The Biosynthesis Pattern and Transcriptome Analysis of Sapindus saponaria Oil
Abstract
1. Introduction
2. Results
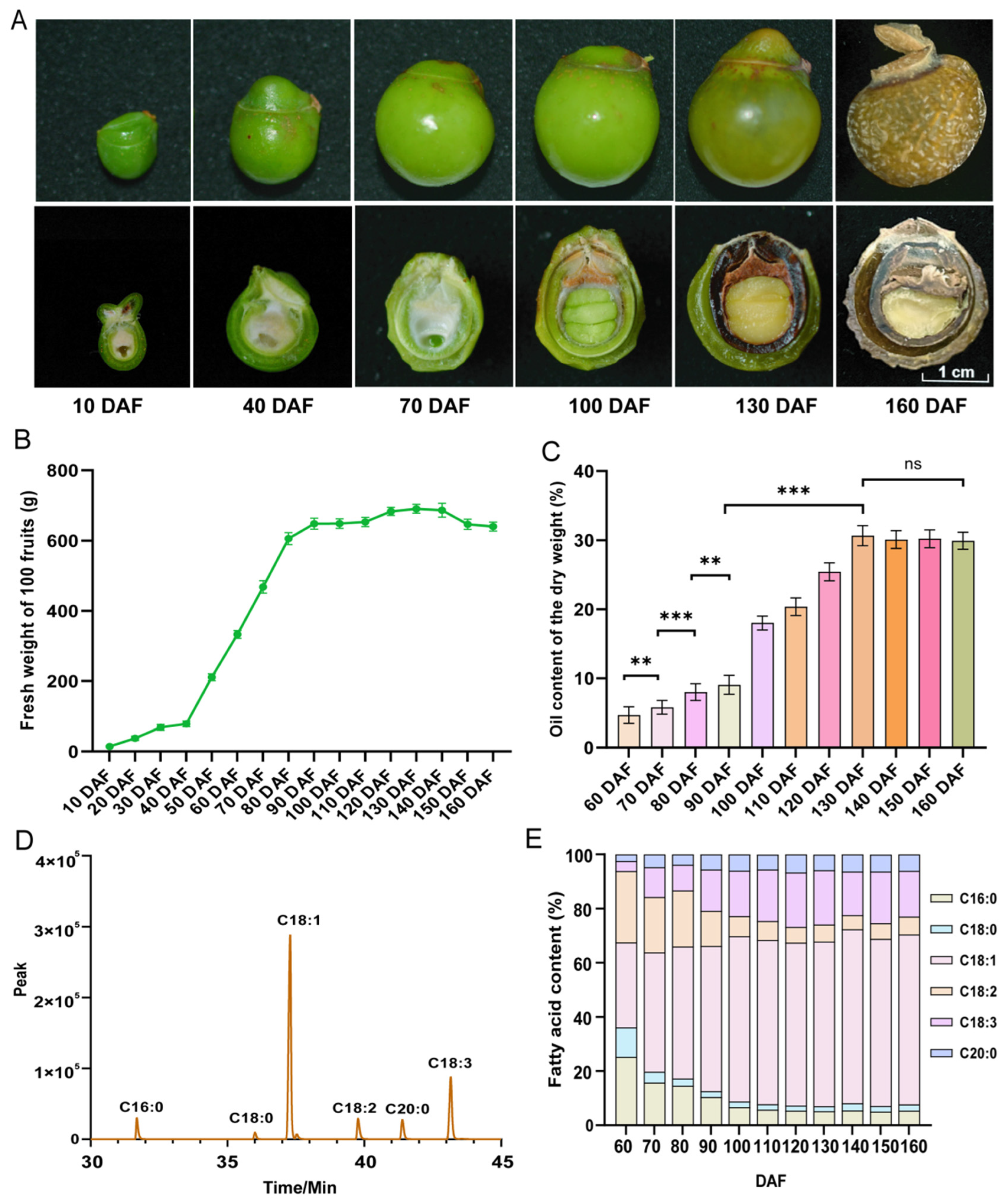
2.1. Dynamic Patterns of Fruit Oil Changes and Development
2.2. Dynamic Content of Fatty Acids
2.3. RNA-Seq and Sequence Alignment
2.4. Transcriptome Annotation and Gene Expression Profiling
2.5. Differential Gene Expression Analysis
2.6. Analysis of Lipid Metabolism DEGs
2.7. Identification and Expression Profiles of Key DEGs for FA Biosynthesis, Elongation and Desaturation
2.8. Identification and Expression Profile of Key DEGs for TAG Assembly
2.9. qRT-PCR
3. Discussion
3.1. S. saponaria Oil Content and Fatty Acid Content
3.2. Fatty Acid Biosynthesis Key Enzyme Genes in S. saponaria Kernels
3.3. S. saponaria TAG Biosynthesis and Oil Accumulation
4. Conclusions
5. Materials and Methods
5.1. Plant Material
5.2. Kernels Oil Extraction
5.3. Gas Chromatography Analysis of Fatty Acid Methyl Esters
5.4. RNA-Seq and Analysis
5.5. qT-PCR Validation of RNA-Seq
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xue, T.; Chen, D.; Zhang, T.Y.; Chen, Y.Q.; Fan, H.H.; Huang, Y.P.; Zhong, Q.L.; Li, B.Y. Chromosome-scale assembly and population diversity analyses provide insights into the evolution of Sapindus mukorossi. Hortic. Res. 2022, 5, uhac012. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.X.; Liu, J.M.; Xu, Y.Y.; Zhao, G.C.; Wang, X.; Rui, X.; Jia, L.M. The correlation between economic traits of soapberry fruit and environmental factors. Chin. J. Ecol. 2021, 40, 373–380. (In Chinese) [Google Scholar] [CrossRef]
- Zhao, G.C.; Gao, Y.H.; Gao, S.L.; Xu, Y.Y.; Liu, J.M.; Sun, C.W.; Gao, Y.; Liu, S.Q.; Chen, Z.; Jia, L.M. The Phenological Growth Stages of Sapindus mukorossi According to BBCH Scale. Forests 2019, 10, 462. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, X.Y.; Ge, F.H. Optimization for Supercritical CO2 Extraction with Response Surface Methodology and Component Analysis of Sapindus mukorossi Oil. Zhong Yao Cai 2012, 35, 300–303. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Baruah, D.C. Production and characterization of biodiesel obtained from Sapindus mukorossi kernel oil. Energy 2013, 60, 159–167. [Google Scholar] [CrossRef]
- Sun, S.; Ke, X.; Cui, L.; Bi, Y.; Song, F.; Xu, X. Enzymatic epoxidation of Sapindus mukorossi seed oil by perstearic acid optimized using response surface methodology. Ind. Crops Prod. 2011, 33, 676–682. [Google Scholar] [CrossRef]
- Chen, C.C.; Nien, C.J.; Chen, L.G.; Huang, K.Y.; Chang, W.J.; Huang, H.M. Effects of Sapindus mukorossi Seed Oil on Skin Wound Healing: In Vivo and in Vitro Testing. Int. J. Mol. Sci. 2019, 20, 2579. [Google Scholar] [CrossRef]
- Shiu, S.T.; Lew, W.Z.; Lee, S.Y.; Feng, S.W.; Huang, H.M. Effects of Sapindus mukorossi Seed Oil on Proliferation, Osteogenetic/Odontogenetic Differentiation and Matrix Vesicle Secretion of Human Dental Pulp Mesenchymal StemCells. Materials 2020, 13, 4063. [Google Scholar] [CrossRef]
- Lin, S.K.; Wu, Y.F.; Chang, W.J.; Feng, S.W.; Huang, H.M. The Treatment Efficiency and Microbiota Analysis of Sapindus mukorossi Seed Oil on the Ligature-Induced Periodontitis Rat Model. Int. J. Mol. Sci. 2022, 23, 8560. [Google Scholar] [CrossRef]
