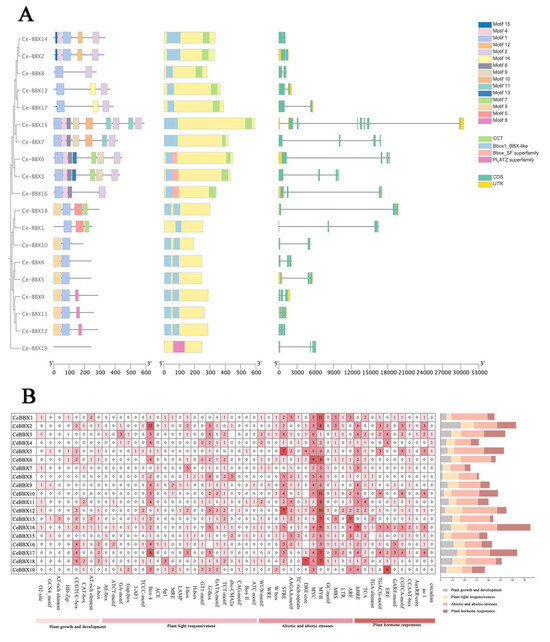
Figure 2.
(A) Gene structures, conserved domains, and motif analysis; (B) Cis-element analysis of CeBBXs.
3.3. Cis-Element Analysis of CeBBXs
A thorough investigation of the promoter regions of the CeBBX gene family in plants was undertaken using the PlantCARE database (Figure 2B) to better understand their regulatory complexity and environmental responsiveness. A total of 800 cis-acting elements were discovered in the promoters of these genes, highlighting the intricate control of gene expression tailored to diverse environmental cues and internal physiological states. The distribution of these elements includes 230 light response elements, showcasing the significant influence of light on the gene expression of this family. Hormonal regulation is also profoundly represented by 202 hormone response elements, underscoring the sensitivity of these genes to hormonal signals. These include a range of specific elements, such as abscisic acid response elements (ABREs), gibberellin response elements (GARE-motifs), and auxin-responsive elements (AuxRR-cores), among others. Notably, salicylic acid response elements and jasmonic acid-responsive elements (MeJA-responsive elements), like the CGTCA motif and TGACG motif, were prevalent. This indicates a strong correlation between CeBBX gene expression and pathways involved in stress and defense mechanisms. In addition, 373 elements related to both abiotic and biotic challenges demonstrate the involvement of CeBBX genes in enhancing plant resistance to environmental pressures. Another 69 elements related to general growth and developmental processes emphasize the involvement of CeBBX genes in various aspects of plant development. Particularly, CeBBX2, CeBBX14, and CeBBX19 had a high concentration of elements related to both light and hormones, suggesting their pivotal roles in light responses mediated by these hormones. CeBBX14, enriched with both light-responsive elements and ABREs, may indicate a dual regulatory mechanism influenced by light and abscisic acid, possibly coordinating photosynthesis with water stress responses. This detailed mapping and categorization of cis-acting elements reveal not only the potential functional roles of each CeBBX gene in response to environmental and developmental signals but also suggest a complex network of gene regulation that allows plants to adapt to their ever-changing environment.
