Unravelling the Phytochemical Composition and Antioxidant Potential of Different Parts of Rumex vesicarius L.: A RP-HPLC-MS-MS/MS, Chemometrics, and Molecular Docking-Based Comparative Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Phytochemical Composition of Rumex vesicarius L. Wildly Grown in Egypt
2.1.1. Phenolic Compounds
Phenolic Acids and Phenols
| Peak No. | RT (min) | [M − H]− | [M + H]+ | (M) | Molecular Formula | Score | Error (ppm) | Main Fragments | DBE | Proposed Compound | Subclass | References | Peak Areas | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Flowers | Leaves | Stems | Roots | |||||||||||||
| 1 | 0.69 | 387.1137 | 388.1209 | C13H24O13 | 96.98 | 2.18 | 341.1073, 179.0566, 119.0342, 89.0244, 71.0139 | 2 | Gluco-hepatonic acid hexoside | Su | [30] | 1.05 × 104 | 1.86 × 104 | 3.78 × 103 | 2.58 × 105 | |
| 2 | 0.76 | 341.1093 | 342.1162 | C12H22O11 | 94.78 | −0.8 | 179.0536 | 2 | Disaccharide | Su | [14,25] | 0 | 0 | 0 | 1.05 × 105 | |
| 3 | 0.82 | 431.1399 | 432.1742 | C15H28O14 | 98.18 | 1.71 | 341.1071, 179.0549, 119.0343, 89.0241, 71.0137 | 2 | Disaccharide glycerol I | Su | [29] | 0 | 0 | 0 | 2.78 × 105 | |
| 4 | 0.90 | 191.0557 | 192.0634 | C7H12O6 | 85.63 | 2.2 | 173.0473, 127.0347 | 2 | Quinic acid | Oa | [25,35] | 8.33 × 104 | 0 | 0 | 0 | |
| 5 | 0.94 | 431.1407 | 432.1742 | C15H28O14 | 99.05 | −0.3 | 341.1068, 179.0562, 119.0340, 89.0243, 71.0147 | 2 | Disaccharide glycerol II | Su | [29] | 0 | 0 | 0 | 4.42 × 105 | |
| 6 | 1.03 | 132.1013 | 131.0949 | C6H13NO2 | 90.3 | 2.8 | N.D. | 1 | Leucine/Isoleucine | Aa | [37,38] | 8.61 × 104 | 0 | 0 | 0 | |
| 7 | 1.80 | 153.0186 | 154.0266 | C7H6O4 | 82.92 | 4.54 | 152.0112, 125.0242, 124.0164, 109.0291, 108.0218, 107.0137 | 5 | Dihydroxybenzoic acid | HB | [21,22] | 1.09 × 105 | 0 | 0 | 0 | |
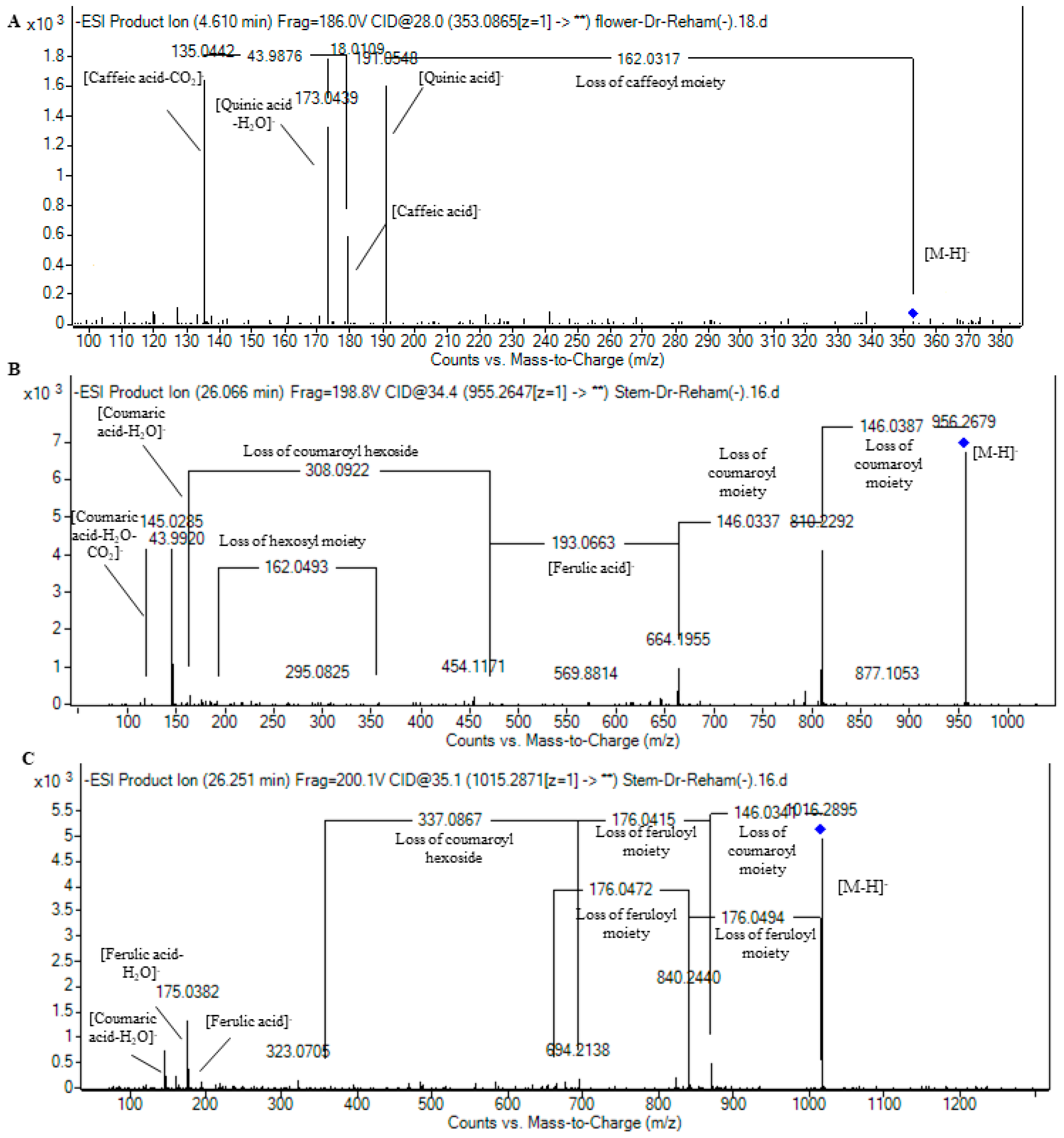
| 8 | 2.09 | 353.087 | 354.0951 | C16H18O9 | 80.92 | 2.22 | 191.0529, 179.0324, 135.0442 | 8 | Caffeoylquinic acid I | HC | [21,22] | 5.03 × 104 | 0 | 0 | 0 | |
| 9 | 3.76 | 353.0866 | 354.0951 | C16H18O9 | 89.66 | 3.33 | 191.0551, 173.0449, 161.0238 | 8 | Caffeoylquinic acid II | HC | [21,22] | 4.63 × 105 | 0 | 0 | 0 | |
| 10 | 4.56 | 353.0863 | 354.0951 | C16H18O9 | 90.71 | 4.67 | 191.0543, 173.0449, 135.0446 | 8 | Caffeoylquinic acid III | HC | [21,22] | 1.92 × 105 | 0 | 0 | 0 | |
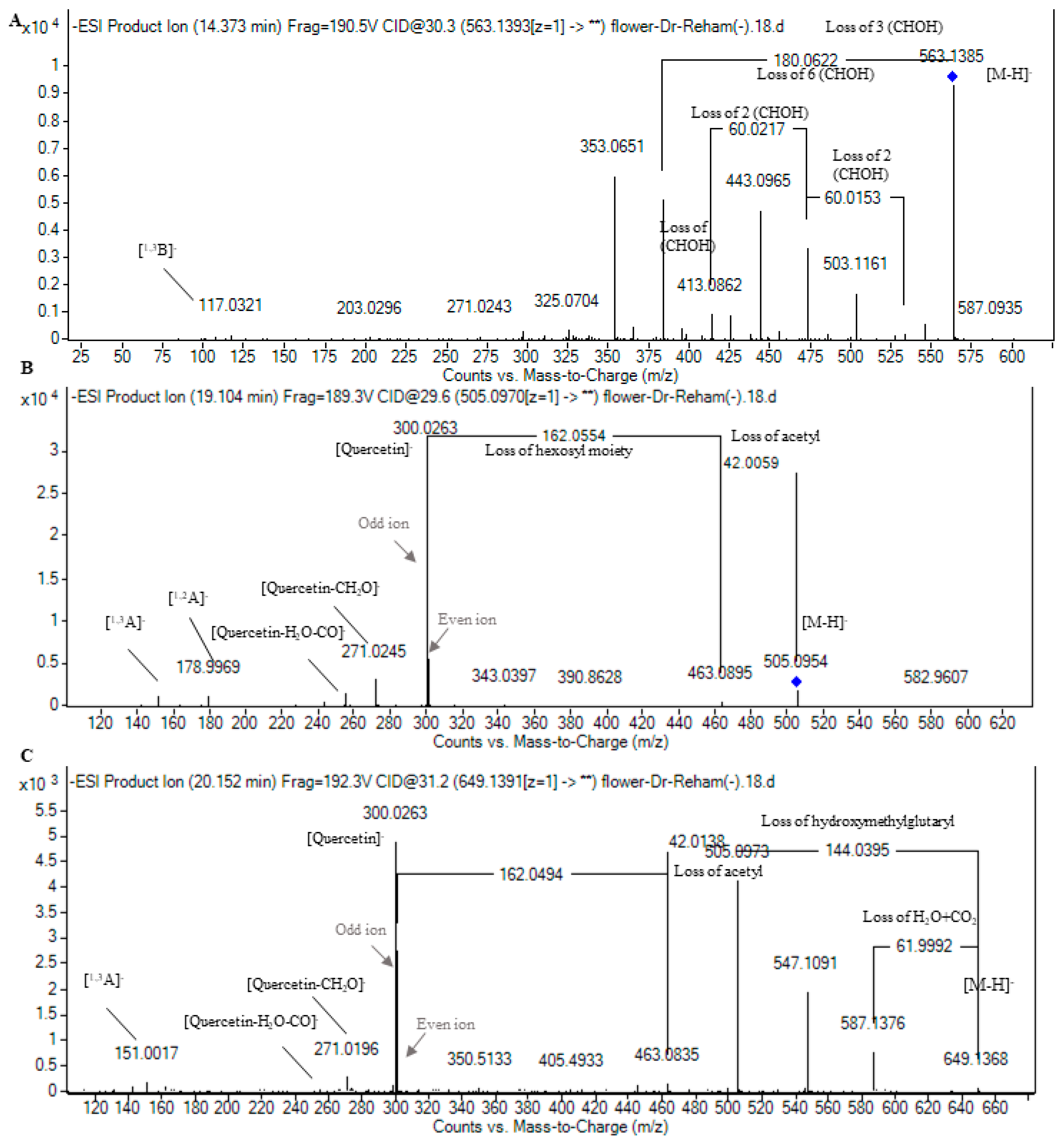
| 11 | 13.73 | 563.1391 | 564.1479 | C26H28O14 | 94.73 | 2.18 | 545.1298, 503.1155, 473.1084, 443.0963, 413.0882, 383.0759, 353.0643 | 13 | Apigenin-C-pentoside-C-hexoside I | Fla | [22,35] | 5.00 × 104 | 1.64 × 104 | 0 | 0 | |
| 12 | 14.06 | 447.0921 | 449.1047 | 448.1006 | C21H20O11 | 96.55 | 2.67 | 357.0597, 327.0495, 297.0394, 151.0380, 133.0290 | 12 | Luteolin-C-hexoside I | Fla | [21,22] | 7.42 × 105 | 0 | 3.82 × 105 | 0 |
| 13 | 14.40 | 563.1391 | 564.1479 | C26H28O14 | 93.05 | 2.22 | 545.1287, 503.1161, 473.1078, 443.0965, 413.0862, 383.0762, 353.0651, 117.0321 | 13 | Apigenin-C-pentoside-C-hexoside II | Fla | [22,35] | 2.29 × 105 | 1.36 × 105 | 0 | 0 | |
| 14 | 14.40 | 447.0931 | 449.1062 | 448.1006 | C21H20O11 | 96.81 | 1.10 | 357.0599, 327.0497, 297.0400 | 12 | Luteolin-C-hexoside II | Fla | [21,22] | 2.41 × 105 | 0 | 1.41 × 105 | 0 |
| 15 | 14.74 | 415.1235 | 416.1309 | C18H24O11 | 79.95 | 2.61 | 207.0629, 193.0443, 192.0415 | 7 | 10-O-acetylgeniposidic acid | Tr | [30] | 0 | 5.94 × 104 | 0 | 0 | |
| 16 | 15.15 | 563.1400 | 564.1479 | C26H28O14 | 91.31 | 0.60 | 545.1366, 503.1144, 473.1106, 443.0963, 413.0834, 383.0775, 353.0653 | 13 | Apigenin-C-pentoside-C-hexoside III | Fla | [22,35] | 5.01 × 104 | 2.10 × 104 | 0 | 0 | |
| 17 | 15.59 | 563.1391 | 564.1479 | C26H28O14 | 92.70 | 2.84 | 545.1250, 503.1098, 473.1091, 443.0967, 413.0783, 383.0747, 353.0636, 117.0324 | 13 | Apigenin-C-pentoside-C-hexoside IV | Fla | [22,35] | 3.40 × 104 | 1.61 × 104 | 0 | 0 | |
