Selection and Validation of Reference Genes in Dendrocalamus brandisii for Quantitative Real-Time PCR
Abstract
1. Introduction
2. Results
2.1. Selection and Amplification Efficiency of Reference Genes
2.2. Ct Value of 21 Reference Genes
2.3. Stability of Gene Expression for Reference Candidates Using Three Different Algorithms
2.3.1. geNorm Analysis
2.3.2. NormFinder Analysis
2.3.3. Delta CT Analysis
2.3.4. Comprehensive Ranking Analysis
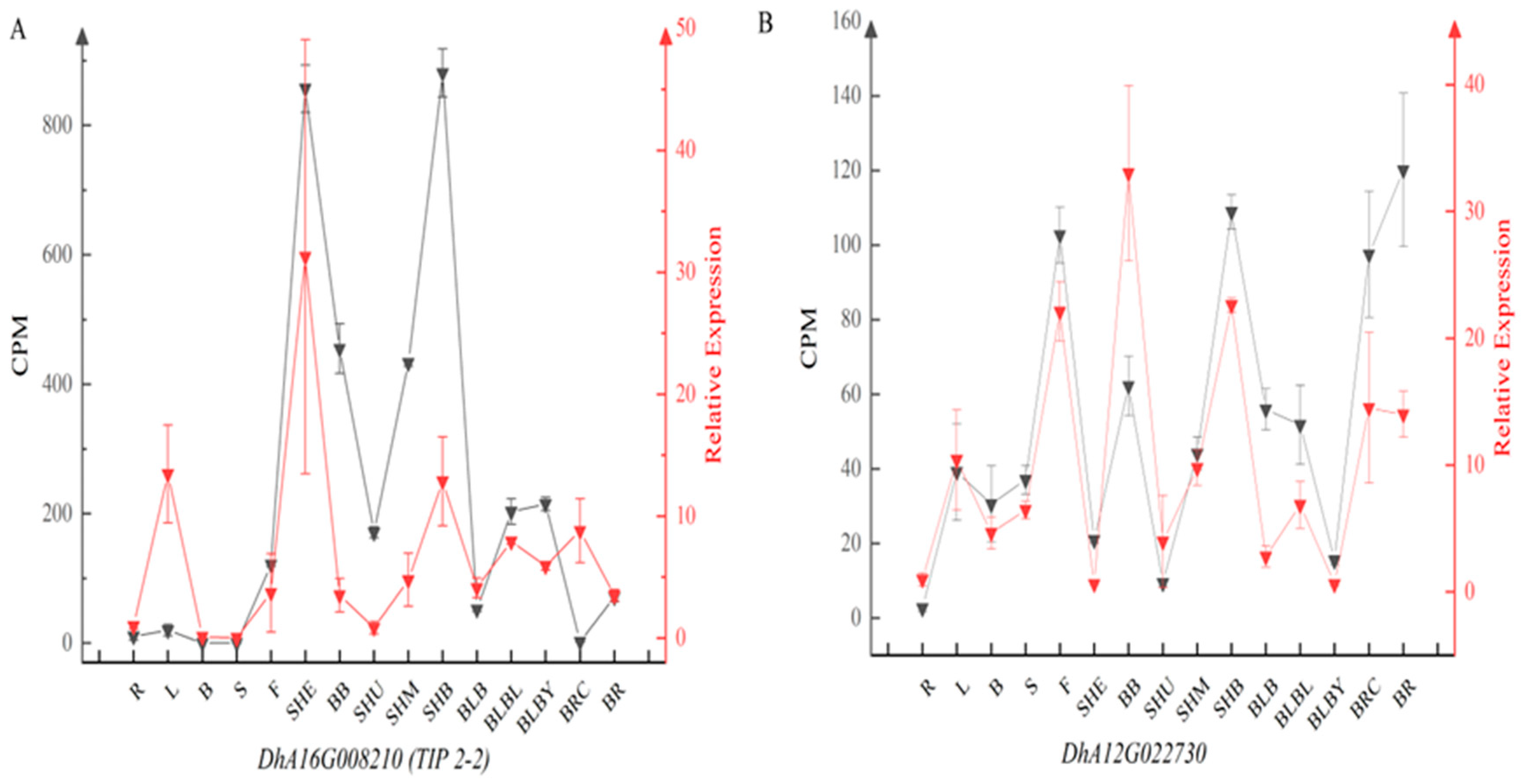
2.4. Validation of Candidate Reference Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Candidate Genes Selection and Primer Design
4.3. RNA Isolation and cDNA Preparation
4.4. qRT-PCR Analyses
4.5. Data Analysis
4.6. Validation of Reference Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seethalakshmi, K.K.; Kumar, M.S.M.; Pillai, K.S.; Sarojam, N. Bamboos of India: A Compendium; Brill: Boston, MA, USA, 1998; Volume 17. [Google Scholar]
- Zhan, H.; Zhang, L.-Y.; Deng, L.; Niu, Z.-H.; Li, M.-B.; Wang, C.-M.; Wang, S. Physiological and Anatomical Response of Foliar Silicon Application to Dendrocalamus Brandisii Plantlet Leaves under Chilling. Acta Physiol. Plant. 2018, 40, 208. [Google Scholar] [CrossRef]
- Li, J.H. Flora of China. In Harvard Papers in Botany 13, no. 2 (2007): 301-02. Available online: http://www.efloras.org/flora_page.aspx?flora_id=2 (accessed on 20 August 2024).
