PagbHLH35 Enhances Salt Tolerance through Improving ROS Scavenging in Transgenic Poplar
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Methods
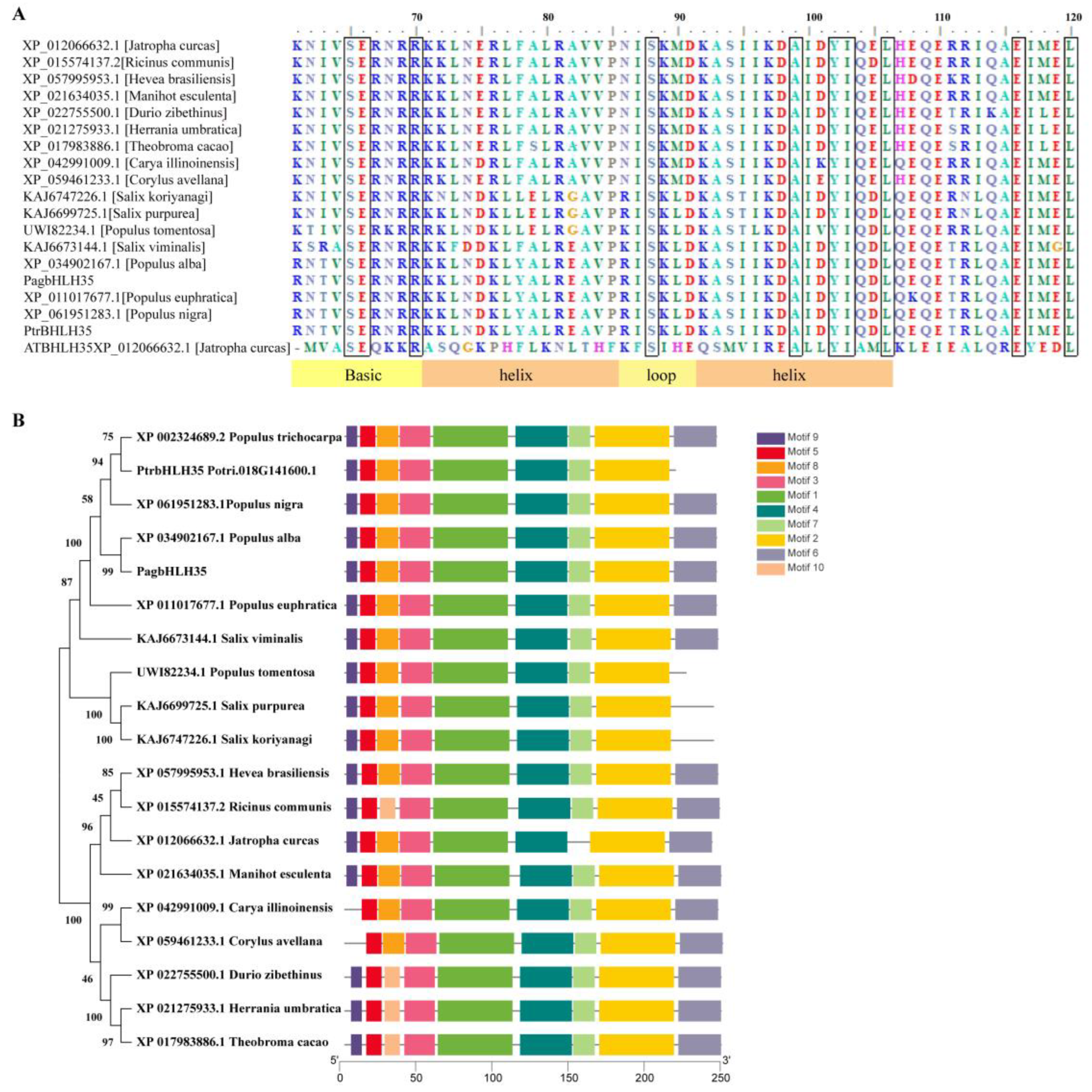
2.2. Sequence Analysis of PagbHLH35
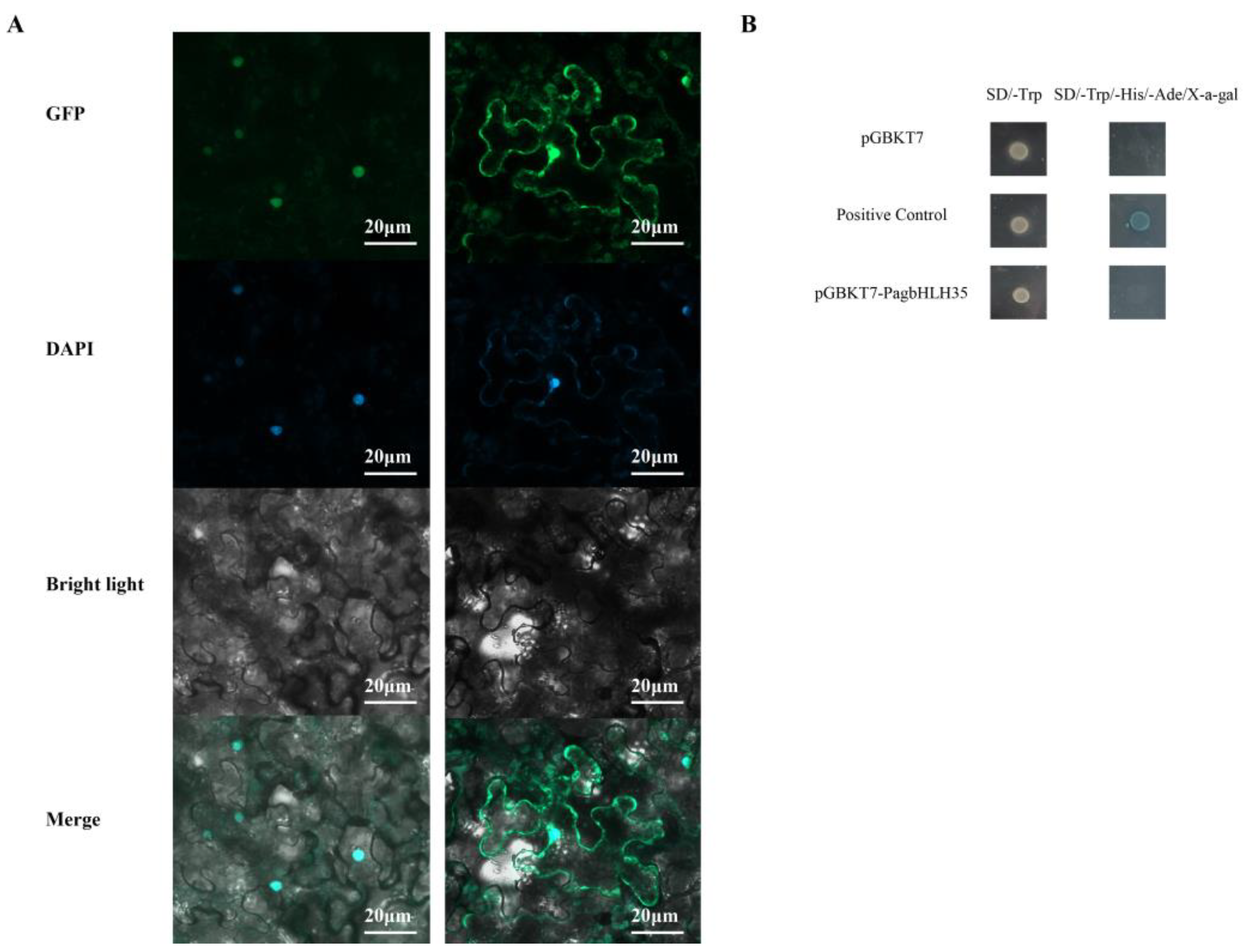
2.3. Subcellular Localization of PagbHLH35
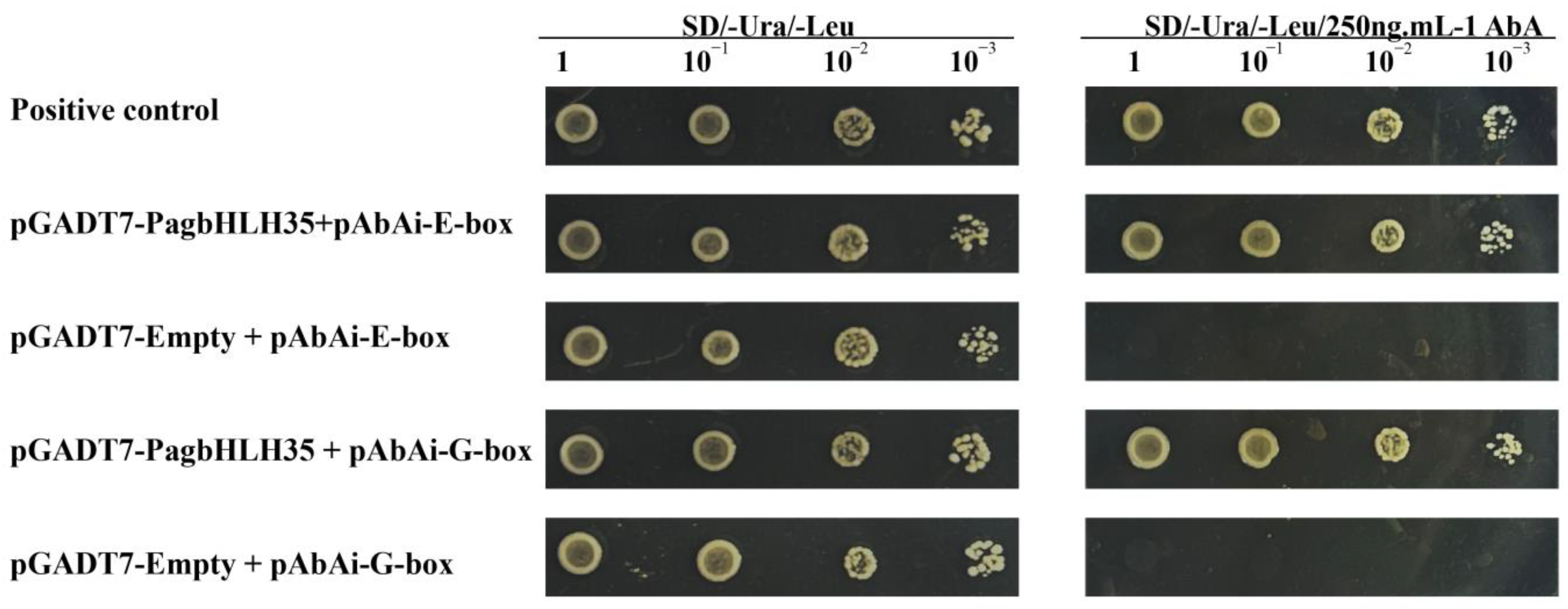
2.4. Yeast Hybrid Assays
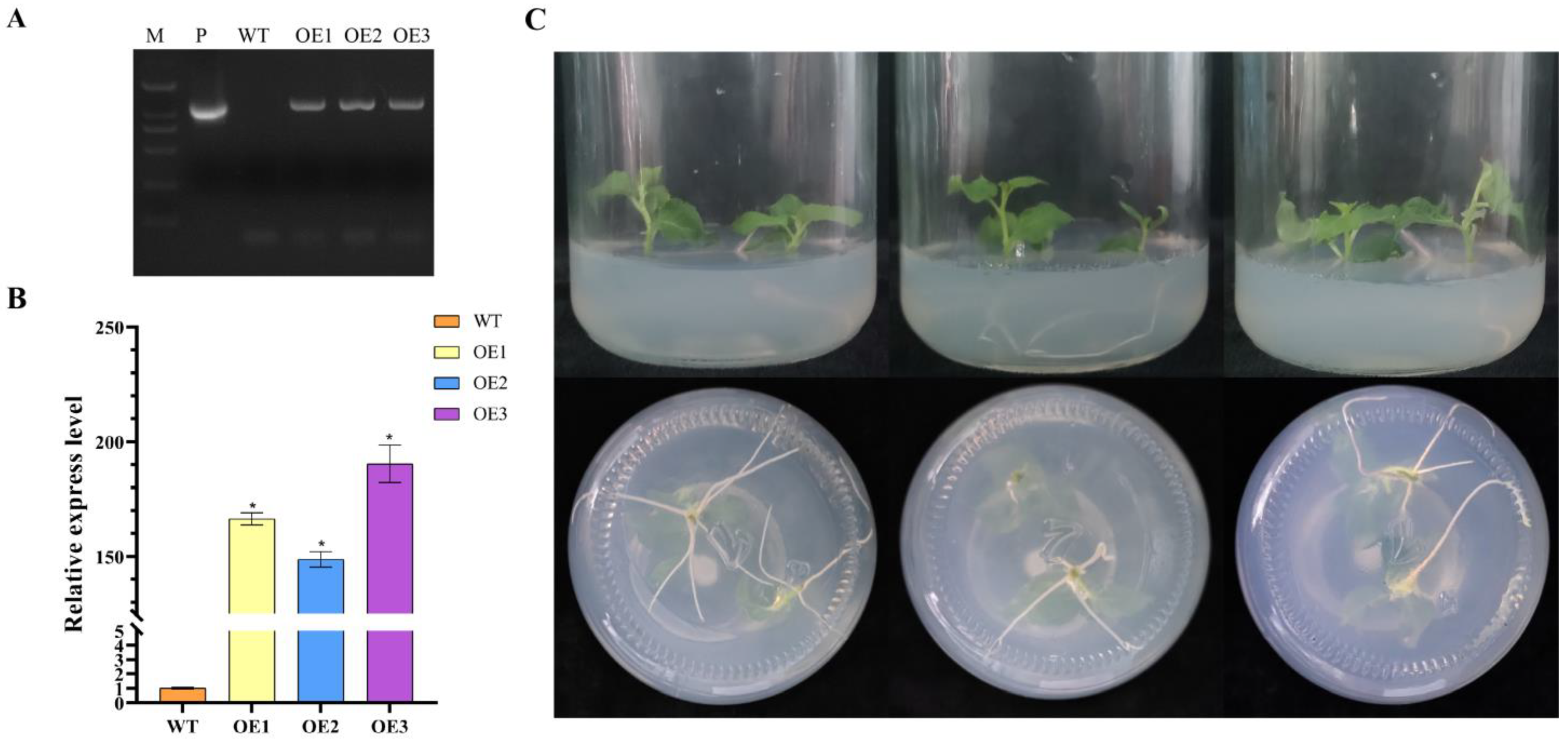
2.5. The Acquisition and Identification of Transgenic Poplar Lines
2.6. Histological Staining and Physiological Measurements
2.7. The Expression Analysis of PagbHLH35 Gene and Antioxidant Genes
2.8. Statistical Analysis
3. Results
3.1. Expression Pattern of PagbHLH35 Gene
3.2. Characterizationof PagbHLH35 Gene
3.3. PagbHLH35 Protein Was Localized in the Nucleus
3.4. Transcriptional Activity of PagbHLH35 Protein
3.5. PagbHLH35 Specifically Binds to G-Box and E-Box Elements
3.6. Molecular Identification of Transgenic Poplar Plants
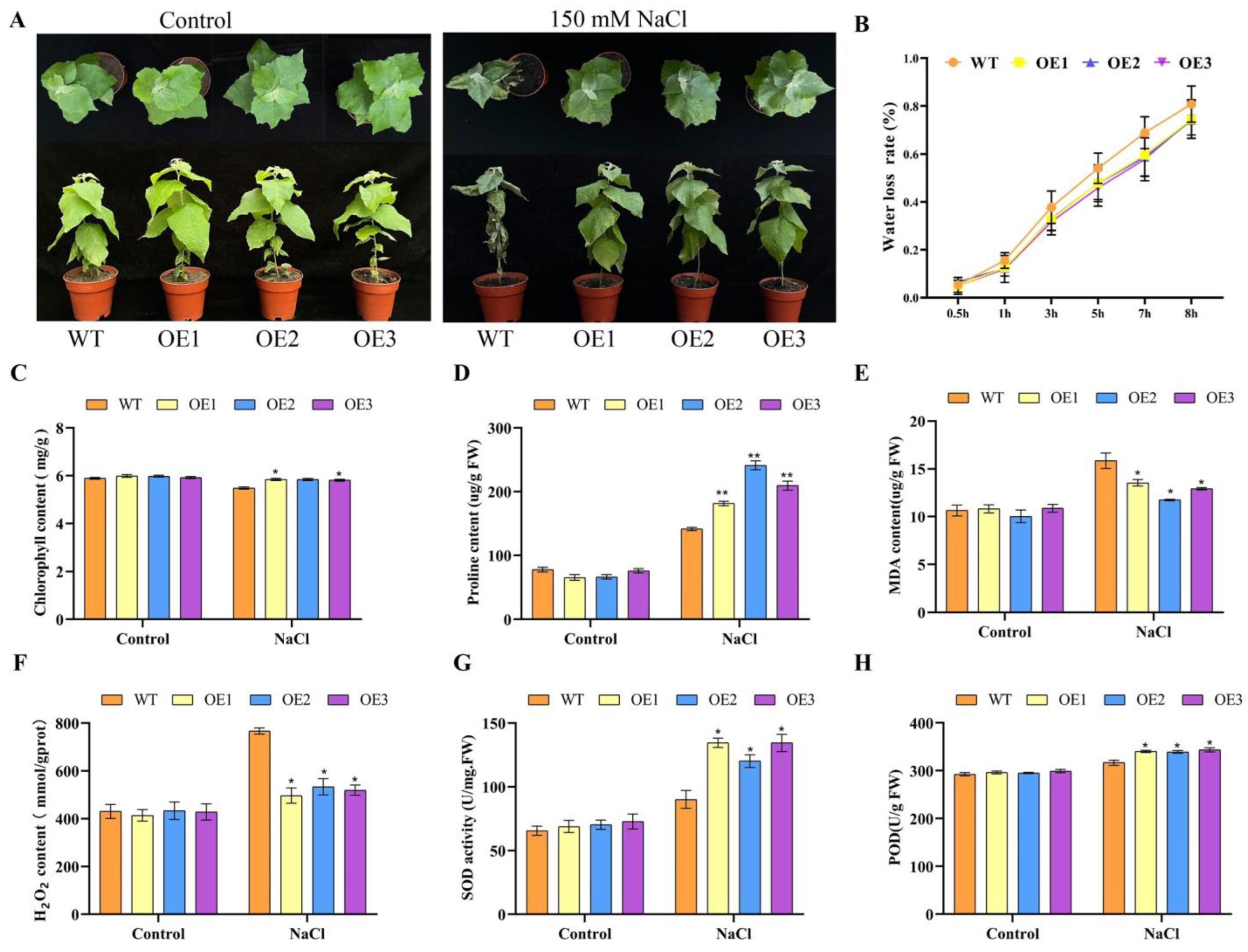
3.7. Salt Tolerance of Transgenic Poplar Overexpressing PagbHLH35 Gene
3.8. Histochemical Staining with NBT and DAB in Transgenic Poplars