3.4. Phylogenetic Analysis and Tertiary Structure Analysis of CeBBXs
Phylogenetic trees were created using the neighbor-joining method to investigate the evolutionary relationships and classification of the BBX gene family. The analysis included species such as C. ensifolium, A. thaliana, and O. sativa. (Figure 3A). The analysis identified 19 CeBBX proteins, 32 AtBBX proteins, and 30 OsBBX proteins, which were categorized into five distinct groups (I–V). Among these, Group II contained the highest number of members, including seven CeBBX proteins. This was followed by Group III with 26 members, Group I with 16 members, Group V with 8 members, and Group IV with 4 members. The congruence between the phylogenetic tree and the clustering results of prior investigations affirms the precision and dependability of the phylogenetic tree [52]. Interestingly, no CeBBX members of C. ensifolium were found in subfamily IV. Meanwhile, BBX proteins from C. ensifolium, Arabidopsis, and rice subfamilies were found to be closely related, indicating a high level of evolutionary conservation within the BBX family and suggesting that they may have comparable biological activities. The phylogenetic tree showed that the BBX protein of C. ensifolium was more closely related to its homology in rice than to its homology in Arabidopsis. The subsequent analysis provides a detailed prediction and examination of the structural prediction of secondary proteins (Table S2) and the tertiary structure of each branch of BBX proteins in C. ensifolium (Figure 3B). The proteins from different branches displayed notable variations in the amounts of alpha helices, extended strands, beta twists, and random coils. Additionally, proteins that clustered together in the evolutionary tree showed similar tertiary structures. The analysis reveals that the numbers of α-helices and β-strands in CeBBX6 and CeBBX16 are greater than those observed in proteins from other classes. CeBBX18 is characterized by a notable number of α-helices and a minimal presence of visible β-strands, suggesting a helix-dominated configuration. Furthermore, the variations in length between CeBBX11 and other Class III proteins could perhaps explain the disparities in their tertiary structures.
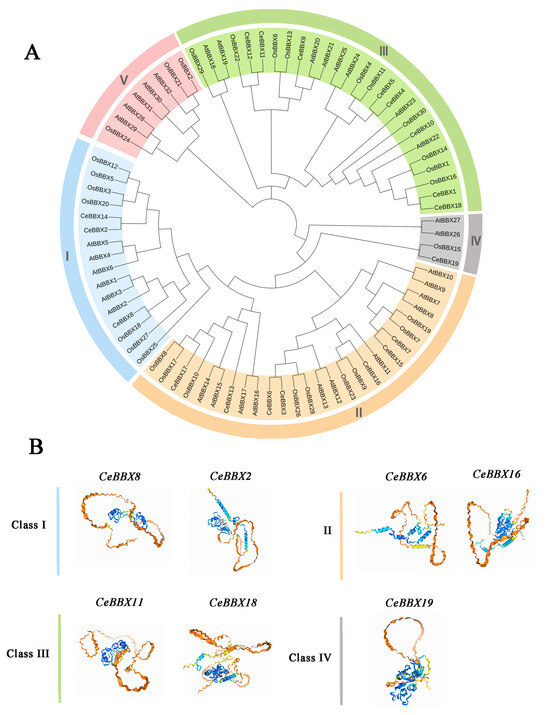
Figure 3.
(A) Phylogenetic trees of BBX gene family in Cymbidium ensifolium, Arabidopsis thaliana, and Oryza sativa; (B) Anticipation of the three-dimensional structure for CeBBX proteins was analyzed. The proteins were categorized into Class I, II, III, and IV. The confidence levels of the protein structures are represented by lines of varying colors and red circles, with the confidence increasing from blue to orange. (Blue: pIDDT > 90; light blue: 90 > pIDDT > 70; yellow: 70 > pIDDT > 50; orange: pIDDT < 50).
3.5. Chromosomal Location and Gene Duplication of CeBBX Gene
An investigation was conducted to determine the specific locations of the 19 CeBBX genes on the chromosomes of the C. ensifolium genome in order to understand how the BBX genes are distributed within the genome (Figure 4A). The 19 CeBBX genes were distributed across 12 out of the 20 C. ensifolium chromosomes, accounting for approximately 60% of the total chromosomes. Notably, chromosome Chr20 harbored the highest number of CeBBX genes, with a total of three. This was followed by chromosomes Chr03, Chr04, Chr07, Chr08, and Chr10, each of which contained two CeBBX genes. The remaining six chromosomes each housed a single CeBBX gene. The overall distribution of CeBBX genes across the C. ensifolium chromosomes was relatively even despite some variability in gene density.