- Li, D.Z. The Biodiversity and Conservation of Bamboos in Yunnan, China. Bamboos 1997, 83–94, Charter 6. [Google Scholar]
- Mackay, I. Real-Time PCR in the Microbiology Laboratory. Clin. Microbiol. Infect. 2004, 10, 190–212. [Google Scholar] [CrossRef] [PubMed]
- Ransbotyn, V.; Reusch, T.B. Housekeeping Gene Selection for Quantitative Real-Time Pcr Assays in the Seagrass Zostera Marina Subjected to Heat Stress. Limnol. Oceanogr. Methods 2006, 4, 367–373. [Google Scholar] [CrossRef]
- Yoo, W.G.; Kim, T.I.; Li, S.; Kwon, O.S.; Cho, P.Y.; Kim, T.-S.; Kim, K.; Hong, S.-J. Reference Genes for Quantitative Analysis on Clonorchis Sinensis Gene Expression by Real-Time Pcr. Parasitol. Res. 2008, 104, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Reddy, D.S.; Bhatnagar-Mathur, P.; Cindhuri, K.S.; Sharma, K.K. Evaluation and Validation of Reference Genes for Normalization of Quantitative Real-Time Pcr Based Gene Expression Studies in Peanut. PLoS ONE 2013, 8, e78555. [Google Scholar] [CrossRef]
- Die, J.V.; Román, B.; Nadal, S.; González-Verdejo, C.I. Evaluation of Candidate Reference Genes for Expression Studies in Pisum Sativum under Different Experimental Conditions. Planta 2010, 232, 145–153. [Google Scholar] [CrossRef]
- Schmidt, G.W.; Delaney, S.K. Stable Internal Reference Genes for Normalization of Real-Time Rt-Pcr in Tobacco (Nicotiana Tabacum) During Development and Abiotic Stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef]
- Deng, Y.; Li, Y.; Sun, H. Selection of Reference Genes for Rt-Qpcr Normalization in Blueberry (Vaccinium Corymbosum× Angustifolium) under Various Abiotic Stresses. FEBS Open Bio 2020, 10, 1418–1435. [Google Scholar] [CrossRef]
- Joseph, J.T.; Poolakkalody, N.J.; Shah, J.M. Plant Reference Genes for Development and Stress Response Studies. J. Biosci. 2018, 43, 173–187. [Google Scholar] [CrossRef]
- Bustin, S.A.; Nolan, T. Pitfalls of Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction. J. Biomol. Tech. JBT 2004, 15, 155. [Google Scholar]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C.; et al. The Lack of a Systematic Validation of Reference Genes: A Serious Pitfall Undervalued in Reverse Transcription-Polymerase Chain Reaction (Rt-Pcr) Analysis in Plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Hu, G.; Zhao, J.; et al. Identification of Reliable Reference Genes for Quantitative Real-Time Pcr Normalization in Pitaya. Plant Methods 2019, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Jiang, J.; Chen, S.; Guan, Z.; Liao, Y.; Chen, F. Reference Genes for Normalizing Transcription in Diploid and Tetraploid Arabidopsis. Sci. Rep. 2014, 4, 6781. [Google Scholar] [CrossRef]
- Su, X.; Fan, B.; Yuan, L.; Cui, X.; Lu, S. Selection and Validation of Reference Genes for Quantitative Rt-Pcr Analysis of Gene Expression in Populus Trichocarpa. Chin. Bull. Bot. 2013, 48, 507. [Google Scholar]