3.9. Relative Expression Levels of POD and SOD-Related Genes in Transgenic Poplars
4. Discussion
4.1. Overexpression of PagbHLH35 Enhances Salt Tolerance
4.2. Overexpression of PagbHLH35 Decreased ROS Levels
4.3. PagbHLH35 May Regulate Target Genes through Specially Binding to G-Box/E-Box
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Chen, K.; Song, M.; Guo, Y.; Liu, L.; Xue, H.; Dai, H.; Zhang, Z. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Peng, T.; Xue, S. Mechanisms of plant saline-alkaline tolerance. J. Plant Physiol. 2023, 281, 153916. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, G.; Gao, M.; Chang, C.; Jia, J.; Li, J. Characteristics and Spatial Variability of Saline-Alkaline Soil Degradation in the Typical Yellow River Delta Area of Kenli County, China. J. Environ. Prot. 2014, 05, 1053–1063. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 16. [Google Scholar] [CrossRef]
- Feng, H.; Ma, N.; Meng, X.; Zhang, S.; Wang, J.; Chai, S.; Meng, Q. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Bioch. 2013, 73, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Riaño-Pachón, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef]
- Zhao, K.; Li, S.; Yao, W.; Zhou, B.; Li, R.; Jiang, T. Characterization of the basic helix-loop-helix gene family and its tissue-differential expression in response to salt stress in poplar. PeerJ 2018, 6, e4502. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genom. 2009, 282, 503–516. [Google Scholar] [CrossRef]
- Sasaki-Sekimoto, Y.; Saito, H.; Masuda, S.; Shirasu, K.; Ohta, H.; Du, J. Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal. Behav. 2014, 1559–2324, e27639. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.; Guo, H.; Xie, D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013, 9, e1003653. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Zhao, M.; Song, A.; Li, P.; Chen, S.; Jiang, J.; Chen, F. A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci. Rep. 2014, 4, 6694. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, F.; Wang, J.L.; Ma, Y.; Chong, K.; Xu, Y.Y. Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J. Plant Physiol. 2009, 166, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, G.; Wang, P.; Guo, Y.; Li, J. A simple and precise method (Y2H-in-frame-seq) improves yeast two-hybrid screening with cDNA libraries. J. Genet. Genom. 2022, 49, 595–598. [Google Scholar] [CrossRef]
