Genome-Wide Identification and Characterization of the GASA Gene Family in Medicago truncatula, and Expression Patterns under Abiotic Stress and Hormone Treatments
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Condition and Treatments
2.2. Identification of GASA Family Member in M. truncatula
2.3. Chromosomal Localization and Gene Duplication
2.4. Sequence Alignment and Polygenetic Analysis
2.5. Tandem Duplication and Synteny Analysis
2.6. Cis-Regulatory Element Analysis
2.7. MtGASA Gene Expression Profiles under Multiple Tissues and Abiotic Stresses
2.8. Quantitative RT-PCR Analysis
3. Results
3.1. Identification and Annotation of GASA Genes in Medicago truncatula
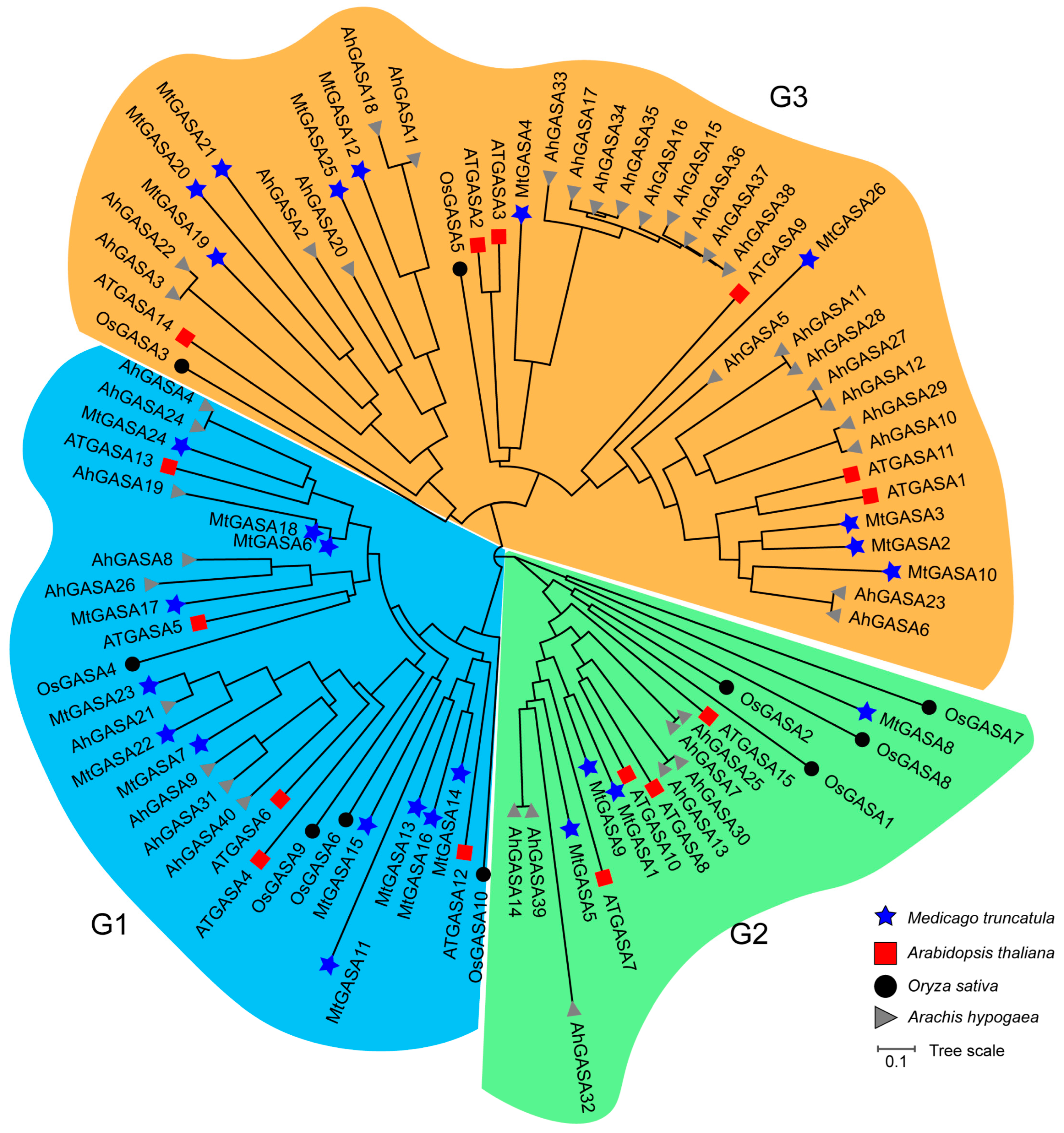
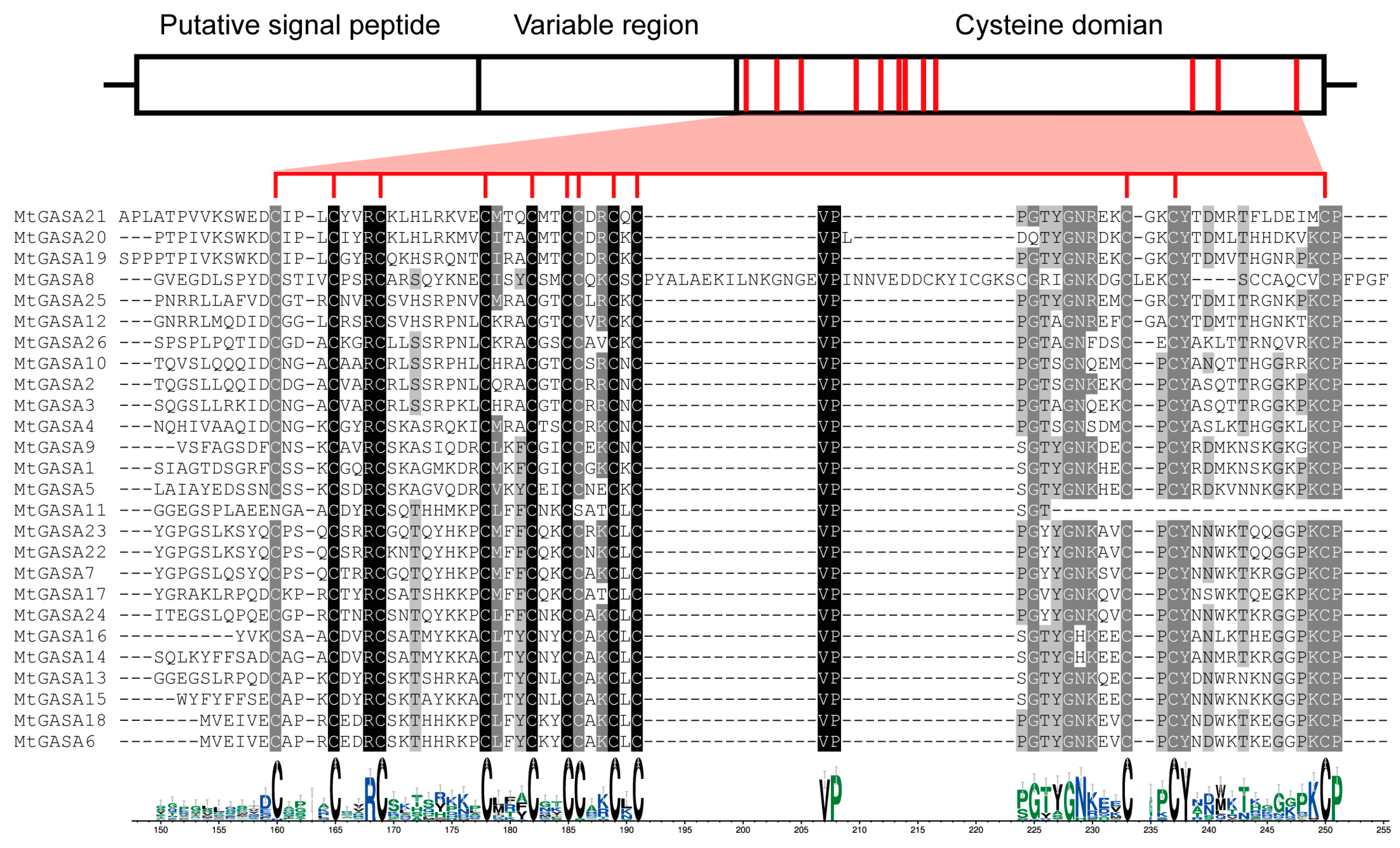
3.2. Systematic Phylogeny, Gene Structure, and Motif Analysis of GASA Genes in M. truncatula
3.3. Chromosomal Localization and Gene Duplication Analysis of MtGASAs
3.4. Promoter Region Analysis of MtGASAs
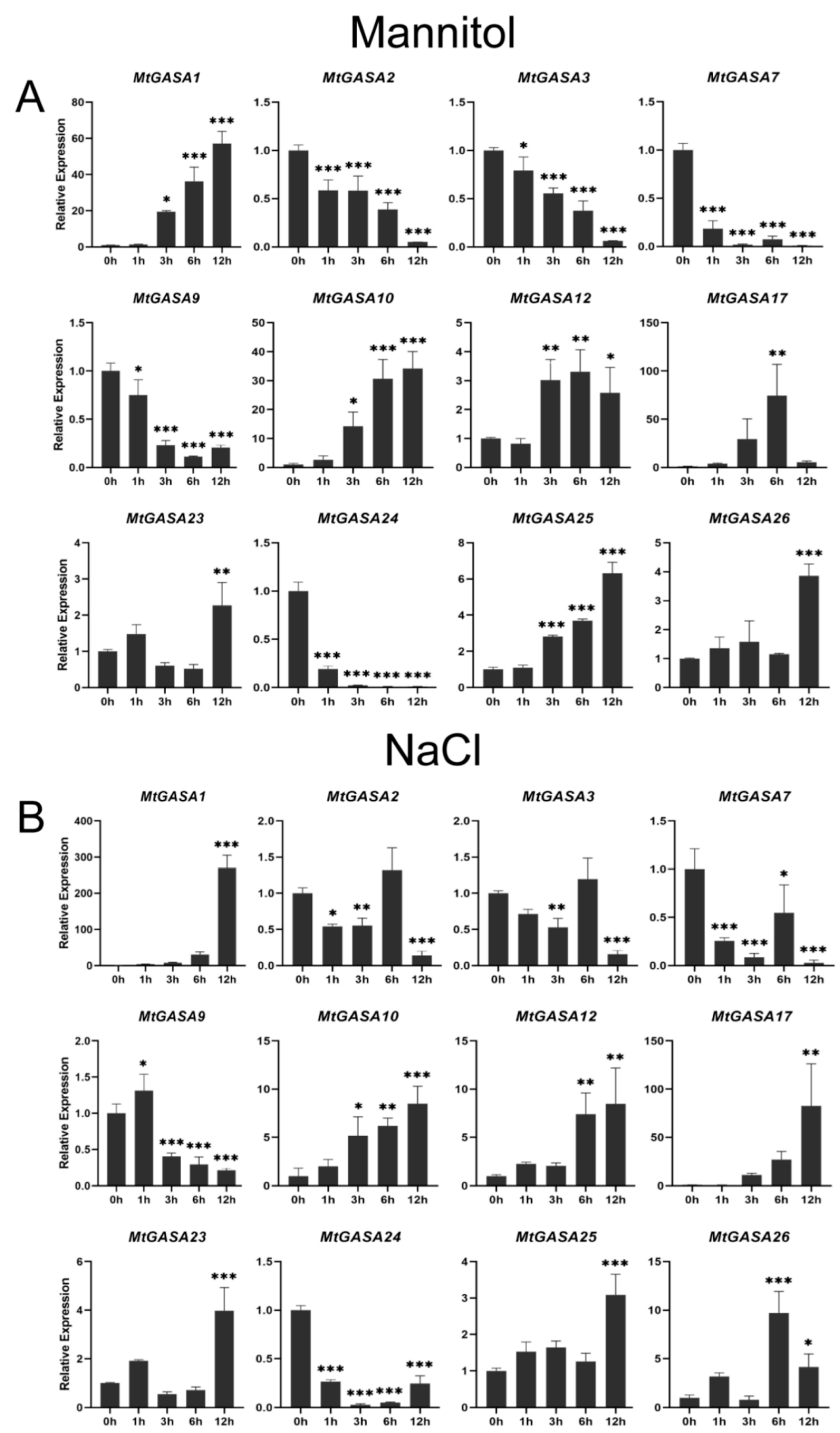
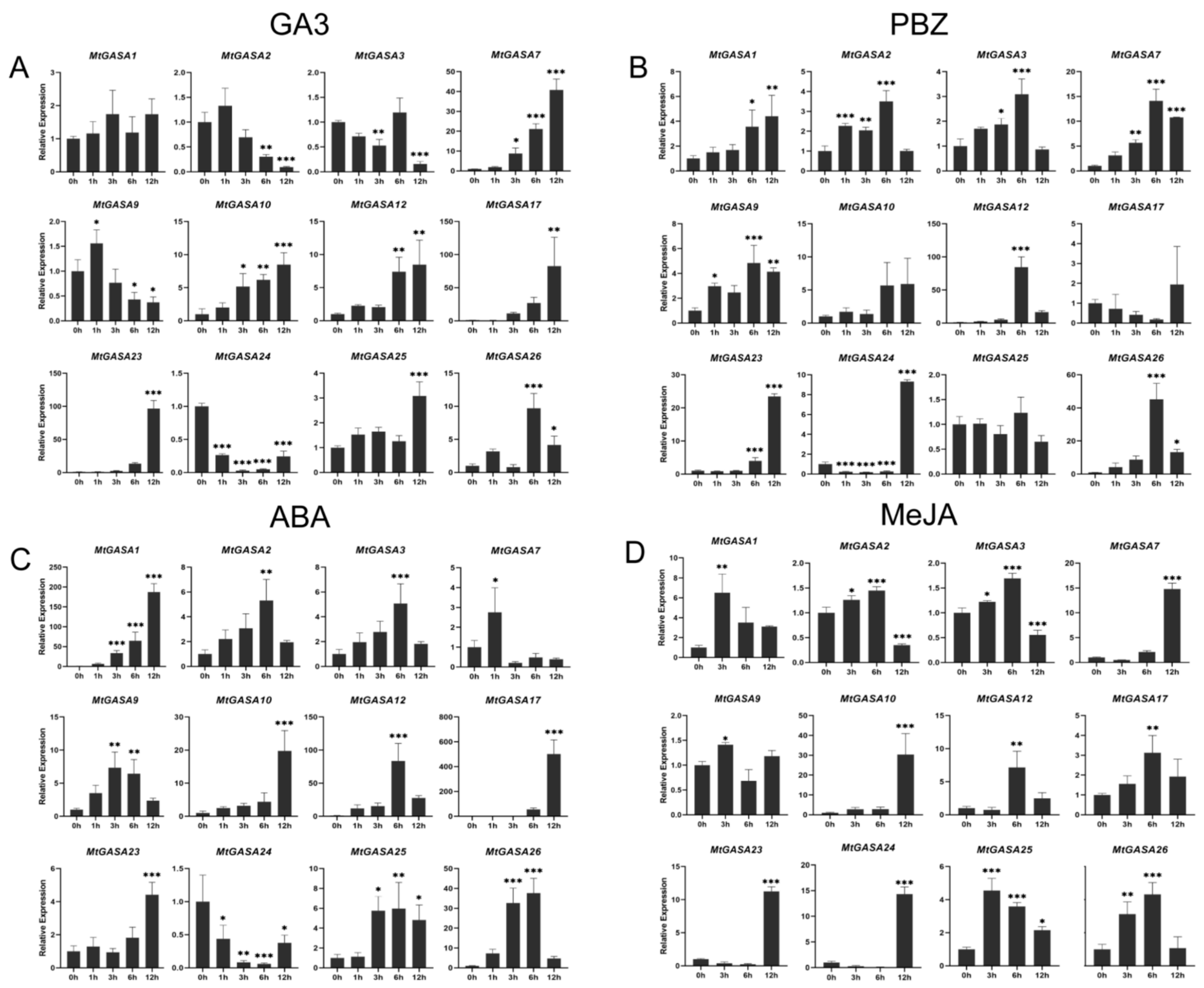
3.5. Expression Patterns of MtGASAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bouteraa, M.T.; Ben Romdhane, W.; Ben Hsouna, A.; Amor, F.; Ebel, C.; Ben Saad, R. Genome-wide characterization and expression profiling of GASA gene family in Triticum turgidum ssp. durum (desf.) husn. (Durum wheat) unveils its involvement in environmental stress responses. Phytochemistry 2023, 206, 113544. [Google Scholar] [CrossRef] [PubMed]
- Herzog, M.; Dorne, A.M.; Grellet, F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol. Biol. 1995, 27, 743–752. [Google Scholar] [CrossRef]