- Wei, Y.D.; Chen, Y.; Guo, H.P.; Xie, H.A.; Zhang, J.F.; Wang, Z.H. Selection of Reference Genes for Real-Time Quantitative Rt-Pcr in Rice (Oryza Sativa L. ssp. Japonica) under Nutrient Deficiency. J. Agric. Biotechnol. 2013, 21, 1302–1312. [Google Scholar]
- González-Agüero, M.; García-Rojas, M.; Di Genova, A.; Correa, J.; Maass, A.; Orellana, A.; Hinrichsen, P. Identification of Two Putative Reference Genes from Grapevine Suitable for Gene Expression Analysis in Berry and Related Tissues Derived from Rna-Seq Data. BMC Genom. 2013, 14, 878. [Google Scholar] [CrossRef]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of Internal Control for Gene Expression Study in Soybean by Quantitative Real-Time Pcr. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative Rt-Pcr Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-Pcr Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An Improvement of the 2ˆ (–Delta Delta Ct) Method for Quantitative Real-Time Polymerase Chain Reaction Data Analysis. Biostat. Bioinform. Biomath. 2013, 3, 71. [Google Scholar]
- Liang, W.; Zou, X.; Carballar-Lejarazú, R.; Wu, L.; Sun, W.; Yuan, X.; Wu, S.; Li, P.; Ding, H.; Ni, L.; et al. Selection and Evaluation of Reference Genes for Qrt-Pcr Analysis in Euscaphis konishii Hayata Based on Transcriptome Data. Plant Methods 2018, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, C.; Yang, H.; Lyu, L.; Li, W.; Wu, W. Selection and Validation of Candidate Reference Genes for Gene Expression Analysis by Rt-Qpcr in Rubus. Int. J. Mol. Sci. 2021, 22, 10533. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of Reference Genes for Quantitative Real-Time Pcr in Bamboo (Phyllostachys Edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef]
- Karuppaiya, P.; Yan, X.X.; Liao, W.; Wu, J.; Chen, F.; Tang, L. Correction: Identification and Validation of Superior Reference Gene for Gene Expression Normalization Via Rt-Qpcr in Staminate and Pistillate Flowers of Jatropha Curcas—A Biodiesel Plant. PLoS ONE 2017, 12, e0177039. [Google Scholar] [CrossRef]
- Li, T.; Wang, J.; Lu, M.; Zhang, T.; Qu, X.; Wang, Z. Selection and Validation of Appropriate Reference Genes for Qrt-Pcr Analysis in Isatis Indigotica Fort. Front. Plant Sci. 2017, 8, 1139. [Google Scholar] [CrossRef]
- Li, J.; Han, X.; Wang, C.; Qi, W.; Zhang, W.; Tang, L.; Zhao, X. Validation of Suitable Reference Genes for Rt-Qpcr Data in Achyranthes Bidentata Blume under Different Experimental Conditions. Front. Plant Sci. 2017, 8, 776. [Google Scholar] [CrossRef]
- Niu, K.; Shi, Y.; Ma, H. Selection of Candidate Reference Genes for Gene Expression Analysis in Kentucky Bluegrass (Poa Pratensis L.) under Abiotic Stress. Front. Plant Sci. 2017, 8, 193. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, X.; Chen, S.; Zheng, L.; He, X.; Liu, M.; Qiao, G.; Wang, Y.; Zhuo, R. Selection of Suitable Reference Genes for Quantitative Real-Time Pcr Gene Expression Analysis in Salix Matsudana under Different Abiotic Stresses. Sci. Rep. 2017, 7, 40290. [Google Scholar] [CrossRef]