- Ji, X.; Wang, L.; Zang, D.; Wang, Y. Transcription Factor-Centered Yeast One-Hybrid Assay. Methods Mol. Biol. 2018, 1794, 183–194. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Cheng, Y.; Wu, J.; Cheng, Q.; Li, W.; Fan, S.; Jiang, L.; Xu, Z.; Kong, F.; Zhang, D.; et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 2015, 66, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Dondurmacolu, F.; Avan, A.N.; Apak, R. Simultaneous detection of superoxide anion radicals and determination of the superoxide scavenging activity of antioxidants using a N,N-dimethyl-p-phenylene diamine/Nafion colorimetric sensor. Anal. Methods 2017, 43, 6202–6212. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, X.; Zhao, K.; Yao, W.; Li, R.; Zhou, B.; Jiang, T. Over-Expression of ERF38 Gene Enhances Salt and Osmotic Tolerance in Transgenic Poplar. Front. Plant Sci. 2019, 10, 1375. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, J.; Chen, X.; Zhou, Y.; Pei, Y.; Chen, L.; Ul Haq, S.; Lu, M.; Gong, H.; Chen, R. Pepper bHLH transcription factorCabHLH035 contributes to salt tolerance by modulating ion homeostasis and proline biosynthesis. Hortic. Res. 2022, 9, uhac203. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, L.; Chen, D.; Liang, M.; Liu, Z.; Shao, H.; Long, X. Salt stress encourages proline accumulation by regulating proline biosynthesis and degradation in Jerusalem artichoke plantlets. PLoS ONE 2013, 8, e62085. [Google Scholar] [CrossRef]
- Verma, D.; Jalmi, S.K.; Bhagat, P.K.; Verma, N.; Sinha, A.K. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 2020, 287, 2560–2576. [Google Scholar] [CrossRef]
- Wang, Y.; Zang, W.; Li, X.; Wang, C.; Wang, R.; Jiang, T.; Zhou, B.; Yao, W. Ectopic Expression of PsnNAC090 Enhances Salt and Osmotic Tolerance in Transgenic Tobacco. Int. J. Mol. Sci. 2023, 24, 8985. [Google Scholar] [CrossRef]
- Bonaventure, G.; Vandoorn, A.; Baldwin, I.T. Herbivore-associated elicitors: FAC signaling and metabolism. Trends Plant Sci. 2011, 16, 294–299. [Google Scholar] [CrossRef]