- Almasia, N.I.; Bazzini, A.A.; Hopp, H.E.; Vazquez-Rovere, C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 2008, 9, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Sakai, H.; Hochholdinger, F. The Gibberellic Acid Stimulated-LikeGene Family in Maize and Its Role in Lateral Root Development. Plant Physiol. 2010, 152, 356–365. [Google Scholar] [CrossRef]
- Nahirñak, V.; Almasia, N.I.; Fernandez, P.V.; Hopp, H.E.; Estevez, J.M.; Carrari, F.; Vazquez-Rovere, C. Potato Snakin-1 Gene Silencing Affects Cell Division, Primary Metabolism, and Cell Wall Composition. Plant Physiol. 2012, 158, 252–263. [Google Scholar] [CrossRef]
- Wu, T.; Zhong, Y.; Chen, M.; Wu, B.; Wang, T.; Jiang, B.; Zhong, G. Analysis of CcGASA family members in Citrus clementina (Hort. ex Tan.) By A Genome-Wide Approach. BMC Plant Biol. 2021, 21, 565. [Google Scholar] [CrossRef]
- Silverstein, K.A.T.; Moskal, W.A.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Gast, R.T.; Gopalraj, M.; Olszewski, N.E. Characterization of a shoot-specific, GA3-regulated and ABA-regulated gene from tomato. Plant J. 1992, 2, 153–159. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Sana, A.; Jamil, A.; Nasir, J.A.; Ahmed, S.; Hameed, M.U.; Abdullah, A. genome-wide approach to the comprehensive analysis of GASA gene family in Glycine max. Plant Mol. Biol. 2019, 100, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, I.; Li, W.-Q.; Jing, X.-Q.; Zhou, M.-R.; Shalmani, A.; Ali, M.; Wei, X.-Y.; Sharif, R.; Liu, W.-T.; Chen, K.-M. A systematic in silico prediction of gibberellic acid stimulated GASA family members: A novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice. J. Plant Physiol. 