- Xiao, Z.; Sun, X.; Liu, X.; Li, C.; He, L.; Chen, S.; Su, J. Selection of Reliable Reference Genes for Gene Expression Studies on Rhododendron Molle G. Don. Front. Plant Sci. 2016, 7, 1547. [Google Scholar] [CrossRef]
- An, H.; Zhu, Q.; Pei, W.; Fan, J.; Liang, Y.; Cui, Y.; Lv, N.; Wang, W. Whole-Transcriptome Selection and Evaluation of Internal Reference Genes for Expression Analysis in Protocorm Development of Dendrobium Officinale Kimura Et Migo. PLoS ONE 2016, 11, e0163478. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, Y.; Hu, B.; Tan, Z.; Huang, B. Identification and Validation of Reference Genes for Quantification of Target Gene Expression with Quantitative Real-Time Pcr for Tall Fescue under Four Abiotic Stresses. PLOS ONE 2015, 10, e0119569. [Google Scholar] [CrossRef]
- Qu, R.; Miao, Y.; Cui, Y.; Cao, Y.; Zhou, Y.; Tang, X.; Yang, J.; Wang, F. Selection of Reference Genes for the Quantitative Real-Time Pcr Normalization of Gene Expression in Isatis indigotica Fortune. BMC Mol. Biol. 2019, 20, 9. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute Quantification of Mrna Using Real-Time Reverse Transcription Polymerase Chain Reaction Assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The Miqe Guidelines: M Inimum I Nformation for Publication of Q Uantitative Real-Time Pcr E Xperiments; Oxford University Press: Oxford, UK, 2009. [Google Scholar]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping Genes as Internal Standards: Use and Limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control Selection for Rna Quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef]
- Duan, M.; Wang, J.; Zhang, X.; Yang, H.; Wang, H.; Qiu, Y.; Song, J.; Guo, Y.; Li, X. Identification of Optimal Reference Genes for Expression Analysis in Radish (Raphanus sativus L.) and Its Relatives Based on Expression Stability. Front. Plant Sci. 2017, 8, 1605. [Google Scholar] [CrossRef]
- Reddy, P.S.; Reddy, D.S.; Sivasakthi, K.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Evaluation of Sorghum [Sorghum bicolor (L.)] Reference Genes in Various Tissues and under Abiotic Stress Conditions for Quantitative Real-Time Pcr Data Normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar]
- Mangeot-Peter, L.; Legay, S.; Hausman, J.F.; Esposito, S.; Guerriero, G. Identification of Reference Genes for Rt-Qpcr Data Normalization in Cannabis Sativa Stem Tissues. Int. J. Mol. Sci. 2016, 17, 1556. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, M.; Meng, Y. Identification and Validation of Reference Genes for Rt-Qpcr Analysis in Switchgrass under Heavy Metal Stresses. Genes 2020, 11, 502. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. Reffinder: A Web-Based Tool for Comprehensively Analyzing and Identifying Reference Genes. Funct. Integr. Genom. 2023, 23, 1–5. [Google Scholar] [CrossRef]