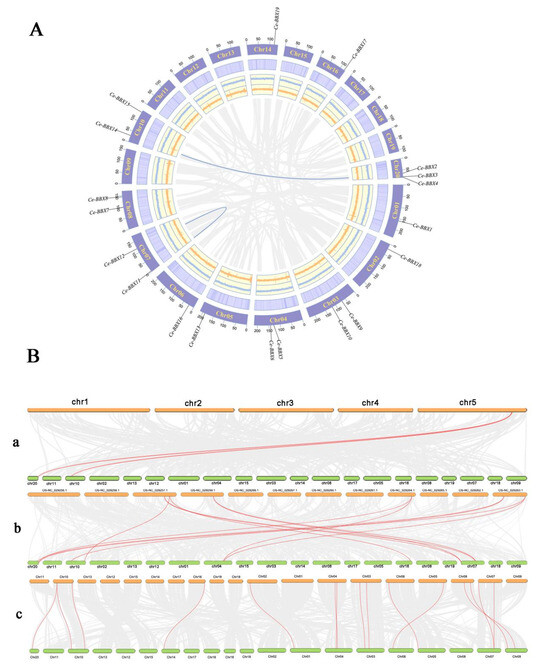
Figure 4.
(A) Chromosomal location and gene duplication of CeBBX gene; (B) Chromosomal localization of 19 CeBBX genes. Collinearity analysis of Cymbidium ensifolium with Arabidopsis thaliana (a), Oryza sativa (b), and Cymbidium goeringii (c).
Gene duplication events are essential for the proliferation of gene families and are a prevalent feature of plant evolution [37]. In the C. ensifolium genome, nineteen CeBBX genes were identified as participating in two segmental duplication events, specifically CeBBX2/CeBBX14 and CeBBX11/CeBBX12 (Figure 4A). All instances of duplicated genes were classified together in the same clade of the evolutionary tree, highlighting gene duplication as the main mechanism responsible for the growth of the CeBBX gene family. In order to obtain a more profound comprehension of the phylogenetic and evolutionary patterns of the CeBBX gene family, a comparative synteny study was performed on C. ensifolium and three other species: A. thaliana, O. sativa (rice), and Cymbidium goeringii (Figure 4B). The analysis reveals homologous gene pair counts of 2, 11, and 15 with C. ensifolium, rice, and C. goeringii, respectively. Notably, the monocotyledonous plant C. ensifolium exhibited a greater number of homologous gene pairs with rice and C. goeringii compared to the dicotyledonous plant A. thaliana, indicating closer phylogenetic relationships within the monocots. Furthermore, several CeBBX genes demonstrated homology with multiple genes in the aforementioned species, suggesting that these genes may play a crucial role in the diversification and functional expansion of the BBX gene family. This comparative genomic approach highlights significant evolutionary relationships and mechanisms that might contribute to the adaptive diversity observed in plant species.
3.6. Expression Characteristics of CeBBX in C. ensifolium under Different Lights
In the initial phase of this research, transcriptome data were collected for various organs of C. ensifolium. These included several vegetative parts, such as leaves, roots, and pseudobulbs, along with different segments of the reproductive structures, including sepals, petals, labellum, and gynandriums. These data enabled us to perform a comprehensive analysis of the transcriptome to assess the expression levels of the CeBBX genes. The investigations focused on the expression patterns of 18 BBX genes in C. ensifolium (Figure 5). Among these, CeBBX9 and CeBBX12 were characterized by low expression levels across all samples, with numerous instances of non-expression. Conversely, CeBBX2 and CeBBX14 showed significantly elevated expression levels relative to their counterparts. The expression of CeBBX genes exhibited notable tissue-specific patterns, with certain genes presenting prominent expression in specific tissues. Specifically, CeBBX10 was highly expressed solely in flowers, while CeBBX8 and CeBBX13 were predominantly expressed in leaves. In contrast, CeBBX11 was uniquely expressed in pseudobulbs. Additionally, the expression dynamics of the BBX gene family revealed a distinct pattern throughout the flowering phase of C. ensifolium. The expression levels of CeBBX2, CeBBX3, CeBBX10, CeBBX16, and CeBBX18 showed a gradual decline as the flowers aged, whereas the expression levels of CeBBX12 and CeBBX17 reached their peak during the decay phase.