2019, 234–235, 117–132. [Google Scholar] [CrossRef]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Ma, C.; Lv, J.; Zhang, C.; Ma, Q.; Fan, S. Identification, characterization, and expression profiles of the GASA genes in cotton. J. Cotton Res. 2021, 4, 7. [Google Scholar] [CrossRef]
- Wu, K.; Qu, Y.; Rong, H.; Han, X.; Tian, Y.; Xu, L. Identification and Expression Analysis of the Populus trichocarpa GASA-Gene Family. Int. J. Mol. Sci. 2022, 23, 1507. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, Z.Q.; Qi, F.Y.; Zhao, M.B.; Dong, W.Z.; Huang, B.Y.; Zheng, Z.; Zhang, X.Y. Comprehensive Analysis of GASA Family Members in the Peanut Genome: Identification, Characterization, and Their Expressions in Response to Pod Development. Agronomy 2022, 12, 3067. [Google Scholar] [CrossRef]
- Li, Z.; Gao, J.; Wang, G.; Wang, S.; Chen, K.; Pu, W.; Wang, Y.; Xia, Q.; Fan, X. Genome-Wide Identification and Characterization of GASA Gene Family in Nicotiana tabacum. Front. Genet. 2022, 12, 768942. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Liu, K.; Yu, Z.; Li, Y.; Zhang, Y.; Zhu, Y.; Wu, Y.; He, H.; Zeng, X.; Chen, H.; et al. Genome-wide characterization of the tomato GASA family identifies SlGASA1 as a repressor of fruit ripening. Hortic. Res. 2023, 10, uhac222. [Google Scholar] [CrossRef]
- Li, K.-L.; Bai, X.; Li, Y.; Cai, H.; Ji, W.; Tang, L.-L.; Wen, Y.-D.; Zhu, Y.-M. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J. Plant Physiol. 2011, 168, 2153–2160. [Google Scholar] [CrossRef]