- Tzen, J.; Cao, Y.; Laurent, P.; Ratnayake, C.; Huang, A. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 1993, 101, 267–276. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Huang, Z.D.; Ao, Y.; Li, W.; Zhang, Z.X. Transcriptome analysis of yellow horn (Xanthoceras sorbifolia Bunge): A potential oil-rich seed tree for biodiesel in China. PLoS ONE 2013, 8, e74441. [Google Scholar] [CrossRef]
- Li, W.J.; Jiang, L.J.; Chen, Y.Z.; Li, C.Z.; Li, P.W.; Yang, Y.; Chen, J.Z.; Liu, Q. Transcriptome Analysis Unveiled the Intricate Interplay between Sugar Metabolism and Lipid Biosynthesis in Symplocos paniculate Fruit. Plants 2023, 12, 2703. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.J.; Zhang, D.F.; Ma, Z. Transcriptome Analysis of Genes Involved in Fatty Acid and Lipid Biosynthesis in Developing Walnut (Juglans regia L.) Seed Kernels from Qinghai Plateau. Plants 2022, 11, 3207. [Google Scholar] [CrossRef] [PubMed]
- Dvorianinova, E.M.; Zinovieva, O.L.; Pushkova, E.N.; Zhernova, D.A.; Rozhmina, T.A.; Povkhova, L.V.; Novakovskiy, R.O.; Sigova, E.A.; Turba, A.A.; Borkhert, E.V.; et al. Key FAD2, FAD3, and SAD Genes Involved in the Fatty Acid Synthesis in Flax Identified Based on Genomic and Transcriptomic Data. Int. J. Mol. Sci. 2023, 24, 14885. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. Interaction of DGAT1 and PDAT1 to enhance TAG assembly in Arabidopsis. Plant Signal. Behav. 2019, 14, 1554467. [Google Scholar] [CrossRef]
- Wang, J.J.; Liu, Z.J.; Liu, H.; Peng, D.S.; Zhang, J.P.; Chen, M.X. Linum usitatissimum FAD2A and FAD3A enhance seed polyunsaturated fatty acid accumulation and seedling cold tolerance in Arabidopsis thaliana. Plant Sci. 2021, 311, 111014. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.Q.; Ao, D.; Wang, L.; Ling, Z.H.; Chen, M.S.; Bai, Y.; Wuyun, T.N.; Chen, J.X.; Zhang, S.N.; Li, F.M. Dynamic transcriptome analysis identifies genes related to fatty acid biosynthesis in the seeds of Prunus pedunculata Pall. BMC Plant Biol. 2021, 21, 152. [Google Scholar] [CrossRef]
- Niu, J.; An, J.Y.; Wang, L.B.; Fang, C.L.; Ha, D.L.; Fu, C.G.; Qiu, L.; Yu, H.Y.; Zhao, H.Y.; Hou, X.Y.; et al. Transcriptomic analysis revealed the mechanism of oil dynamic accumulation during developing Siberian apricot (Prunus sibirica L.) seed kernels for the development of woody biodiesel. Biotechnol. Biofuels 2015, 22, 8–29. [Google Scholar] [CrossRef]
- Zhang, L.L.; Wu, P.; Lu, W.Y.; Lü, S.Y. Molecular mechanism of the extended oil accumulation phase contributing to the high seed oil content for the genotype of tung tree (Vernicia fordii). BMC Plant Biol. 2018, 18, 248. [Google Scholar] [CrossRef]
- Nishimura, N.; Yoshida, T.; Kitahata, N.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007, 50, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Gibellini, F.; Smith, T.K. The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 2010, 62, 414–428. [Google Scholar] [CrossRef] [PubMed]