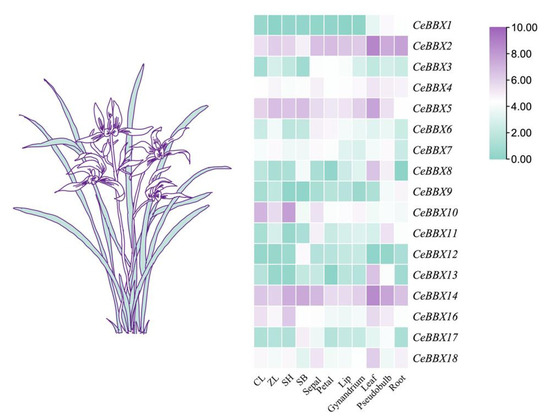
Figure 5.
Heatmap of the expression patterns of the BBX gene family in wild C. ensifolium with different organs and different periods. Note: CL: initial bud period (1–5 mm); ZL: middle bud period (6–10 mm); SH: Full flowering period; SB: Flower decay period.
3.7. Expression Profiles of Cymbidium ensifolium BBX Genes with Blue Light Treatment
Seven CeBBX genes (CeBBX2, CeBBX4, CeBBX5, CeBBX9, CeBBX14, CeBBX16, and CeBBX18) were selected based on their positions in the phylogenetic tree and their relatively high expression levels in leaves. Leaf samples were obtained at six different time points, 0, 1, 3, 5, 7, and 15 days following exposure to blue light. The relative expression levels of these seven CeBBX genes before and after treatment at different time points were analyzed using RT-qPCR (Figure 6). The results indicate that the expression levels of CeBBX4, CeBBX14, CeBBX16, and CeBBX18 significantly decreased within the first five days, with all genes exhibiting lower expression levels compared to the control group. However, expression levels increased with prolonged blue light exposure. On the 15th day, the expression levels of CeBBX2 and CeBBX18 remained lower than those of the control group, while the expression levels of CeBBX9 and CeBBX16 were approximately the same as those of the control group, consistent with the transcriptome data. Furthermore, the expression level of CeBBX5 was elevated compared to the control group on the 15th day, while the expression level of CeBBX14 was notably reduced. Given that blue light influences plant morphogenesis and photoperiod, it is plausible that these changes in BBX gene expression are a direct response to blue light signaling, thereby regulating related biological processes.
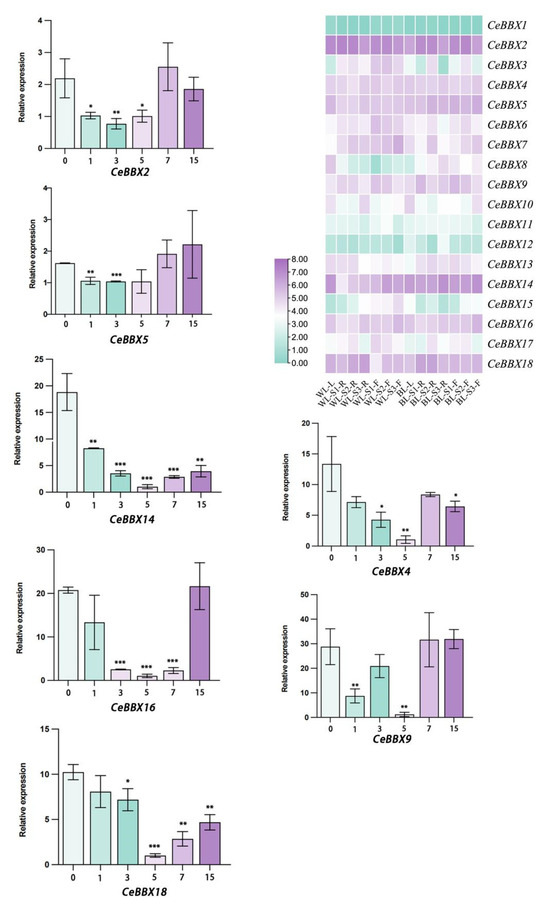
Figure 6.