- Wang, H.; Wei, T.; Wang, X.; Zhang, L.; Yang, M.; Chen, L.; Song, W.; Wang, C.; Chen, C. Transcriptome Analyses from Mutant Salvia miltiorrhiza Reveals Important Roles for SmGASA4 during Plant Development. Int. J. Mol. Sci. 2018, 19, 2088. [Google Scholar] [CrossRef]
- Chen, B.; Sun, Y.; Tian, Z.; Fu, G.; Pei, X.; Pan, Z.; Nazir, M.F.; Song, S.; Li, H.; Wang, X.; et al. GhGASA10-1 promotes the cell elongation in fiber development through the phytohormones IAA-induced. BMC Plant Biol. 2021, 21, 448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X. Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J. Plant Physiol. 2011, 168, 2093–2101. [Google Scholar] [CrossRef]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.-C. Molecular and cellular characterization of GA-Stimulated Transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.L.; Opsahl-Sorteberg, H.G. GASA4, One of the 14-Member Arabidopsis GASA Family of Small Polypeptides, Regulates Flowering and Seed Development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, C.; Peng, J.; Sun, S.; Wang, X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 2009, 69, 745–759. [Google Scholar] [CrossRef]
- Han, M.; Jin, X.; Yao, W.; Kong, L.; Huang, G.; Tao, Y.; Li, L.; Wang, X.; Wang, Y. A Mini Zinc-Finger Protein (MIF) from Gerbera hybrida Activates the GASA Protein Family Gene, GEG, to Inhibit Ray Petal Elongation. Front. Plant Sci. 2017, 8, 1649. [Google Scholar] [CrossRef]