- Chang, Y.; Hu, S.; Xu, J.; Gong, H.; Guo, X.; Song, Q.; Gong, W.; Yuan, D. Identification of Reference Genes Provides Insights into the Determinants of Self-Incompatibility in Camellia Oleifera. Sci. Hortic. 2023, 321, 112301. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, R.; Hu, X.; Li, J.; Zhao, P.; Guo, F.; Zhao, H.; Wang, P.; Wang, Y.; Ni, D. Reference Gene Selection for Qrt-Pcr Analysis in the Shoots and Roots of Camellia Sinensis Var. Sinensis under Nutritional Stresses. Sci. Hortic. 2023, 320, 112237. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, G.; Rao, Y.; Wang, B.; Tian, R.; Tan, Y.; Peng, T. Identification and Validation of Reference Genes for Qrt-Pcr Analyses under Different Experimental Conditions in Allium Wallichii. J. Plant Physiol. 2023, 281, 153925. [Google Scholar] [CrossRef]
- Liu, M.; Jiang, J.; Han, X.; Qiao, G.; Zhuo, R. Validation of Reference Genes Aiming Accurate Normalization of Qrt-Pcr Data in Dendrocalamus Latiflorus Munro. PLoS ONE 2014, 9, e87417. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Z.; Bai, Y.; Wang, X.; Dou, Y.; Geng, R.; Wu, C.; Zhang, H.; Lu, C.; Gu, L. Chromosomal-Level Genome and Metabolome Analyses of Highly Heterozygous Allohexaploid Dendrocalamus Brandisii Elucidate Shoot Quality and Developmental Characteristics. J. Integr. Plant Biol. 2024, 66, 1087–1105. [Google Scholar] [CrossRef]
- Trakunram, K.; Champoochana, N.; Chaniad, P.; Thongsuksai, P.; Raungrut, P. Microrna Isolation by Trizol-Based Method and Its Stability in Stored Serum and Cdna Derivatives. Asian Pac. J. Cancer Prev. APJCP 2019, 20, 1641. [Google Scholar] [CrossRef][Green Version]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of Reliable Reference Genes for Gene Expression Studies in Peach Using Real-Time Pcr. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]

| Genes | Forward Primer | Reverse Primer | Length (bp) | Efficiency (%) | Correlation Coefficient (R2) |
|---|---|---|---|---|---|
| 18SrRNA1 | AATGGGTGGGGAAAGATGT | TGGCTCAGTTGGAAGCTCT | 216 | 114.7 | 0.9903 |
| 18SrRNA2 | AGAAATGGGTGGGAAAAGA | TGGCTCAGTTGGAAGCTCT | 219 | 110.5 | 0.9855 |
| ACTIN-1 | TGGCAGATTGATGCTAAGA | TCAGGTCGTGAACTGCAAG | 192 | 106.0 | 0.9812 |
| ACTIN-2 | GGACGCAAGTGGAAACTTA | AAAAGGCCAAGCATAGCA | 206 | 92.9 | 0.983 |
| CYP-1 | CAGTGATCAAAGGATGGATG | ACTGGGCGCTGAACTTCA | 171 | 97.9 | 0.9793 |
| CYP-2 | ACTTCAAGCACAGCACACG | TCGATGGCCTGACAACAT | 169 | 101.2 | 0.9765 |
| EF-1-α-1 | AAGCCTGGCATGGTTGTTAC | TCATCCTTGGAGTGGAAGC | 179 | 111.4 | 0.9969 |
| EF-1-α-2 | GCTTCCAACTCCAAGGATGA | CCAGCTCAGCAAACTGACA | 151 | 109.6 | 0.9904 |
| GAPDH-1 | GAATGCTAGCTGCACCACAA | AGCTTGCCATTCAAATCAGG | 240 | 113.6 | 0.975 |