Seven CeBBX genes were analyzed using RT-qPCR. The transcriptome data of these genes were collected throughout four distinct growth stages of C. ensifolium under both white and blue light treatments. WL: white light, BL: blue light, F: flower, R: rhizome, L: leaf, S1: the flower bud phase, S2: the complete flowering time, and S3: the period when the flowers wither. The color scale indicates the logarithm base 2 of the normalized counts per million kilobases of reading. The data shown are the means ± standard error (SE) of three independent measurements. The statistical significance levels are represented as follows: a p-value less than 0.05 is designated by *, a p-value less than 0.01 is denoted by **, and a p-value less than 0.001 is denoted by ***.
4. Discussions
The BBX transcription factor family, ubiquitously present in plants, plays a vital part in numerous physiological processes, such as the control of blooming time, light signal transduction, and stress responses [53]. Over the past decade, advancements in genomic data and bioinformatics have facilitated the identification of BBX transcription factors across numerous species, with notable interspecies variation in the number of BBX family members [54,55]. In this study, we conducted a comprehensive analysis of the BBX gene family in the genome of C. ensifolium, identifying a total of 19 BBX members (Table S1). Following the established classification framework for BBX genes in C. ensifolium, we grouped these genes into four distinct clades, with Group II showing the highest number of members. This suggests a possible expansion and functional diversification within this clade specific to C. ensifolium.
Gene duplication events are pivotal in the evolutionary dynamics of plant genomes, with both small-scale duplications (e.g., tandem gene duplication) and large-scale duplications (e.g., whole-genome and segmental duplications) driving these processes [56]. The relatively lower number of BBX genes in C. ensifolium compared to other species may be attributable to two whole genome duplication events within the orchid lineage [57]. Our findings suggest that both small-scale duplications and large-scale duplications have significantly influenced the evolution and diversification of the BBX gene family in C. ensifolium, reflecting evolutionary patterns observed in other plant species.
BBX genes play diverse functional roles in various biological processes within plants [34]. Mounting evidence suggests that the biological functions of BBX genes are strongly linked to their particular expression patterns. Gene expression is controlled by cis-regulatory elements located in promoter regions. These elements function as molecular switches that allow transcripts to be involved in intricate regulatory networks [58]. Our examination of cis-acting elements in the BBX promoters indicates a significant concentration of photoresponsive elements in the promoter regions of CeBBX genes. Additionally, plant hormone-responsive cis-elements were widely distributed, indicating that BBX gene expression is modulated not only by light but also by internal hormone levels, enabling the plant to respond to external environmental cues and developmental processes.
Additionally, differential expression analysis under blue light treatment underscored the functional diversity of CeBBX genes. CeBBX8 and CeBBX14 were significantly upregulated under blue light, while CeBBX3 and CeBBX15 were significantly downregulated, suggesting their involvement in the blue-mediated pathway and their potential roles in stress response and light morphogenesis. Interestingly, the upregulation of CeBBX14 in response to blue light was not observed in the fluorescence quantitative experiments, possibly due to circadian rhythm influences, which aligns with findings from FvCO studies [35]. Furthermore, CeBBX14 is a homologous protein to RoCO, which regulates the flowering time in roses, thereby emphasizing the evolutionary conservation and functional significance of BBX genes across species [59].