- Cui, J.W.; Wang, X.S.; Wei, Z.W.; Jin, B. Medicago truncatula (model legume), Medicago sativa (alfalfa), Medicago polymorpha (bur clover), and Medicago ruthenica. Trends Genet. 2022, 38, 782–783. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.J.; Liu, Y.; Zhang, J.; Song, L.L.; Guo, C.H. Genome-Wide Analysis of the AP2/ERF Superfamily Genes and their Responses to Abiotic Stress in Medicago truncatula. Front. Plant Sci. 2016, 6, 1247. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.F.; Ma, L.; Jiang, W.B.; Yao, Y.; Tang, Y.H.; Pang, Y.Z. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Bioch 2021, 158, 21–33. [Google Scholar] [CrossRef]
- Zhu, T.T.; Herrfurth, C.; Xin, M.M.; Savchenko, T.; Feussner, I.; Goossens, A.; De Smet, I. Warm temperature triggers JOX and ST2A-mediated jasmonate catabolism to promote plant growth. Nat. Commun. 2021, 12, 4804. [Google Scholar] [CrossRef]
- Li, X.; Hou, Y.Y.; Zhang, F.; Li, M.N.; Yi, F.Y.; Kang, J.M.; Yang, Q.C.A.; Long, R.C. Identification and characterization of stress responsive homeodomain leucine zipper transcription factors in Medicago truncatula. Mol. Biol. Rep. 2022, 49, 3569–3581. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.P.; Cui, H.T.; Yang, W.L.; Yu, B.; Zhang, C.; Wen, J.Q.; Kang, J.M.; Wang, Z.; Yang, Q.C. The UDP-glycosyltransferase MtUGT84A1 regulates anthocyanin accumulation and plant growth via JA signaling in Medicago truncatula. Environ. Exp. Bot. 2022, 201, 104972. [Google Scholar] [CrossRef]
- Zhang, S.C.; Wang, X.J. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin. Sci. Bull. 2008, 53, 3839–3846. [Google Scholar] [CrossRef]