| 18 | 15.71 | 431.0978 | 433.1131 | 432.1056 | C21H20O10 | 96.05 | 1.21 | 341.0636, 311.0557, 117.0319 | 12 | Apigenin-C-hexoside I | Fla | [30] | 9.58 × 105 | 0 | 1.83 × 105 | 0 |
| 19 | 15.90 | 431.0979 | 433.1124 | 432.1056 | C21H20O10 | 98.57 | 1.42 | 341.0658, 311.0549, 117.0343 | 12 | Apigenin-C-hexoside II | Fla | [30] | 8.29 × 105 | 0 | 5.14 × 105 | 0 |
| 20 | 15.98 | 463.0868 | 464.0941 | C21H20O12 | 84.12 | 1.98 | 301.0359, 300.0260, 271.0224, 255.0285, 178.9977, 151.0028 | 14 | Quercetin-O-hexoside | Flo | [21,22,28] | 9.23 × 104 | 0 | 0 | 0 | |
| 21 | 16.57 | 447.0931 | 448.1006 | C21H20O11 | 96.81 | 1.10 | 285.0396 | 12 | Luteolin-O-hexoside | Fla | [25,30] | 9.70 × 104 | 0 | 4.34 × 104 | 0 | |
| 22 | 17.23 | 187.0967 | 188.1049 | C9H16O4 | 96.23 | 4.4 | 125.0966, 97.0656 | 2 | Azelaic acid | Oa | [24,25] | 5.20 × 104 | 4.36 × 105 | 0 | 0 | |
| 23 | 17.49 | 607.1305 | 608.1366 | C27H28O16 | 84.12 | 1.98 | 505.0970, 463.0873, 301.0336, 300.0264, 271.0257, 255.0277, 178.9982 | 14 | Quercetin-O-hydroxymethylglutaryl-hexoside | Flo | [14] | 8.73 × 104 | 0 | 0 | 0 | |
| 24 | 17.95 | 505.0966 | 529.0934 * | 506.105 | C23H22O13 | 71.34 | 3.58 | 463.0895, 301.0341, 300.0263, 271.0245, 255.0285, 178.9969, 151.0021 | 13 | Quercetin-O-acetylhexoside I | Flo | [30] | 3.27 × 104 | 0 | 0 | 0 |
| 25 | 18.38 | 431.0973 | 432.1056 | C21H20O10 | 81.61 | 2.36 | 269.0443, 268.0367 | 12 | Apigenin-O-hexoside I | Fla | [22,30] | 3.48 × 104 | 3.85 × 104 | 2.83 × 104 | 0 | |
| 26 | 19.02 | 649.1401 | 650.1467 | C29H30O17 | 83.5 | 1.07 | N.D. | 15 | Quercetin-O-hydroxymethylglutaryl acetylhexoside I | Flo | 6.02 × 104 | 0 | 0 | 0 | ||
| 27 | 19.08 | 505.0979 | 506.105 | C23H22O13 | 97.66 | 1.95 | 463.0920, 301.0337, 300.0266, 271.0244, 255.0299, 151.0015 | 13 | Quercetin-O-acetylhexoside II | Flo | [30] | 3.70 × 105 | 8.40 × 103 | 0 | 0 | |
| 28 | 19.59 | 431.0969 | 432.1056 | C21H20O10 | 94.62 | 3.25 | 269.0441, 268.0369 | 12 | Apigenin-O-hexoside II | Fla | [22,30] | 1.36 × 105 | 0 | 9.39 × 103 | 0 | |
| 29 | 20.19 | 649.139 | 650.1467 | C29H30O17 | 85.47 | 2.53 | 587.1376, 505.0973, 463.0835, 301.0341, 300.0263, 271.0196, 255.0196, 178.9968, 151.0017 | 15 | Quercetin-O-hydroxymethylglutaryl acetylhexoside II | Flo | 6.93 × 104 | 0 | 0 | 0 | ||
| 30 | 20.97 | 649.1389 | 650.1467 | C29H30O17 | 86.02 | 3.81 | 587.1365, 505.0968, 463.0818, 301.0327, 300.0261, 271.0219, 255.0196, 178.9967 | 15 | Quercetin-O-hydroxymethylglutaryl acetylhexoside III | Flo | 7.53 × 104 | 0 | 0 | 0 | ||
| 31 | 21.90 | 285.0392 | 286.0469 | C15H10O6 | 79.66 | 4.0 | 151.0009, 133.0289 | 11 | Luteolin | Fla | [21] | 4.47 × 104 | 0 | 4.66 × 104 | 0 | |
| 32 | 22.59 | 431.0972 | 432.1056 | C21H20O10 | 94.74 | −0.9 | 269.0441, 268.0369, 255.0620, 225.0554 | 12 | Apigenin-O-hexoside III | Fla | [22,30] | 3.63 × 105 | 3.11 × 104 | 2.21 × 105 | 4.60 × 104 | |
| 33 | 22.61 | 735.2125 | 736.2198 | C39H40O19 | 80.22 | 2.81 | 560.1761, 367.1231, 193.0506, 175.0382 | 15 | Helonioside B | HC | [31] | 0 | 0 | 4.00 × 104 | 0 | |
| 34 | 22.86 | 517.0997 | 518.1046 | C24H22O13 | 82.33 | 0.17 | N.D. | 14 | Quercetin-O-diacetylpentoside I | Flo | [39] | 1.51 × 104 | 0 | 0 | 0 | |
| 35 | 23.42 | 389.1448 | 413.1406 * | 390.1516 | C17H26O10 | 82.94 | 1.28 | 227.0728, 189.0752, 145.0495, 127.0395, 83.0499 | 5 | Loganin | Tr | [30] | 7.80 × 104 | 4.44 × 104 | 1.46 × 105 | 1.07 × 104 |
| 36 | 23.51 | 517.0973 | 518.1046 | C24H22O13 | 86.03 | 1.47 | 475.0975, 301.03327, 300.0261, 271.0201, 255.0266, 178.9967 | 14 | Quercetin-O-diacetylpentoside II | Flo | [39] | 1.05 × 105 | 0 | 1.83 × 104 | 1.72 × 103 | |
| 37 | 23.74 | 269.0442 | 270.0528 | C15H10O5 | 96.2 | 4.6 | 225.0509, 151.0038, 117.0344 | 11 | Apigenin | Fla | [22,28] | 2.43 × 104 | 0 | 3.59 × 104 | 0 | |