- Liu, D.; Li, Y.; Zhou, Z.; Xiang, X.; Liu, X.; Wang, J.; Hu, Z.; Xiang, S.; Li, W.; Xiao, Q.; et al. Tobacco transcription factor bHLH123 improves salt tolerance by activating NADPH oxidaseNtRbohE expression. Plant Physiol. 2021, 186, 1706–1720. [Google Scholar] [CrossRef]
- Fang, X.; Mo, J.; Zhou, H.; Shen, X.; Xie, Y.; Xu, J.; Yang, S. Comparative transcriptome analysis of gene responses of salt-tolerant and salt-sensitive rice cultivars to salt stress. Sci. Rep. 2023, 13, 19065. [Google Scholar] [CrossRef]
- Zang, D.; Wang, C.; Ji, X.; Wang, Y. Tamarix hispida zinc finger protein ThZFP1 participates in salt and osmotic stress tolerance by increasing proline content and SOD and POD activities. Plant Sci. 2015, 235, 111–121. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Q.; Yan, C.; Sun, Q.; Wang, J.; Li, C.; Yuan, C.; Mou, Y.; Shan, S. The bHLH transcription factor AhbHLH121 improves salt tolerance in peanut. Int. J. Biol. Macromol. 2024, 256, 128492. [Google Scholar] [CrossRef]
- Yang, T.; Yao, S.; Hao, L.; Zhao, Y.; Lu, W.; Xiao, K. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep. 2016, 35, 2309–2323. [Google Scholar] [CrossRef]