- Pecrix, Y.; Staton, S.E.; Sallet, E.; Lelandais-Brère, C.; Moreau, S.; Carrère, S.; Blein, T.; Jardinaud, M.F.; Latrasse, D.; Zouine, M.; et al. Whole-genome landscape of Medicago truncatula symbiotic genes. Nat. Plants 2018, 4, 1017–1025. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.N.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lee, T.H.; Tang, H.B.; Wang, X.Y.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant -acting regulatory elements and a portal to tools for analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Yang, J.; Song, J.X.; Feng, Y.Y.; Cao, Y.M.; Fu, B.Z.; Zhang, Z.Q.; Ma, N.; Li, Q.; Hu, T.M.; Wang, Y.F.; et al. Osmotic stress-induced lignin synthesis is regulated at multiple levels in alfalfa (Medicago sativa L.). Int. J. Biol. Macromol. 2023, 246, 125501. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, X.; Zhang, Y.; Miao, Z.; Xie, C.; Meng, X.; Deng, J.; Wen, J.; Mysore, K.S.; Frugier, F.; et al. Opposing Control by Transcription Factors MYB61 and MYB3 Increases Freezing Tolerance by Relieving C-Repeat Binding Factor Suppression. Plant Physiol. 2016, 172, 1306–1323. [Google Scholar] [CrossRef]
- Aubert, D.; Chevillard, M.; Dorne, A.-M.; Arlaud Ge Herzog, M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 1998, 36, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Nahirnak, V.; Rivarola, M.; Almasia, N.I.; Barrios Baron, M.P.; Hopp, H.E.; Vile, D.; Paniego, N.; Vazquez Rovere, C. Snakin-1 affects reactive oxygen species and ascorbic acid levels and hormone balance in potato. PLoS ONE 2019, 14, e0214165. [Google Scholar] [CrossRef] [PubMed]