5. Conclusions
In this study, we discovered and examined the BBX gene family within the genome of C. ensifolium. Detailed investigations were conducted to elucidate the gene structures, conserved patterns, regulatory elements, chromosomal placements, and similarities with genes from different species. Our results highlight, under artificial control, the strong response of BBX genes to blue light during meristem induction in C. ensifolium. This study establishes a foundational framework for understanding the meristem induction of C. ensifolium by artificial light control, contributing to a broader comprehension of gene functions in orchids. These investigations will provide deeper insights into their contributions to the phenotypic traits of C. ensifolium and related species, thereby enhancing our ability to utilize these genes for horticultural and agricultural advancements.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13172375/s1, Table S1 BBX gene family identified in Cymbidium ensifolium; Table S2 Prediction of protein secondary structure of BBX gene.
Author Contributions
Conceptualization, K.Z. and Y.Z.; methodology, Y.P.; software, X.C.; formal analysis, X.C.; investigation, M.C.; resources, D.P.; data curation, X.W.; writing—original draft preparation, X.C.; writing—review and editing, K.Z.; visualization, X.C., M.N. and R.Z.; supervision, D.P., K.Z. and Y.Z.; funding acquisition, D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Project of National Key R & D Program (2023YFD1600504) and National Natural Science Foundation of China (Grant No. 32101583).
Data Availability Statement
All data generated or analyzed during this study are included in this published article (Supplementary Materials) and available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bula, R.J.; Morrow, R.C.; Tibbitts, T.W.; Barta, D.J.; Martin, T.S. Light-emitting Diodes as a Radiation Source for Plants. HortScience A Publ. Am. Soc. Hortic. Sci. 1991, 26, 203–205. [Google Scholar] [CrossRef]
- Jacob-Lopes, E.; Zepka, L.Q.; Queiroz, M.I. Light-Emitting Diodes: Progress in Plant Micropropagation. InTech 2017, 6, 93–103. [Google Scholar]
- Reza, R.H.; Mahdi, B.; Mansour, G. The growth, nutrient uptake and fruit quality in four strawberry cultivars under different Spectra of LED supplemental light. BMC Plant Biol. 2024, 24, 179. [Google Scholar]
- Yang, J.; Song, J.; Jeong, B.R. The flowering of SDP chrysanthemum in response to intensity of supplemental or night-interruptional blue light is modulated by both photosynthetic carbon assimilation and photoreceptor-mediated regulation. Front. Plant Sci. 2022, 13, 981143. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Runkle, E.S. The role of blue light in night-interruption lighting of petunia. Acta Hortic. 2015, 1107, 101–106. [Google Scholar] [CrossRef]
- Meng, Q.; Runkle, E.S. Low-intensity blue light in night-interruption lighting does not influence flowering of herbaceous ornamentals. Sci. Hortic. 2015, 186, 230–238. [Google Scholar] [CrossRef]
- Rantanen, M.; Kurokura, T.; Mouhu, K.; Pinho, P.; Tetri, E.; Halonen, L.; Palonen, P.; Elomaa, P.; HytöNen, T. Light quality regulates flowering in FvFT1/FvTFL1 dependent manner in the woodland strawberry Fragaria vesca. Front. Plant Sci. 2014, 5, 271. [Google Scholar] [CrossRef]
- Yoshida, H.; Hikosaka, S.; Goto, E.; Takasuna, H.; Kudou, T. Effects of light quality and light period on flowering of everbearing strawberry in a closed plant production system. Acta Hortic. 2012, 956, 107–112. [Google Scholar] [CrossRef]