- McMaster, C.R. From yeast to humans-roles of the Kennedy pathway for phosphatidylcholine synthesis. FEBS Lett. 2018, 592, 1256–1272. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.H.; Dutta, K.; Deka, D.C. Fatty acid composition of Sapindus mukorossi seed oil. Adv. Appl. Sci. Res. 2014, 5, 43–50. [Google Scholar]
- Sengupta, A.; Basu, S.P.; Saha, S. Triglyceride composition of Sapindus mukorossi seed oil. Lipids 1975, 10, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Liu, J.N.; Fang, H.C.; Dong, Y.H.; Wang, C.X.; Bao, Y.; Hou, W.R.; Zhou, R.; Ma, X.M.; Gai, S.S.; et al. Genomic and transcriptomic analyses provide insights into valuable fatty acid biosynthesis and environmental adaptation of yellow horn. Front. Plant Sci. 2022, 13, 991197. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Gao, W.Y.; Li, X.L.; Sun, S.L.; Xu, J.; Shi, X.Q.; Guo, H.Y. Regulatory mechanisms of fatty acids biosynthesis in Armeniaca sibirica seed kernel oil at different developmental stages. PeerJ 2022, 10, e14125. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Nikolau, B.J.; Ohlrogge, J.B.; Wurtele, E.S. Plant biotin-containing carboxylases. Arch. Biochem. Biophys. 2003, 414, 211–222. [Google Scholar] [CrossRef]
- Roesler, K.; Shintani, D.; Savage, L.; Boddupalli, S.; Ohlrogge, J. Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol. 1997, 113, 75–81. [Google Scholar] [CrossRef]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef]
- Domann, B.; Kridl, J.C.; Ohlrogge, J.B. Cloning and expression in Escherich ia coli of a cDNA coding for the oleoyl acyl carrier protein thioesterase from coriander (Corianum sativum L). Biochim. Biophys. Acta 1994, 1212, 134–136. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.J.; Ohlrogge, J.B. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 2002, 403, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.B.; Liu, Y.L.; Xiong, B.; Jiang, X.G.; Zhang, Z.X. Transcriptomic Analysis of a Potential Bioenergy Tree, Pistacia chinensis Bunge, and Identification of Candidate Genes Involved in the Biosynthesis of Oil. BioEnergy Res. 2016, 9, 740–749. [Google Scholar] [CrossRef]
- Knutzon, D.S.; Thompson, G.A.; Radke, S.E.; Johnson, W.B.; Knauf, V.C.; Kridl, J.C. Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proc. Natl. Acad. Sci. USA 1992, 89, 2624–2628. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, A.A.; Kezimana, P.; Rozhmina, T.A.; Zhuchenko, A.A.; Povkhova, L.V.; Pushkova, E.N.; Novakovskiy, R.O.; Pavelek, M.; Vladimirov, G.N.; Nikolaev, E.N.; et al. Genetic diversity of SAD and FAD genes responsible for the fatty acid composition in flax cultivars and lines. BMC Plant Biol. 2020, 14, 301. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Du, Z.L.; Li, Y.; Lu, S.P.; Tang, S.; Guo, L. Improving linolenic acid content in rapeseed oil by overexpression of CsFAD2 and CsFAD3 genes. Mol. Breed. 2024, 29, 9. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.H.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book. 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.D.; Browse, J. The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 2012, 3, 147. [Google Scholar] [CrossRef]