| GAPDH-2 | GAATGCTAGCTGCACCACAA | TGCTGCTGGGAATGATGTTA | 188 | 118.5 | 0.9673 |
| NAC-2 | ATTTGCTCCCTGGGATCTTC | CTTTAGGTGCCTTGCCTTCA | 227 | 110.4 | 0.9882 |
| NTB-2 | GCTTTGCATGGAAGGAACAT | GAAGCTCCAGGTTGTTGGAA | 206 | 131.1 | 0.9370 |
| RPL-2 | CCAACAAGCTCTCCAGATCA | AAAGAGAGCTGGTGCTGGTT | 184 | 128.0 | 0.9922 |
| TEF-1 | ACTGTCTTTTCCTGCCCATTC | CACACTCGTCAATCCATTCG | 172 | 108.3 | 0.9857 |
| TEF-2 | TTCCTGGAGATGGACAAGGA | ACGTTGTTGTCTTGCAAATC | 198 | 109.1 | 0.9886 |
| TUBULIN-1 | ACCATTGGAGGAGGTGATGA | AAGTTGTTGGCTGCATCCTC | 188 | 121.3 | 0.971 |
| TUBULIN-2 | CGACCACAAGTTTGACCTCAT | CCTTGACAAAGCACCAGAT | 189 | 187.0 | 0.9347 |
| UB2C-1 | TTGAAGGACCTGCAGAAGGA | TTCGGTGGCTTGAAAGGATA | 170 | 120.2 | 0.9926 |
| UB2C-2 | TTGAAGGACCTGCAGAAGGA | TTCGGTGGCTTGAAAGGATA | 195 | 128.2 | 0.9681 |
| UBC-1 | GGTGGCATTCAAGACAAAGG | ATGTGAGCAATCTCCGGAAC | 180 | 119.2 | 0.9934 |
| UBC-2 | GCGATTTGTTTCTCGGATGT | ATTCCCGCTTGTTCTCACTG | 203 | 109.4 | 0.9857 |
| Genes | Tissues | Leaves | Culms | Roots |
|---|---|---|---|---|
| 18S-1 | 0.798 | 0.764 | 0.541 | 0.897 |
| 18S-2 | 0.936 | 0.731 | 0.376 | 0.578 |
| ACTIN-1 | 1.268 | 0.551 | 0.218 | 0.165 |
| ACTIN-2 | 1.145 | 0.597 | 0.589 | 0.86 |
| CYP-1 | 0.868 | 0.857 | 0.609 | 0.611 |
| CYP-2 | 1.325 | 0.911 | 0.726 | 0.796 |
| EF-1-α-1 | 0.322 | 0.186 | 0.145 | 0.264 |
| EF-1-α-2 | 0.322 | 0.227 | 0.145 | 0.29 |
| GAPDH-1 | 0.463 | 0.263 | 0.314 | 0.165 |
| GAPDH-2 | 0.521 | 0.352 | 0.277 | 0.213 |
| NAC-2 | 1.079 | 0.205 | 0.566 | 0.832 |
| NTB-2 | 0.757 | 0.443 | 0.796 | 0.958 |
| RPL-2 | 0.593 | 0.631 | 0.428 | 0.341 |
| TEF-1 | 0.992 | 0.186 | 0.51 | 0.466 |
| TEF-2 | 0.694 | 0.31 | 0.674 | 0.498 |
| TUBULIN-1 | 1.597 | 0.801 | 0.235 | 0.419 |
| TUBULIN-2 | 1.209 | 0.829 | 0.646 | 0.719 |
| UB2C-1 | 1.405 | 0.388 | 0.699 | 0.675 |
| UB2C-2 | 1.471 | 0.694 | 0.758 | 0.756 |
| UBC-1 | 1.53 | 0.661 | 0.835 | 0.532 |
| UBC-2 | 1.035 | 0.487 | 0.474 | 0.376 |
| Genes | Tissues | Leaves | Culms | Roots |
|---|---|---|---|---|
| 18Sr-1 | 1.003 | 0.888 | 0.592 | 1.136 |
| 18S-2 | 1.21 | 0.791 | 0.325 | 0.774 |
| ACTIN-1 | 1.754 | 0.583 | 0.36 | 0.341 |
| ACTIN-2 | 1.264 | 0.59 | 0.58 | 0.987 |
| CYP-1 | 1.057c | 0.977 | 0.663 | 0.783 |
| CYP-2 | 1.55 | 1.341 | 0.826 | 0.992 |
| EF-1-α-1 | 0.651 | 0.158 | 0.361 | 0.34 |
| EF-1-α-2 | 0.505 | 0.084 | 0.376 | 0.29 |
| GAPDH-1 | 0.776 | 0.422 | 0.704 | 0.424 |
| GAPDH-2 | 0.743 | 0.63 | 0.515 | 0.489 |
| NAC-2 | 0.85 | 0.189 | 0.417 | 0.945 |
| NTB-2 | 0.602 | 0.428 | 1.01 | 1.413 |
| RPL-2 | 0.916 | 0.593 | 0.467 | 0.58 |
| TEF-1 | 0.867 | 0.278 | 0.317 | 0.254 |
| TEF-2 | 0.418 | 0.46 | 0.641 | 0.383 |
| TUBULIN-1 | 1.994 | 0.972 | 0.326 | 0.415 |
| TUBULIN-2 | 1.311 | 0.991 | 0.691 | 0.795 |
| UB2C-1 | 1.642 | 0.646 | 0.739 | 0.793 |
| UB2C-2 | 1.729 | 0.933 | 0.995 | 0.895 |
| UBC-1 | 1.731 | 0.653 | 1.082 | 0.648 |
| UBC-2 | 0.793 | 0.606 | 0.507 | 0.368 |
| Genes | Tissues | Leaves | Culms | Roots |