- Jeong, S.W.; Park, S.; Jin, J.S.; Seo, O.N.; Kim, G.-S.; Kim, Y.-H.; Bae, H.; Lee, G.; Kim, S.T.; Lee, W.S.; et al. Influences of Four Different Light-Emitting Diode Lights on Flowering and Polyphenol Variations in the Leaves of Chrysanthemum (Chrysanthemum morifolium). J. Agric. Food Chem. 2012, 60, 9793–9800. [Google Scholar] [CrossRef]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Deng, Z.; Yang, Z.; Liu, X.; Dai, X.; Zhang, J.; Deng, K. Genome-Wide Identification and Expression Analysis of C3H Zinc Finger Family in Potato (Solanum tuberosum L.). Int. J. Mol. Sci. 2023, 24, 1288. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Bian, Y.; Liu, J.; Sun, Y.; Xu, D. B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 2020, 62, 1293–1309. [Google Scholar] [CrossRef]
- Torok, M.; Etkin, L.D. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation 2001, 67, 63–71. [Google Scholar] [CrossRef]
- Kwan, Y.S.; Sook, C.K.; Joonki, K.; Hwan, L.J.; Myun, H.S.; Jeon, Y.S.; Yeon, Y.S.; Seob, L.J.; Hoon, A.J. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005, 139, 770–778. [Google Scholar]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef]
- Holm, M.; Hardtke, C.S.; Gaudet, R.; Deng, X.W. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001, 20, 118–127. [Google Scholar] [CrossRef]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef]
- Federico, V. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 2011, 62, 2453–2463. [Google Scholar]
- Datta, S.; Hettiarachchi, G.H.C.M.; Deng, X.W.; Holm, M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 2006, 18, 70–84. [Google Scholar] [CrossRef]
- Zhang, X.; Shang, F.; Huai, J.; Xu, G.; Tang, W.; Jing, Y.; Lin, R. A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Holm, M.; Deng, X. The B-Box Domain Protein BBX21 Promotes Photomorphogenesis1. Plant Physiol. 2017, 176, 2365–2375. [Google Scholar] [CrossRef]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.A.; Preuss, S. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX Protein BBX25 Interacts with HY5, Negatively Regulating BBX22 Expression to Suppress Seedling Photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Jiang, Y.; Li, J.; Yan, T.; Fan, L.; Liang, J.; Chen, Z.J.; Xu, D. Deng B-BOX DOMAIN PROTEIN28 Negatively Regulates Photomorphogenesis by Repressing the Activity of Transcription Factor HY5 and Undergoes COP1-Mediated Degradation. Plant Cell 2018, 30, 2006–2019. [Google Scholar] [CrossRef]
- Yadav, A.; Bakshi, S.; Yadukrishnan, P.; Lingwan, M.; Dolde, U.; Wenkel, S.; Masakapalli, S.K.; Datta, S. The B-Box-Containing MicroProtein miP1a/BBX31 Regulates Photomorphogenesis and UV-B Protection. Plant Physiol. 2019, 179, 1876–1892. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Jaina, M.; Sara, C.; Lowri, W.; Matloob, Q.; Gustavoa, S.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Lisanna, P.; Shriya, R.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2010, 43, 758–768. [Google Scholar] [CrossRef]
- Moon, Y.H. EMF1 Interacts with EIP1, EIP6 or EIP9 Involved in the Regulation of Flowering Time in Arabidopsis. Plant Cell Physiol. 2011, 52, 1376–1388. [Google Scholar]
- Talar, U.; Kiebowicz-Matuk, A. Beyond Arabidopsis: BBX Regulators in Crop Plants. Int. J. Mol. Sci. 2021, 22, 2906. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Liu, Y.; Li, X.; Chen, Q.; Zhang, Y.; Luo, Y.; Liu, Z.; Wang, Y.; Lin, Y.; Zhang, Y.; et al. Transcriptome Profile Analysis of Strawberry Leaves Reveals Flowering Regulation under Blue Light Treatment. Int. J. Genom. 2021, 2021, 5572076. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jin, F.; Jin, L.; Jackson, A.; Ma, X.; Shu, X.; Wu, D.; Jin, G. Characterization and comparative profiling of the small RNA transcriptomes in two phases of flowering in Cymbidium ensifolium. BMC Genom. 2015, 16, 622. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Z.; Lian, H. Studies on Cymbidium ensifolium susin clonal propagation and floral bud differentiation by means of tissue culture. Hortic. Res. 1988, 8, 15. [Google Scholar]