| 38 | 24.20 | 779.2185 | 780.2251 | C39H40O19 | 95.07 | 1.44 | 634.1851, 633.1764, 488.1500, 487.1413, 163.0387, 145.0287 | 20 | Hydropiperoside | HC | [32] | 0 | 0 | 6.07 × 104 | 0 | |
| 39 | 24.31 | 285.0390 | 286.0469 | C15H10O6 | 91.06 | 5.5 | 257.0450, 227.0341 | 11 | Kaempferol | Flo | [22,28] | 1.23 × 105 | 0 | 0 | 0 | |
| 40 | 25.75 | 187.1333 | 188.1405 | C10H20O3 | 97.44 | 3.74 | 168.7586, 125.2039 | 1 | Hydroxydecanoic acid | Fa | [30] | 0 | 7.95 × 105 | 0 | 0 | |
| 41 | 26.07 | 955.2654 | 956.2717 | C49H48O20 | 82.94 | 1.28 | 810.2292, 664.1955, 471.1293, 356.1108, 193.0473, 163.0371, 145.0285, 119.0451 | 26 | Vanicoside B I | HC | [34] | 0 | 0 | 1.85 × 105 | 0 | |
| 42 | 26.13 | 315.0502 | 316.0572 | C16H12O7 | 90.09 | 3.37 | 300.0249, 271.0239, 255.0215 | 11 | Isorhamnetin | Flo | [28,35] | 3.94 × 104 | 0 | 0 | 0 | |
| 43 | 26.17 | 985.275 | 986.2825 | C50H50O21 | 92.79 | 2.2 | 810.2335, 664.2372, 502.0949, 193.0443, 175.0392, 145.0303, 119.0451 | 26 | Lapathoside A I | HC | [33] | 0 | 0 | 1.18 × 105 | 0 | |
| 44 | 26.29 | 1015.289 | 1016.2925 | C51H52O22 | 91.96 | 1.03 | 870.2553, 840.2440, 694.2138, 664.1926, 357.1271, 193.0485, 175.0382, 145.0274 | 26 | Lapathoside B I | HC | [33] | 0 | 0 | 9.33 × 104 | 0 | |
| 45 | 26.41 | 955.2657 | 956.2717 | C49H48O20 | 79.61 | 2.47 | 810.2354, 664.2075, 163.0393, 145.0269, 119.0451 | 26 | Vanicoside B II | HC | [34] | 0 | 0 | 2.87 × 104 | 0 | |
| 46 | 26.41 | 985.2775 | 986.2825 | C50H50O21 | 89.36 | 1.06 | N.D. | 26 | Lapathoside A II | HC | [33] | 0 | 0 | 2.66 × 104 | 0 | |
| 47 | 26.50 | 1015.285 | 1016.2925 | C51H52O22 | 86.13 | 1.5 | N.D. | 26 | Lapathoside B II | HC | [33] | 0 | 0 | 2.87 × 104 | 0 | |
| 48 | 27.39 | 293.1748 | 295.1913 | 294.1818 | C17H26O4 | 91.2 | 3.6 | 277.2879, 221.1539, 177.01912 | 5 | 6-Gingerol | Ph | [30,36] | 2.01 × 105 | 1.27 × 105 | 2.55 × 105 | 1.91 × 105 |
| 49 | 27.59 | 213.1489 | 214.1562 | C12H22O3 | 71.11 | 4.2 | 153.192 | 2 | Hexanoic anhydride | Fa | [30] | 0 | 0 | 0 | 5.33 × 104 | |
| 50 | 27.96 | 395.2197 | 394.2124 | C25H30O4 | 99.8 | −2.2 | N.D. | Kazinol A | Hf | [30] | 0 | 0 | 0 | 1.60 × 105 | ||
| 51 | 28.46 | 269.0442 | 270.0528 | C15H10O5 | 96.2 | 4.6 | 241.0492, 225.0544 | 11 | Galangin | Flo | [30] | 4.14 × 105 | 1.33 × 104 | 5.88 × 105 | 4.92 × 105 | |
| 52 | 28.75 | 293.2105 | 294.2211 | C18H30O3 | 90.5 | 5.2 | 275.2024, 231.2131 | 4 | Hydroxylinolenic acid I | Fa | [30] | 1.43 × 105 | 8.59 × 105 | 6.80 × 104 | 0 | |
| 53 | 29.26 | 295.2269 | 296.2364 | C18H32O3 | 95.3 | 3.3 | 277.2158, 232.8021 | 3 | Hydroxylinoleic acid I | Fa | [30,40] | 6.40 × 105 | 1.44 × 106 | 4.09 × 105 | 0 | |
| 54 | 29.3 | 345.2048 * | 322.2153 | C19H30O4 | 98.8 | 2.2 | 279.2286, 179.0477 | 5 | 8-Gingerol | Ph | [30,36] | 8.91 × 104 | 7.02 × 104 | 0 | 7.72 × 104 | |
| 55 | 29.37 | 297.2427 | 298.2520 | C18H34O3 | 96.7 | 3.1 | 279.2303, 233.181 | 2 | Hydroxyoleic acid I | Fa | [24,30] | 0 | 1.37 × 105 | 0 | 0 | |
| 56 | 29.56 | 293.2118 | 317.2079 * | 294.2211 | C18H30O3 | 80.9 | 1.8 | 249.2155 | 4 | Hydroxylinolenic acid II | Fa | [30] | 2.13 × 105 | 3.87 × 105 | 1.52 × 105 | 0 |
| 57 | 29.56 | 297.2427 | 298.2520 | C18H34O3 | 97.6 | 2.8 | 279.2313 | 2 | Hydroxyoleic acid II | Fa | [24,30] | 1.10 × 105 | 7.65 × 105 | 5.82 × 104 | 0 | |
| 58 | 29.74 | 293.2118 | 317.2088 * | 294.2211 | C18H30O3 | 80.9 | 1.8 | 249.2094 | 4 | Hydroxylinolenic acid III | Fa | [30] | 0 | 0 | 2.29 × 105 | 0 |
| 59 | 29.81 | 297.2425 | 298.2520 | C18H34O3 | 90.6 | 4.3 | 279.2310, 233.1859 | 2 | Hydroxyoleic acid III | Fa | [24,30] | 5.04 × 105 | 6.40 × 105 | 2.33 × 105 | 0 | |
| 60 | 30.17 | 295.2266 | 319.2224 * | 296.2364 | C18H32O3 | 94.0 | 3.8 | 277.2160, 261.0815, 232.92, 199.1686 | 3 | Hydroxylinoleic acid II | Fa | [30,40] | 2.73 × 105 | 1.48 × 105 | 7.01 × 104 | 0 |
 highest value.