- Chen, H.C.; Hsieh-Feng, V.; Liao, P.C.; Cheng, W.H.; Liu, L.Y.; Yang, Y.W.; Lai, M.H.; Chang, M.C. The function of OsbHLH068 is partially redundant with its homolog, AtbHLH112, in the regulation of the salt stress response but has opposite functions to control flowering in Arabidopsis. Plant Mol. Biol. 2017, 94, 531–548. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Dong, L.; Yao, W.; Jiang, T. PagbHLH35 Enhances Salt Tolerance through Improving ROS Scavenging in Transgenic Poplar. Plants 2024, 13, 1835. https://doi.org/10.3390/plants13131835
Wang S, Dong L, Yao W, Jiang T. PagbHLH35 Enhances Salt Tolerance through Improving ROS Scavenging in Transgenic Poplar. Plants. 2024; 13(13):1835. https://doi.org/10.3390/plants13131835
Chicago/Turabian StyleWang, Shuang, Liben Dong, Wenjing Yao, and Tingbo Jiang. 2024. "PagbHLH35 Enhances Salt Tolerance through Improving ROS Scavenging in Transgenic Poplar" Plants 13, no. 13: 1835. https://doi.org/10.3390/plants13131835
APA StyleWang, S., Dong, L., Yao, W., & Jiang, T. (2024). PagbHLH35 Enhances Salt Tolerance through Improving ROS Scavenging in Transgenic Poplar. Plants, 13(13), 1835. https://doi.org/10.3390/plants13131835