- Poole, R.L. The TAIR Database; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Chandran, A.K.N.; Jung, K. Resources for systems biology in rice. J. Plant Biol. 2014, 57, 80–92. [Google Scholar] [CrossRef]
- Ai, Y.; Li, Z.; Sun, W.H.; Chen, J.; Zhang, D.; Ma, L.; Zhang, Q.H.; Chen, M.K.; Zheng, Q.D.; Liu, J.F. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 15. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Elisabeth, G.; Alexandre, G.; Christine, H.; Ivan, I.; Appel, R.D.; Amos, B. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar]
- Yu, C.S.; Hwang, J.K. Prediction of Protein Subcellular Localizations. In Proceedings of the Intelligent Systems Design and Applications, Kaohsiung, Taiwan, 26–28 November 2008. [Google Scholar]
- Peng, Y.; Zhao, K.; Zheng, R.; Chen, J.; Zhu, X.; Xie, K.; Huang, R.; Zhan, S.; Su, Q.; Shen, M.; et al. A Comprehensive Analysis of Auxin Response Factor Gene Family in Melastoma dodecandrum Genome. Int. J. Mol. Sci. 2024, 25, 806. [Google Scholar] [CrossRef]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Jingyuan, R.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Kohli, D.K.; Bachhawat, A.K. CLOURE: Clustal Output Reformatter, a program for reformatting ClustalX/ClustalW outputs for SNP analysis and molecular systematics. Nucleic Acids Res. 2003, 31, 3501–3502. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, G.Z.; Huang, J.; Liu, D.K.; Xue, F.; Chen, X.L.; Chen, S.Q.; Liu, C.G.; Liu, H.; Ma, H. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res. 2021, 1, 10. [Google Scholar] [CrossRef]
- Chen, G.Z.; Huang, J.; Zhou, X.Q.; Hao, Y.; Chen, J.L.; Zhou, Y.Z.; Ahmad, S.; Lan, S.; Liu, Z.J.; Peng, D.H. Comprehensive Analysis for GRF Transcription Factors in Sacred Lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2022, 23, 6673. [Google Scholar] [CrossRef]
- Shi, Z.; Zhao, W.; Li, C.; Tan, W.; Zhu, Y.; Han, Y.; Ai, P.; Li, Z.; Wang, Z. Overexpression of the Chrysanthemum lavandulifolium ROS1 gene promotes flowering in Arabidopsis thaliana by reducing the methylation level of CONSTANS. Plant Sci. Int. J. Exp. Plant Biol. 2024, 342, 112019. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, X.; Aiwaili, P.; Mu, X.; Zhao, M.; Zhao, J.; Cheng, L.; Ma, C.; Gao, J.; Hong, B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020, 103, 1783–1795. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The Rice B-Box Zinc Finger Gene Family: Genomic Identification, Characterization, Expression Profiling and Diurnal Analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.; Sheng, F.; Peng, J.; Guofang, L.; Izhar, M.; Youmei, L.; Rahat, S.; Feng, D.; Xiya, Z.; Ke, L. Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules 2018, 23, 1763. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, M.; Qiao, X.; Cheng, Y.; Li, X.; Zhang, H.; Wu, J. A New Insight into the Evolution and Functional Divergence of SWEET Transporters in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef]
- Unruh, S.A.; McKain, M.R.; Lee, Y.-I.; Yukawa, T.; McCormick, M.K.; Shefferson, R.P.; Smithson, A.; Leebens-Mack, J.H.; Pires, J.C. Phylotranscriptomic analysis and genome evolution of the Cypripedioideae (Orchidaceae). Am. J. Bot. 2018, 105, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin, G.; Yin, C.; Fang, Y. B-box transcription factor 28 regulates flowering by interacting with constans. Sci. Rep. 2020, 10, 17789. [Google Scholar] [CrossRef]
- Xian, H. Chinese Rose CONSTANS (RoCO) Gene Cloning and Function Identification. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2017. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