highest value.2.1.2. Flavonoids
Flavones
Flavonols
2.1.3. Terpenes
2.1.4. Fatty Acids
2.1.5. Amino Acids, Organic Acids, and Sugars
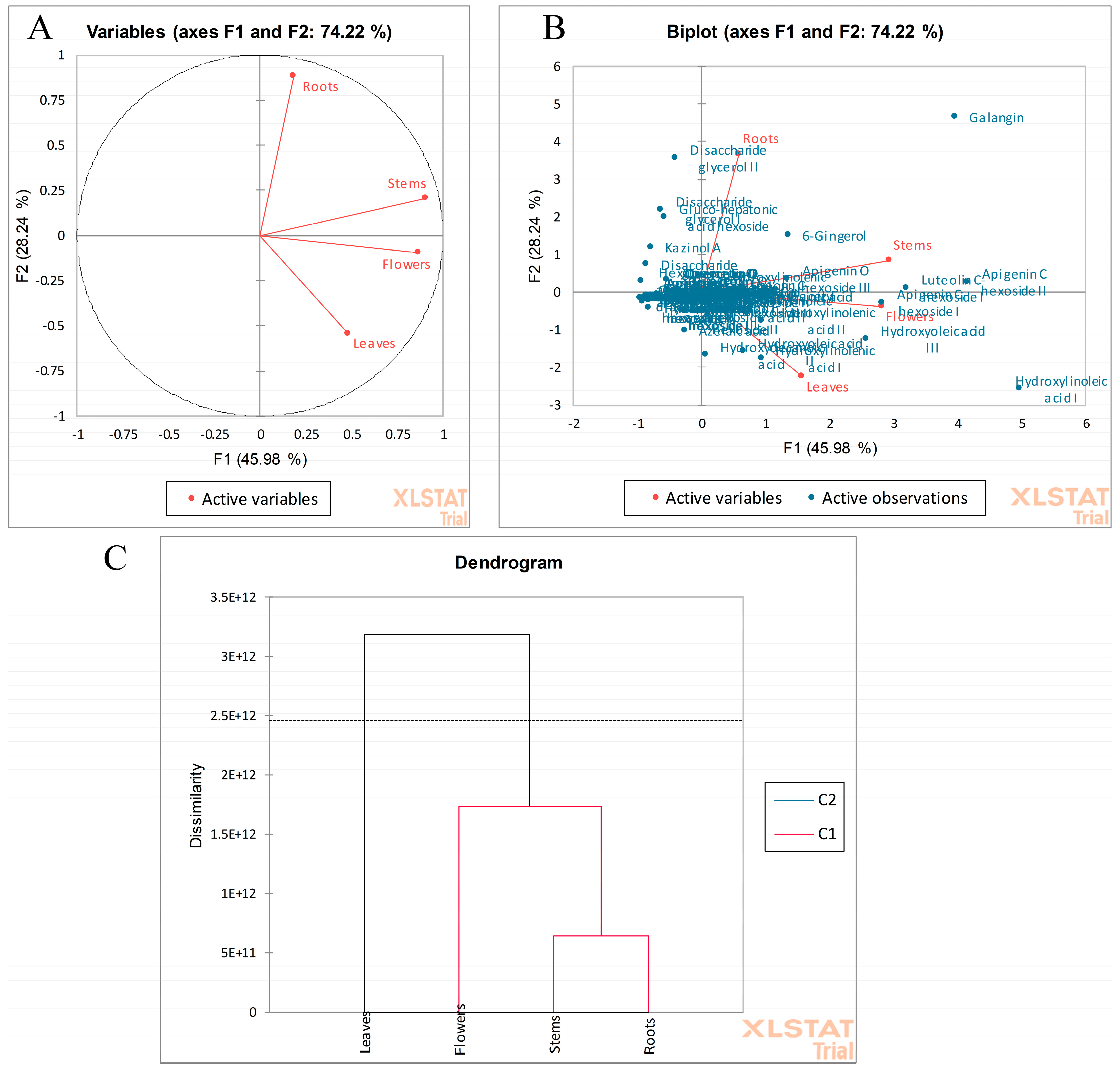
2.2. Comparison of the R. vesicarius Different Parts by Multivariate Data Analysis
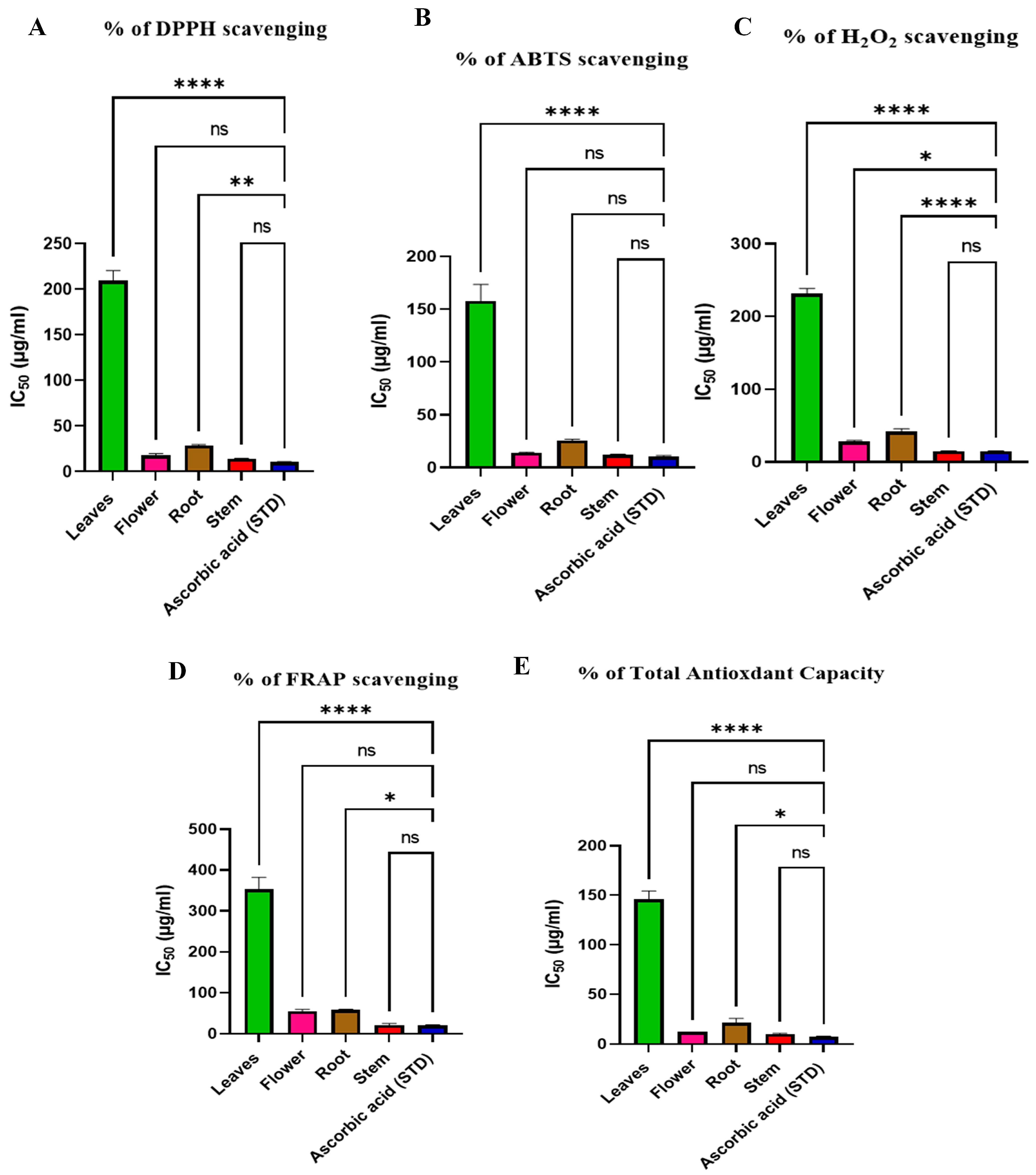
2.3. Antioxidant Evaluation of Different Parts of Rumex vesicarius Extracts
2.4. Docking against Antioxidant Molecular Targets
2.4.1. Interactions with the NADPH Receptor
2.4.2. Interactions with the Human Peroxiredoxin 5 Protein (PRDX5)
2.5. ADME Predictions of the Selected Compounds
3. Materials and Methods
3.1. Plant Material
3.1.1. Plant Collection
3.1.2. Preparation of Different Parts Extracts
3.2. RP-HPLC-ESI-MS and Tandem MS/MS Analysis
3.3. Determination of the Antioxidant Activity
3.3.1. Evaluation of the Antioxidant Activity Using DPPH Scavenging
3.3.2. Evaluation of the Antioxidant Activity Using ABTS Radical Scavenging
3.3.3. Evaluation of the Antioxidant Activity Using H2O2 Scavenging Assay
3.3.4. Evaluation of the Antioxidant Activity Using FRAP Scavenging
3.3.5. Determination of the Total Antioxidant Capacity (TAC)
3.4. Statistical Analysis
3.5. In Silico Docking Study and ADME Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nandi, S.; Khatua, S.; Nag, A.; Sen, S.; Chakraborty, N.; Naskar, A.; Acharya, K.; Hassan Mekky, R.; del Mar Contreras, M.; Calina, D.; et al. Dolastatins and their analogues present a compelling landscape of potential natural and synthetic anticancer drug candidates. Curr. Res. Biotechnol. 2023, 7, 100167. [Google Scholar] [CrossRef]
- Najahi, A.; Alaya, A.; Mufti, A.; Tir, M.; Contreras, M.d.M.; Feriani, A.; Harrath, A.H.; Hfaiedh, N.; Tlili, N. HPLC-QTOF-MS analysis of Polygonum maritimum aerial parts extract and focus on the therapeutic potential against ethylene glycol-induced lithiasis in rats. Food Biosci. 2024, 57, 103481. [Google Scholar] [CrossRef]
- Paunovic, D.; Rajkovic, J.; Novakovic, R.; Grujic-Milanovic, J.; Mekky, R.H.; Popa, D.; Calina, D.; Sharifi-Rad, J. The potential roles of gossypol as anticancer agent: Advances and future directions. Chin. Med. 2023, 18, 163. [Google Scholar] [CrossRef]
- Idris, O.A.; Wintola, O.A.; Afolayan, A.J. Phytochemical and antioxidant activities of Rumex crispus L. in treatment of gastrointestinal helminths in Eastern Cape Province, South Africa. Asian Pac. J. Trop. Biomed. 2017, 7, 1071–1078. [Google Scholar] [CrossRef]
- Faysal, M.; Dehbia, Z.; Zehravi, M.; Sweilam, S.H.; Haque, M.A.; Kumar, K.P.; Chakole, R.D.; Shelke, S.P.; Sirikonda, S.; Nafady, M.H.; et al. Flavonoids as Potential Therapeutics Against Neurodegenerative Disorders: Unlocking the Prospects. Neurochem. Res. 2024. [Google Scholar] [CrossRef]
- Elshnoudy, I.A.; Elkhouly, A.M.; Masoud, M.; Rabea, H.A.; Mansour, F.R. Medicinal plants cultivated in Egypt with anticancer potential; a systematic review. Phytochem. Rev. 2024. [Google Scholar] [CrossRef]
- Ammar, S.; Abidi, J.; Vlad Luca, S.; Boumendjel, M.; Skalicka-Wozniak, K.; Bouaziz, M. Untargeted metabolite profiling and phytochemical analysis based on RP-HPLC-DAD-QTOF-MS and MS/MS for discovering new bioactive compounds in Rumex algeriensis flowers and stems. Phytochem. Anal. 2020, 31, 616–635. [Google Scholar] [CrossRef]
- Vasas, A.; Orbán-Gyapai, O.; Hohmann, J. The Genus Rumex: Review of traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2015, 175, 198–228. [Google Scholar] [CrossRef]
- Shah, A.; Singh, T.; Vijayvergia, R. In vitro antioxidant properties and total phenolic and flavonoid contents of Rumex vesicaius L. Int. J. Pharm. Pharm. Sci. 2015, 7, 81–84. [Google Scholar]
- Li, J.J.; Li, Y.X.; Li, N.; Zhu, H.T.; Wang, D.; Zhang, Y.J. The genus Rumex (Polygonaceae): An ethnobotanical, phytochemical and pharmacological review. Nat. Prod. Bioprospect. 2022, 12, 21. [Google Scholar] [CrossRef]
- Mishra, A.P.; Sharifi-Rad, M.; Shariati, M.A.; Mabkhot, Y.N.; Al-Showiman, S.S.; Rauf, A.; Salehi, B.; Župunski, M.; Gusain, P.; Sharifi-Rad, J. Bioactive compounds and health benefits of edible Rumex species-A review. Cell Mol. Biol. 2018, 6, 27–34. [Google Scholar] [CrossRef]