|---|---|---|---|---|
| 18S-1 | 1.48 | 1.05 | 0.81 | 1.27 |
| 18S-2 | 1.6 | 0.98 | 0.69 | 0.99 |
| ACTIN-1 | 1.99 | 0.86 | 0.67 | 0.76 |
| ACTIN-2 | 1.7 | 0.87 | 0.84 | 1.18 |
| CYP-1 | 1.51 | 1.15 | 0.87 | 1.01 |
| CYP-2 | 1.89 | 1.42 | 0.98 | 1.17 |
| EF-1-α-1 | 1.27 | 0.66 | 0.67 | 0.75 |
| EF-1-α-2 | 1.21 | 0.65 | 0.68 | 0.73 |
| GAPDH-1 | 1.32 | 0.71 | 0.88 | 0.78 |
| GAPDH-2 | 1.31 | 0.86 | 0.76 | 0.82 |
| NAC-2 | 1.45 | 0.68 | 0.75 | 1.15 |
| NTB-2 | 1.3 | 0.79 | 1.14 | 1.54 |
| RPL-2 | 1.42 | 0.88 | 0.74 | 0.87 |
| TEF-1 | 1.44 | 0.78 | 0.69 | 0.75 |
| TEF-2 | 1.24 | 0.78 | 0.86 | 0.8 |
| TUBULIN-1 | 2.24 | 1.12 | 0.66 | 0.8 |
| TUBULIN-2 | 1.73 | 1.14 | 0.87 | 1 |
| UB2C-1 | 1.95 | 0.88 | 0.9 | 1.01 |
| UB2C-2 | 2.02 | 1.08 | 1.1 | 1.07 |
| UBC-1 | 2.05 | 0.93 | 1.21 | 0.91 |
| UBC-2 | 1.42 | 0.89 | 0.77 | 0.77 |
| Genes | Tissues | Leaves | Culms | Roots |
|---|---|---|---|---|
| 18S-1 | 10.38 | 9.66 | 11.72 | 12.45 |
| 18S-2 | 9.07 | 7.75 | 4.28 | 6.45 |
| ACTIN-1 | 18.19 | 8.38 | 4.12 | 2.99 |
| ACTIN-2 | 15.31 | 7.54 | 11.72 | 18.74 |
| CYP-1 | 10.93 | 15.7 | 11.98 | 8.44 |
| CYP-2 | 8.12 | 21 | 16.9 | 18.69 |
| EF-1-α-1 | 3.66 | 2.74 | 3.41 | 3.94 |
| EF-1-α-2 | 2.06 | 2.58 | 3.94 | 3.31 |
| GAPDH-1 | 6.24 | 6.79 | 13.24 | 4.12 |
| GAPDH-2 | 5.32 | 10.8 | 9.49 | 6.64 |
| NAC-2 | 8.49 | 4.33 | 7.61 | 16.71 |
| NTB-2 | 4.92 | 7.64 | 19.75 | 20.75 |
| RPL-2 | 8.78 | 8.7 | 7.2 | 9.4 |
| TEF-1 | 7.19 | 4 | 3.16 | 3.71 |
| TEF-2 | 2.91 | 7.84 | 7.21 | 8.24 |
| TUBULIN-1 | 21 | 13.07 | 3.13 | 8.9 |
| TUBULIN-2 | 15.7 | 14.43 | 14.74 | 14.93 |
| UB2C-1 | 16.13 | 12.41 | 11.02 | 14.48 |
| UB2C-2 | 15.97 | 17.16 | 18.2 | 16.48 |
| UBC-1 | 18.67 | 12.87 | 21 | 8.54 |
| UBC-2 | 6.05 | 11.45 | 10.83 | 6.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Mu, C.; Bai, Y.; Cheng, W.; Geng, R.; Xu, J.; Dou, Y.; Cheng, Z.; Gao, J. Selection and Validation of Reference Genes in Dendrocalamus brandisii for Quantitative Real-Time PCR. Plants 2024, 13, 2363. https://doi.org/10.3390/plants13172363
Jiang J, Mu C, Bai Y, Cheng W, Geng R, Xu J, Dou Y, Cheng Z, Gao J. Selection and Validation of Reference Genes in Dendrocalamus brandisii for Quantitative Real-Time PCR. Plants. 2024; 13(17):2363. https://doi.org/10.3390/plants13172363
Chicago/Turabian StyleJiang, Jutang, Changhong Mu, Yucong Bai, Wenlong Cheng, Ruiman Geng, Junlei Xu, Yuping Dou, Zhanchao Cheng, and Jian Gao. 2024. "Selection and Validation of Reference Genes in Dendrocalamus brandisii for Quantitative Real-Time PCR" Plants 13, no. 17: 2363. https://doi.org/10.3390/plants13172363
APA StyleJiang, J., Mu, C., Bai, Y., Cheng, W., Geng, R., Xu, J., Dou, Y., Cheng, Z., & Gao, J. (2024). Selection and Validation of Reference Genes in Dendrocalamus brandisii for Quantitative Real-Time PCR. Plants, 13(17), 2363. https://doi.org/10.3390/plants13172363