- Almasia, N.I.; Nahirnak, V.; Hopp, H.E.; Vazquez-Rovere, C. Potato Snakin-1: An antimicrobial player of the trade-off between host defense and development. Plant Cell Rep. 2020, 39, 839–849. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, C.L.; Li, H.L.; Wang, G.L.; You, C.X. Apple SINA E3 ligase MdSINA3 negatively mediates JA-triggered leaf senescence by ubiquitinating and degrading the MdBBX37 protein. Plant J. 2022, 111, 457–472. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, C.; Li, Z.; Zhang, H.; Li, C.; Sun, H.; Li, S.; Ma, N.; Qi, X.; Cui, Y.; Yang, P.; et al. Genome-Wide Identification and Characterization of the GASA Gene Family in Medicago truncatula, and Expression Patterns under Abiotic Stress and Hormone Treatments. Plants 2024, 13, 2364. https://doi.org/10.3390/plants13172364
Gao C, Li Z, Zhang H, Li C, Sun H, Li S, Ma N, Qi X, Cui Y, Yang P, et al. Genome-Wide Identification and Characterization of the GASA Gene Family in Medicago truncatula, and Expression Patterns under Abiotic Stress and Hormone Treatments. Plants. 2024; 13(17):2364. https://doi.org/10.3390/plants13172364
Chicago/Turabian StyleGao, Cai, Zhongxing Li, Hanwen Zhang, Chun Li, Haoyang Sun, Shuo Li, Nan Ma, Xiangyu Qi, Yilin Cui, Peizhi Yang, and et al. 2024. "Genome-Wide Identification and Characterization of the GASA Gene Family in Medicago truncatula, and Expression Patterns under Abiotic Stress and Hormone Treatments" Plants 13, no. 17: 2364. https://doi.org/10.3390/plants13172364
APA StyleGao, C., Li, Z., Zhang, H., Li, C., Sun, H., Li, S., Ma, N., Qi, X., Cui, Y., Yang, P., & Hu, T. (2024). Genome-Wide Identification and Characterization of the GASA Gene Family in Medicago truncatula, and Expression Patterns under Abiotic Stress and Hormone Treatments. Plants, 13(17), 2364. https://doi.org/10.3390/plants13172364