- Khare, C.P. Indian Medicinal Plants: An Illustrated Dictionary; Springer Science & Business Media: Berlin, Germany, 2008. [Google Scholar]
- Fatima, N.; Zia, M.; Rehman, R.; Rizvi, Z.F.; Ahmad, S.; Mirza, B.; Chaudhary, M.F. Biological activities of Rumex dentatus L: Evaluation of methanol and hexane extracts. Afr. J. Biotechnol. 2009, 8, 6945–6951. [Google Scholar]
- Abidi, J.; Ammar, S.; Ben Brahim, S.; Skalicka-Wozniak, K.; Ghrabi-Gammar, Z.; Bouaziz, M. Use of ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry system as valuable tool for an untargeted metabolomic profiling of Rumex tunetanus flowers and stems and contribution to the antioxidant activity. J. Pharm. Biomed. Anal. 2019, 162, 66–81. [Google Scholar] [CrossRef]
- El-Hawary, S.A.; Sokkar, N.M.; Ali, Z.Y.; Yehia, M.M. A profile of bioactive compounds of Rumex vesicarius L. J. Food Sci. 2011, 76, C1195–C1202. [Google Scholar] [CrossRef]
- Davella, R.; Mamidala, E. Molecular Docking and Dynamic Simulation Studies of compounds from Rumex vesicarius against Maltase-Glucoamylase to treat type 2 Diabetes. Ann. Rom. Soc. Cell Biol. 2021, 25, 21062–21077. [Google Scholar]
- Hasan, M.M.; Tasmin, M.S.; El-Shehawi, A.M.; Elseehy, M.M.; Reza, M.A.; Haque, A. R. vesicarius L. exerts nephroprotective effect against cisplatin-induced oxidative stress. BMC Complement. Med. Ther. 2021, 21, 225. [Google Scholar] [CrossRef]
- Jerezano Alberto, V.; Ríos Saúl, A.; Tepancal-Gomez, E.; Salas-Mendosa, E.; Villanueva, L. Some traditional medicinal plants of North region from Puebla, Mexico: Uses and potential pharmacological activity of Rumex spp. Nat. Prod. Chem. Res. 2016, 4, 2. [Google Scholar]
- Elbermawi, A.; Darwish, M.S.; El-Awady, A.A.; Zaki, A.A.; Qiu, L.; Samra, R.M. Isolation and biological activities of compounds from Rumex vesicarius L. and their use as a component of a synbiotic preparation. Food Chem. X 2022, 14, 100306. [Google Scholar] [CrossRef]
- El-Bakry, A.; Mostafa, H.; Eman, A.A. Antibacterial and antioxidant activities of seedlings of Rumex vesicarius L. (Polygonaceae). J. Med. Plan. Res. 2013, 7, 1754. [Google Scholar]
- Mekky, R.H.; Abdel-Sattar, E.; Segura-Carretero, A.; Contreras, M.d.M. Metabolic Profiling of the Oil of Sesame of the Egyptian Cultivar ‘Giza 32′ Employing LC-MS and Tandem MS-Based Untargeted Method. Foods 2021, 10, 298. [Google Scholar] [CrossRef]
- Mekky, R.H.; Abdel-Sattar, E.; Segura-Carretero, A.; Contreras, M.d.M. A comparative study on the metabolites profiling of linseed cakes from Egyptian cultivars and antioxidant activity applying mass spectrometry-based analysis and chemometrics. Food Chem. 2022, 395, 133524. [Google Scholar] [CrossRef]
- Mekky, R.H.; Thabet, M.M.; Rodríguez-Pérez, C.; Elnaggar, D.M.Y.; Mahrous, E.A.; Segura-Carretero, A.; Abdel-Sattar, E. Comparative metabolite profiling and antioxidant potentials of seeds and sprouts of three Egyptian cultivars of Vicia faba L. Food Res. Int. 2020, 136, 109537. [Google Scholar] [CrossRef]
- Tej, A.; Mekky, R.H.; Contreras, M.d.M.; Feriani, A.; Tir, M.; L’Taief, B.; Alshaharni, M.O.; Faidi, B.; Mnafgui, K.; Abbes, Z.; et al. Eucalyptus torquata seeds: Investigation of phytochemicals profile via LC-MS and its potential cardiopreventive capacity in rats. Food Biosci. 2024, 59, 103666. [Google Scholar] [CrossRef]
- Saeed, N.M.; Ramadan, L.A.; El-Sabbagh, W.A.; Said, M.A.; Abdel-Rahman, H.M.; Mekky, R.H. Exploring the anti-osteoporosis potential of Petroselinum crispum (Mill.) Fuss extract employing experimentally ovariectomized rat model and network pharmacology approach. Fitoterapia 2024, 175, 105971. [Google Scholar] [CrossRef]
- ChemSpider. Available online: http://www.chemspider.com (accessed on 15 May 2024).
- Egyptian-Knowledge-Bank. Available online: https://www.ekb.eg/ (accessed on 15 May 2024).
- KNApSAcK-Core-System. Available online: http://www.knapsackfamily.com/knapsack_jsp/top.html (accessed on 15 May 2024).
- PubChem. Available online: http://pubchem.ncbi.nlm.nih.gov (accessed on 15 May 2024).
- Reaxys. Available online: http://www.reaxys.com (accessed on 15 May 2024).