- Guo, Z.H.; Lung, S.C.; Hamdan, M.F.; Chye, M.L. Interactions between plant lipid-binding proteins and their ligands. Prog. Lipid Res. 2022, 86, 101156. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Ståhl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid: Diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.K. Assessing compartmentalized flux in lipid metabolism with isotopes. Biochim. Biophys. Acta 2016, 1861 Pt B, 1226–1242. [Google Scholar] [CrossRef]
- Bates, P.D. Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim. Biophys. Acta 2016, 1861 Pt B, 1214–1225. [Google Scholar] [CrossRef]
- Bates, P.D.; Johnson, S.R.; Cao, X.; Li, J.; Nam, J.W.; Jaworski, J.G.; Ohlrogge, J.B.; Browse, J. Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc. Natl. Acad. Sci. USA 2014, 111, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.T.; Ji, H.Y.; Yang, Z.L. Functional Characterization of Three Novel Genes Encoding Diacylglycerol Acyltransferase (DGAT) from Oil-Rich Tubers of Cyperus esculentus. Plant Cell Physiol. 2020, 61, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Li, Q.; Lin, Y.Q.; Wang, Y.; Shi, H.B.; Xiang, H.; Zhu, J.J. Diacylglycerol acyltransferase 2 promotes the adipogenesis of intramuscular preadipocytes in goat. Anim. Biotechnol. 2023, 34, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- State Food and Drug Administration. National Food Safety Standard. In Determination of Fats in Foods; China Standards Press: Beijing, China, 2016; Volume GB 5009, p. 6. [Google Scholar]
- Zhang, L.H.; Zhou, X.; Chen, Y.Z.; Li, P.W.; Yang, Y.; Li, C.Z.; Chen, J.Z. Fruit Morphology and Fatty Acid Composition of Swida wilsoniana Populations Collected from Different Regions in Southern China. Forests 2022, 13, 1811. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]




| Category | Value |
|---|---|
| RawData (bp) | 61,500,521,100 |
| RawDatas | 410,003,474 |
| CleanData (bp) | 60,716,969,523 |
| CleanDatas | 407,463,420 |
| CleanData GC (%) | 45.91% |
| CleanData Q20 (%) | 97.51% |
| CleanData Q30 (%) | 93.01% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Jiang, L.; Li, P.; Chen, J.; Chen, Y.; Yang, Y.; Zhang, L.; Ji, Y.; Xiao, Z.; Sheng, K.; et al. The Biosynthesis Pattern and Transcriptome Analysis of Sapindus saponaria Oil. Plants 2024, 13, 1781. https://doi.org/10.3390/plants13131781
Zhou X, Jiang L, Li P, Chen J, Chen Y, Yang Y, Zhang L, Ji Y, Xiao Z, Sheng K, et al. The Biosynthesis Pattern and Transcriptome Analysis of Sapindus saponaria Oil. Plants. 2024; 13(13):1781. https://doi.org/10.3390/plants13131781
Chicago/Turabian StyleZhou, Xiao, Lijuan Jiang, Peiwang Li, Jingzhen Chen, Yunzhu Chen, Yan Yang, Luhong Zhang, Yuena Ji, Zhihong Xiao, Kezhai Sheng, and et al. 2024. "The Biosynthesis Pattern and Transcriptome Analysis of Sapindus saponaria Oil" Plants 13, no. 13: 1781. https://doi.org/10.3390/plants13131781
APA StyleZhou, X., Jiang, L., Li, P., Chen, J., Chen, Y., Yang, Y., Zhang, L., Ji, Y., Xiao, Z., Sheng, K., Sheng, X., Yao, H., Liu, Q., & Li, C. (2024). The Biosynthesis Pattern and Transcriptome Analysis of Sapindus saponaria Oil. Plants, 13(13), 1781. https://doi.org/10.3390/plants13131781