- Sun, X.; Zimmermann, M.L.; Campagne, J.-M.; Sneden, A.T. New Sucrose Phenylpropanoid Esters from Polygonum perfoliatum. J. Nat. Prod. 2000, 63, 1094–1097. [Google Scholar] [CrossRef]
- Takasaki, M.; Kuroki, S.; Kozuka, M.; Konoshima, T. New Phenylpropanoid Esters of Sucrose from Polygonum lapathifolium. J. Nat. Prod. 2001, 64, 1305–1308. [Google Scholar] [CrossRef]
- Fan, P.; Terrier, L.; Hay, A.E.; Marston, A.; Hostettmann, K. Antioxidant and enzyme inhibition activities and chemical profiles of Polygonum sachalinensis F.Schmidt ex Maxim (Polygonaceae). Fitoterapia 2010, 81, 124–131. [Google Scholar] [CrossRef]
- Shin, H.; Jang, J.; Lee, M.K.; Lee, K.Y. Cell extraction method coupled with LC-QTOF MS/MS analysis for predicting neuroprotective compounds from Polygonum tinctorium. J. Pharm. Biomed. Anal. 2022, 220, 114988. [Google Scholar] [CrossRef]
- Mekky, R.H.; Abdel-Sattar, E.; Segura-Carretero, A.; Contreras, M.d.M. Phenolic Compounds from Sesame Cake and Antioxidant Activity: A New Insight for Agri-Food Residues’ Significance for Sustainable Development. Foods 2019, 8, 432. [Google Scholar] [CrossRef]
- Asamenew, G.; Kim, H.-W.; Lee, M.-K.; Lee, S.-H.; Kim, Y.J.; Cha, Y.-S.; Yoo, S.M.; Kim, J.-B. Characterization of phenolic compounds from normal ginger (Zingiber officinale Rosc.) and black ginger (Kaempferia parviflora Wall.) using UPLC–DAD–QToF–MS. Eur. Food Res. Technol. 2018, 245, 653–665. [Google Scholar] [CrossRef]
- Santos-Fandila, A.; Zafra-Gomez, A.; Barranco, A.; Navalon, A.; Rueda, R.; Ramirez, M. Quantitative determination of beta-hydroxymethylbutyrate and leucine in culture media and microdialysates from rat brain by UHPLC-tandem mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 2863–2872. [Google Scholar] [CrossRef]
- Sun, L.; Jiao, H.; Gao, B.; Yuanzi, Q.; Zhang, H.; Wang, Y.; Ou, N.; Yan, Z.; Zhou, H. Hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry method for the simultaneous determination of l-valine, l-leucine, l-isoleucine, l-phenylalanine, and l-tyrosine in human serum. J. Sep. Sci. 2015, 38, 3876–3883. [Google Scholar] [CrossRef]
- Chin, Y.W.; Kim, J. Three new flavonol glycosides from the aerial parts of Rodgersia podophylla. Chem. Pharm. Bull. 2006, 54, 234–236. [Google Scholar] [CrossRef]
- Fayek, N.M.; Mekky, R.H.; Dias, C.N.; Kropf, M.; Heiss, A.G.; Wessjohann, L.A.; Farag, M.A. UPLC-MS Metabolome-Based Seed Classification of 16 Vicia Species: A Prospect for Phyto-Equivalency and Chemotaxonomy of Different Accessions. J. Agric. Food Chem. 2021, 69, 5252–5266. [Google Scholar] [CrossRef]
- Cuyckens, F.; Claeys, M. Mass spectrometry in the structural analysis of flavonoids. J. Mass Spectro. 2004, 39, 1–15. [Google Scholar] [CrossRef]
- Sweilam, S.H.; Abdel Bar, F.M.; Foudah, A.I.; Alqarni, M.H.; Elattal, N.A.; El-Gindi, O.D.; El-Sherei, M.M.; Abdel-Sattar, E. Phytochemical, Antimicrobial, Antioxidant, and In Vitro Cytotoxicity Evaluation of Echinops erinaceus Kit Tan. Separations 2022, 9, 447. [Google Scholar] [CrossRef]
- Gad, H.; Al-Sayed, E.; Ayoub, I. Phytochemical discrimination of Pinus species based on GC–MS and ATR-IR analyses and their impact on Helicobacter pylori. Phytochem. Anal. 2021, 32, 820–835. [Google Scholar] [CrossRef]
- Abd Elhafeez, M.S. Alternative natural therapeutic plants and diabetes mellitus. ERU Res. J. 2024, 3, 871–885. [Google Scholar] [CrossRef]
- Nassar, A.Y.; Meligy, F.Y.; Abd-Allah, G.M.; Khallil, W.A.; Sayed, G.A.; Hanna, R.T.; Nassar, G.A.; Bakkar, S.M. Oral acetylated whey peptides (AWP) as a potent antioxidant, anti-inflammatory, and chelating agent in iron-overloaded rats’ spleen. J. Funct. Foods 2023, 102, 105444. [Google Scholar] [CrossRef]
- Abd El Hafeez, M.S.; El Gindi, O.; Hetta, M.H.; Aly, H.F.; Ahmed, S.A. Quality Control, Anti-Hyperglycemic, and Anti-Inflammatory Assessment of Colvillea racemosa Leaves Using In Vitro, In Vivo Investigations and Its Correlation with the Phytoconstituents Identified via LC-QTOF-MS and MS/MS. Plants 2022, 11, 830. [Google Scholar] [CrossRef]
- Elhawary, E.A.; Korany, D.A.; Eldahshan, O.A.; Singab, A.N.B. Insights on Dietary Anticancer Products: Food Supplements, Prebiotics, and Probiotics. In Interdisciplinary Cancer Research; Springer: Cham, Switzerland, 2024. [Google Scholar]
- Beddou, F.; Bekhechi, C.; Ksouri, R.; Chabane Sari, D.; Atik Bekkara, F. Potential assessment of Rumex vesicarius L. as a source of natural antioxidants and bioactive compounds. J. Food Sci. Technol. 2015, 52, 3549–3560. [Google Scholar] [CrossRef][Green Version]
- Salama, S.A.; Al-Faifi, Z.E.; Masood, M.F.; El-Amier, Y.A. Investigation and biological assessment of Rumex vesicarius L. extract: Characterization of the chemical components and antioxidant, antimicrobial, cytotoxic, and anti-dengue vector activity. Molecules 2022, 27, 3177. [Google Scholar] [CrossRef]
- Mostafa, H.; Elbakry, A.; Eman, A.A. Evaluation of antibacterial and antioxidant activities of different plant parts of Rumex vesicarius L. (Polygonaceae). Int. J. Pharm. Pharm. Sci. 2011, 3, 109–118. [Google Scholar]
- Panday, A.; Sahoo, M.K.; Osorio, D.; Batra, S. NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015, 12, 5–23. [Google Scholar] [CrossRef]
- Sweilam, S.H.; Abdel Bar, F.M.; Foudah, A.I.; Alqarni, M.H.; El-Gindi, O.D.; El-Sherei, M.M.; Abdel-Sattar, E. Phytochemical Investigation, Antiulcer, Cyclooxygenase-2, and 15-Lipoxygenase Inhibitory Activities of Echinops erinaceus Kit Tan. Separations 2023, 10, 76. [Google Scholar] [CrossRef]
- Vermot, A.; Petit-Härtlein, I.; Smith, S.M.E.; Fieschi, F. NADPH Oxidases (NOX): An Overview from Discovery, Molecular Mechanisms to Physiology and Pathology. Antioxidants 2021, 10, 890. [Google Scholar] [CrossRef]
- Poncin, M.A.; Van Meerbeeck, P.; Simpson, J.D.; Clippe, A.; Tyckaert, F.; Bouillenne, F.; Degand, H.; Matagne, A.; Morsomme, P.; Knoops, B.; et al. Role of the Redox State of Human Peroxiredoxin-5 on Its TLR4-Activating DAMP Function. Antioxidants 2021, 10, 1902. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Al Zahrani, N.A.; El-Shishtawy, R.M.; Elaasser, M.M.; Asiri, A.M. Synthesis of novel chalcone-based phenothiazine derivatives as antioxidant and anticancer agents. Molecules 2020, 25, 4566. [Google Scholar] [CrossRef]
- Sánchez, C.S.; González, A.T.; García-Parrilla, M.; Granados, J.Q.; De La Serrana, H.L.G.; Martínez, M.L. Different radical scavenging tests in virgin olive oil and their relation to the total phenol content. Anal. Chim. Acta 2007, 593, 103–107. [Google Scholar] [CrossRef]
- Ling, L.T.; Yap, S.-A.; Radhakrishnan, A.K.; Subramaniam, T.; Cheng, H.M.; Palanisamy, U.D. Standardised Mangifera indica extract is an ideal antioxidant. Food Chem. 2009, 113, 1154–1159. [Google Scholar] [CrossRef]
- Ruch, R.J.; Cheng, S.-j.; Klaunig, J.E. Prevention of cytotoxicity and inhibition of intercellular communication by antioxidant catechins isolated from Chinese green tea. Carcinogenesis 1989, 10, 1003–1008. [Google Scholar] [CrossRef]
- Firuzi, O.; Lacanna, A.; Petrucci, R.; Marrosu, G.; Saso, L. Evaluation of the antioxidant activity of flavonoids by “ferric reducing antioxidant power” assay and cyclic voltammetry. BBA Gen. Subj. 2005, 1721, 174–184. [Google Scholar] [CrossRef]
- Oyaizu, M. Studies on products of browning reaction antioxidative activities of products of browning reaction prepared from glucosamine. Jpn. J. Nutr. Diet. 1986, 44, 307–315. [Google Scholar] [CrossRef]
- Prieto, P.; Pineda, M.; Aguilar, M. Spectrophotometric quantitation of antioxidant capacity through the formation of a phosphomolybdenum complex: Specific application to the determination of vitamin E. Anal. Biochem. 1999, 269, 337–341. [Google Scholar] [CrossRef]
- Bouzabata, A.; Montoro, P.; Gil, K.A.; Piacente, S.; Youssef, F.S.; Al Musayeib, N.M.; Cordell, G.A.; Ashour, M.L.; Tuberoso, C.I.G. HR-LC-ESI-Orbitrap-MS-Based Metabolic Profiling Coupled with Chemometrics for the Discrimination of Different Echinops spinosus Organs and Evaluation of Their Antioxidant Activity. Antioxidants 2022, 11, 453. [Google Scholar] [CrossRef]
- Lountos, G.T.; Jiang, R.; Wellborn, W.B.; Thaler, T.L.; Bommarius, A.S.; Orville, A.M. The crystal structure of NAD(P)H oxidase from Lactobacillus sanfranciscensis: Insights into the conversion of O2 into two water molecules by the flavoenzyme. Biochemistry 2006, 45, 9648–9659. [Google Scholar] [CrossRef]
- Declercq, J.P.; Evrard, C.; Clippe, A.; Stricht, D.V.; Bernard, A.; Knoops, B. Crystal structure of human peroxiredoxin 5, a novel type of mammalian peroxiredoxin at 1.5 A resolution. J. Mol. Biol. 2001, 311, 751–759. [Google Scholar] [CrossRef]
- Noumi, E.; Snoussi, M.; Anouar, E.H.; Alreshidi, M.; Veettil, V.N.; Elkahoui, S.; Adnan, M.; Patel, M.; Kadri, A.; Aouadi, K.; et al. HR-LCMS-Based Metabolite Profiling, Antioxidant, and Anticancer Properties of Teucrium polium L. Methanolic Extract: Computational and In Vitro Study. Antioxidants 2020, 9, 1089. [Google Scholar] [CrossRef]


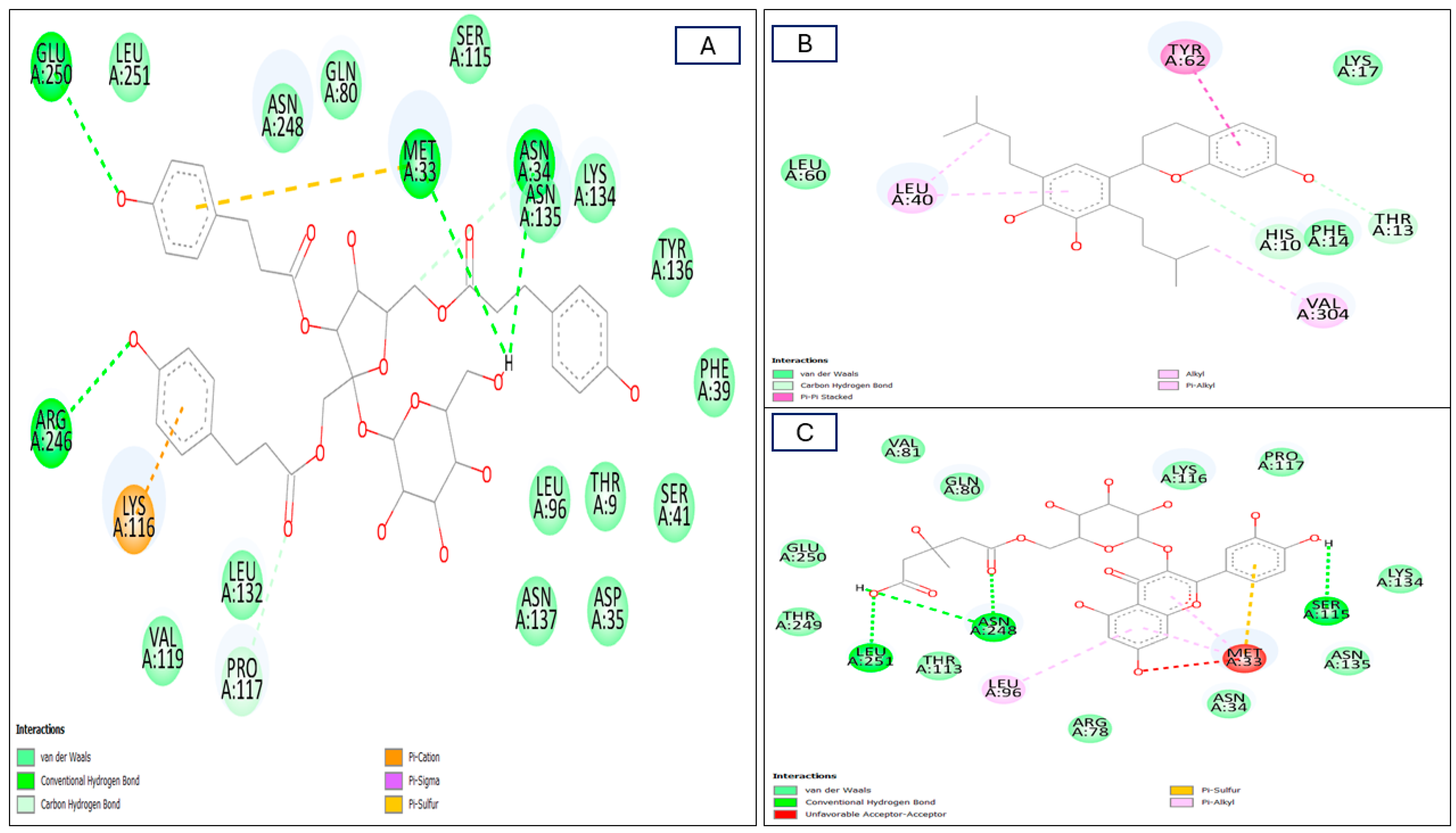
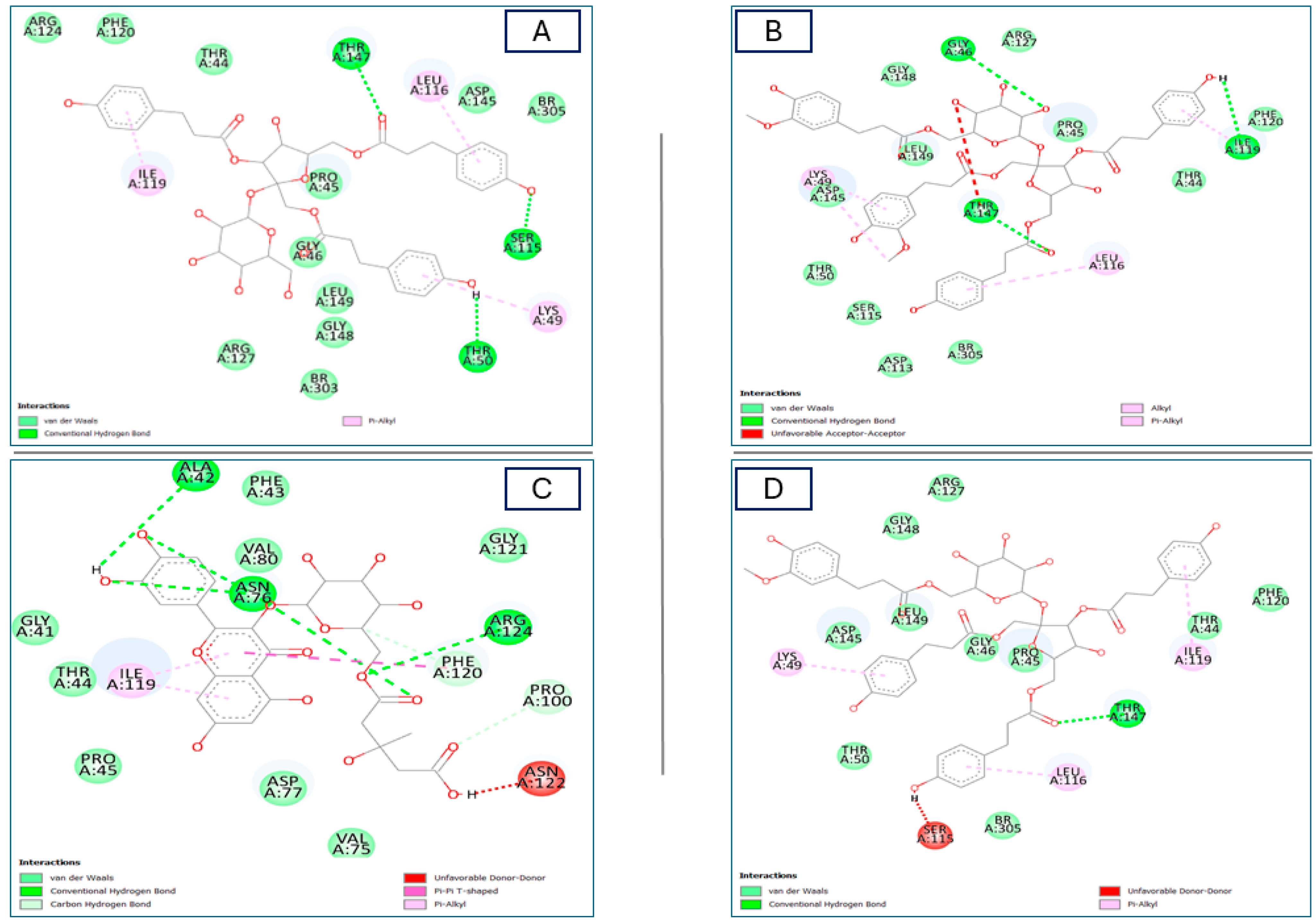

| No | Components | 2CDU (BA) | No. of Formed Bonds/AA-Residues | 1HD2 (BA) | No. of Formed Bonds/AA-Residues |
|---|---|---|---|---|---|
| 1 | Gentiobiosylglycerol | −6.3 | 2 */Asn34, Asn135, Asn248, Gln80, Glu250, Leu251, Lys116, Lys134, Met33, Pro117, Ser115, Thr113, Thr249 | −4.6 | 2/Arg127, Gly46, Pro45, Thr147 |
| 2 | Helonioside B | −7.0 | 5 */Asn34, Asn36, Asn134, Asn135, Asn137, Asn248, Asp138, Gln80, Glu32, Ile37, Leu132, Leu251, Lys116, Met33, Pro117, Ser41, Thr9, Thr82, Thr113, Tyr136, Val81, Val119 | −6.0 | 3/Arg124, Arg127, Asp145, Br303, Gly46, Gly148, Ile119, Leu116, Leu149, Phe43, Phe120, Pro45, Thr44, Thr147 |
| 3 | Hydropiperoside | −7.5 | 6/Arg246, Asn34, Asn135, Asn137, Asn248, Asp35, Gln80, Glu250, Leu96, Leu132, Leu251, Lys116, Lys134, Met33, Phe39, Pro117, Ser41, Ser115, Thr9, Tyr136, Val119 | −6.8 | 3/Arg124, Arg127, Asp145, Br303, Br305, Gly46, Gly148, Ile119, Leu116, Leu149, Lys49, Phe120, Pro45, Ser115, Thr44, Thr50, Thr147 |
| 4 | Isoorientin | −6.8 | 3 */Asn34, Asn36, Asn135, Asn248, Ile37, Lys116, Lys134, Met33, Phe39, Pro117, Ser41, Thr9, Tyr136 | −6.0 | 4/Ala42, Arg124, Asn76, Gly41, Ile119, Phe43, Phe120, Pro45, Thr44, Val80 |
| 5 | Isovitexin | −6.5 | 3 */Asn34, Asn135, Asn248, Leu132, Lys116, Lys134, Met33, Phe39, Pro117, Ser115, Thr113, Tyr136 | −5.9 | 4/Arg124, Gly41, Ile119, Phe43, Phe120, Pro45, Thr44, Val80 |
| 6 | Kazinol A | −7.7 | 5/His10, Leu40, Leu60, Lys17, Phe14, Thr13, Tyr62, Val304 | −6.1 | 5/Arg127, Asp145, Br305, Gly46, Gly148, Leu112, Leu116, Leu149, Ile119, Pro45, Ser115, Thr147 |
| 7 | Lapathoside A | −6.7 | 6/Arg78, Asn34, Asn135, Asn137, Asn248, Asp35, Asp138, Gln80, Glu250, Leu96, Leu132, Lys116, Lys134, Met33, Phe39, Pro117, Ser41, Ser115, Thr9, Tyr136, Val119 | −6.4 | 4 */Arg127, Asp113, Asp145, Br305, Gly46, Gly148, Ile119, Leu116, Leu149, Lys49, Phe120, Pro45, Ser115, Thr44, Thr50, Thr147 |
| 8 | Lapathoside B | −7.1 | 5/Arg78, Asn34, Asn36, Asn135, Asn137, Asn248, Asp35, Asp138, Gln80, Glu250, Ile37, Leu96, Leu132, Leu251, Lys116, Lys134, Met33, Phe39, Pro117, Ser38, Ser41, Ser115, Thr9, Thr82, Thr113, Tyr136, Val81, Val119 | −6.1 | 4 */Arg127, Asp145, Br303, Br305, Gly46, Gly148, Ile119, Leu116, Leu149, Lys49, Phe120, Pro45, Thr44, Thr50, Thr147 |
| 9 | Orientin | −6.2 | 4 */Asn34, Asn135, Asp35, Asp138, Leu132, Met33, Pro117, Ser115 | −5.1 | 2 */Arg127, Br303, Gly46, Gly148, Leu149, Pro45 |
| 10 | Quercetin_3-O-6″-(3-hydroxyl-3-methylglutaryl)-D-glucopyranoside | −7.6 | 5 */Arg78, Asn34, Asn135, Asn248, Gln80, Glu250, Leu96, Leu251, Lys116, Lys134, Met33, Pro117, Ser115, Thr113, Thr249, Val81 | −6.9 | 5 */Ala42, Arg124, Asn76, Asp77, Gly41, Gly121, Ile119, Phe43, Phe120, Pro45, Pro100, Thr44, Val75, Val80, |
| 11 | Vanicoside B | −6.7 | 4/Asn34, Asn36, Asn135, Asn248, Gln80, Glu250, Leu96, Leu132, Lys116, Lys134, Met33, Phe39, Pro117, Ser41, Thr9, Tyr136, Val119 | −6.2 | 3 */Arg127, Asp145, Br305, Gly46, Gly148, Ile119, Leu116, Leu149, Lys49, Phe120, Pro45, Thr44, Thr50, Thr147 |
| 12 | Vitexin | −6.3 | 5/Asn34, Asn135, Asn248, Gln80, Glu32, Glu250, Leu132, Leu251, Lys116, Lys134, Met33, Pro117, Ser115, Val81, Val119 | −5.2 | 4/Arg127, Gly46, Gly148, Ile119, Leu116, Leu149, Lys49, Pro45, Thr147 |
| FAD | −7.4 | 6/Arg78, Asn34, Asn135, Asn137, Asn248, Asp35, Asp138, Gln80, Leu96, Leu132, Lys116, Lys134, Met33, Pro117 | −6.1 | 5 */Ala42, Arg124, Arg127, Gly46, Ile119, Pro45, Thr44, Thr147 | |
| Ben | −4 | 3/Arg127, Gly46, Gly148, Leu149, Thr147 | |||
| Predictive Parameters | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADME Prediction | ||||||||||||
| Physicochemical parameters | ||||||||||||
| TPSA (Å): | 250.22 Å2 | 266.66 Å2 | 268.43 Å2 | 201.28 Å2 | 181.05 Å2 | 69.92 Å2 | 313.19 Å2 | 322.42 Å2 | 201.28 Å2 | 274.11 Å2 | 303.96 Å2 | 181.05 Å2 |
| MR | 86.06 | 173.62 | 192.27 | 108.63 | 106.61 | 118.59 | 246.63 | 253.12 | 108.63 | 142.09 | 240.14 | 106.61 |
| Drug likeness Prediction | ||||||||||||
| Bioavailability Value | 0.17 | 0.55 | 0.17 | 0.55 | ||||||||
| Synthetic accessibility | 5.97 | 6.7 | 6.86 | 5.04 | 4.99 | 4.34 | 7.95 | 8.12 | 5.17 | 6.1 | 7.77 | 5.12 |
| Absorption Prediction | ||||||||||||
| Log S (ESOL) | 1.55 | −3.2 | −5.01 | −2.7 | −2.84 | −6.42 | −6.97 | −7.07 | −2.7 | −2.84 | −6.88 | −2.84 |
| Consensus Log Po/w | −4.82 | −0.3 | 1.5 | −0.24 | −0.24 | 5.11 | 2.61 | 2.61 | −0.41 | −0.57 | −0.57 | −0.07 |
| Solubility class | Highly soluble | Soluble | Moderately soluble | Soluble | Soluble | Poorly soluble | Poorly soluble | Poorly soluble | Soluble | Soluble | Poorly soluble | Soluble |
| Pharmacokinetics | ||||||||||||
| Log Kp (skin permeation, cm/s) | −13.01 | −11.08 | −9.69 | −9.14 | −8.79 | −4.02 | −9.75 | −9.95 | −9.14 | −10.45 | −9.55 | −8.79 |
| GI absorption | Low | High | Low | |||||||||
| BBB permeant | No | |||||||||||
| Metabolism Estimation | ||||||||||||
| P-gp substrate | Yes | Yes | Yes | No | No | No | Yes | Yes | No | Yes | Yes | No |
| CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4 inhibitors | No | All No, except Yes (CYP3A4, CYP2C9) | No | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sweilam, S.H.; Abd El Hafeez, M.S.; Mansour, M.A.; Mekky, R.H. Unravelling the Phytochemical Composition and Antioxidant Potential of Different Parts of Rumex vesicarius L.: A RP-HPLC-MS-MS/MS, Chemometrics, and Molecular Docking-Based Comparative Study. Plants 2024, 13, 1815. https://doi.org/10.3390/plants13131815
Sweilam SH, Abd El Hafeez MS, Mansour MA, Mekky RH. Unravelling the Phytochemical Composition and Antioxidant Potential of Different Parts of Rumex vesicarius L.: A RP-HPLC-MS-MS/MS, Chemometrics, and Molecular Docking-Based Comparative Study. Plants. 2024; 13(13):1815. https://doi.org/10.3390/plants13131815
Chicago/Turabian StyleSweilam, Sherouk Hussein, Mohamed S. Abd El Hafeez, Mahmoud A. Mansour, and Reham Hassan Mekky. 2024. "Unravelling the Phytochemical Composition and Antioxidant Potential of Different Parts of Rumex vesicarius L.: A RP-HPLC-MS-MS/MS, Chemometrics, and Molecular Docking-Based Comparative Study" Plants 13, no. 13: 1815. https://doi.org/10.3390/plants13131815
APA StyleSweilam, S. H., Abd El Hafeez, M. S., Mansour, M. A., & Mekky, R. H. (2024). Unravelling the Phytochemical Composition and Antioxidant Potential of Different Parts of Rumex vesicarius L.: A RP-HPLC-MS-MS/MS, Chemometrics, and Molecular Docking-Based Comparative Study. Plants, 13(13), 1815. https://doi.org/10.3390/plants13131815










