Genetic Diversity and Population Structure of a Large USDA Sesame Collection
Abstract
1. Introduction
2. Results and Discussion
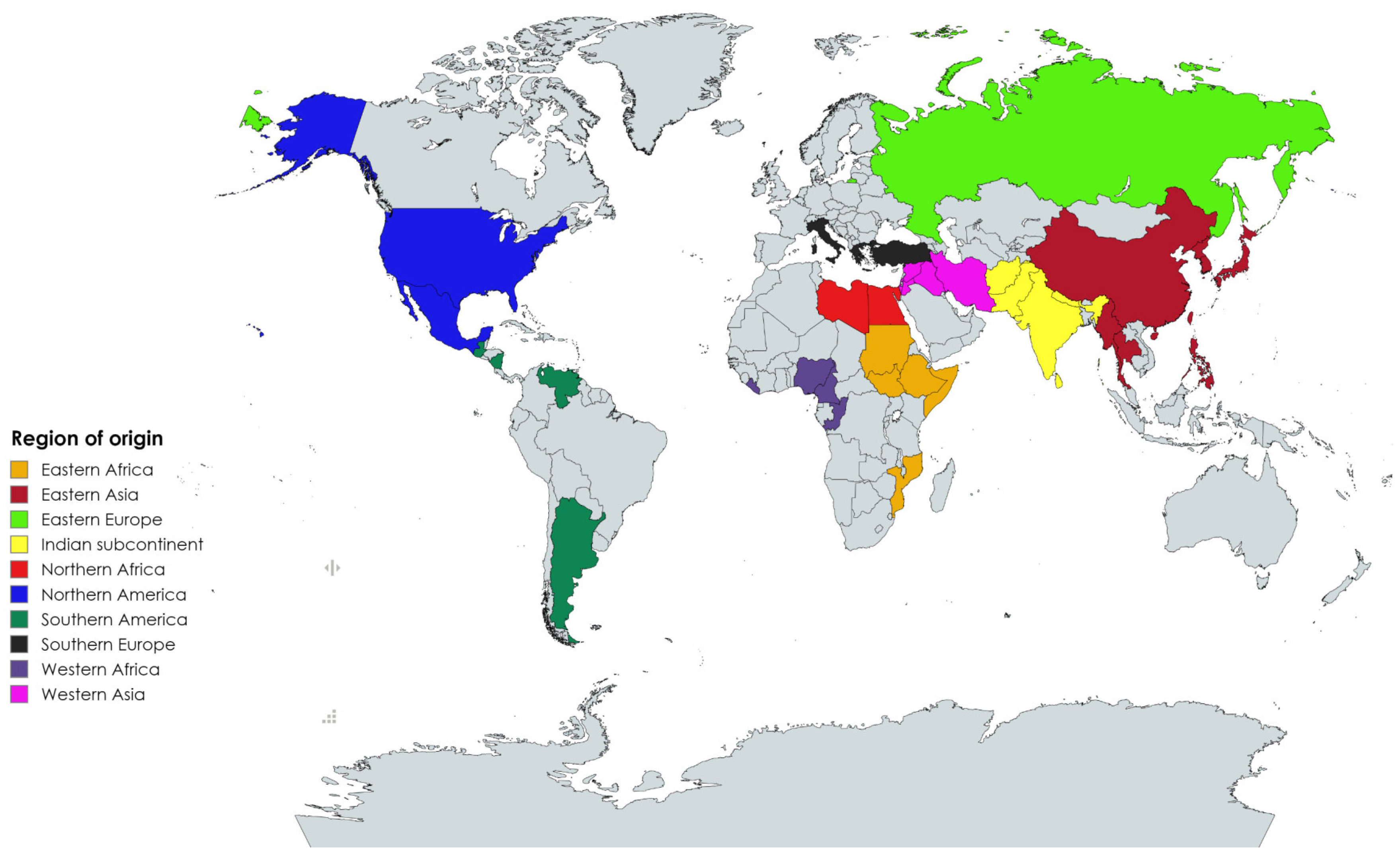
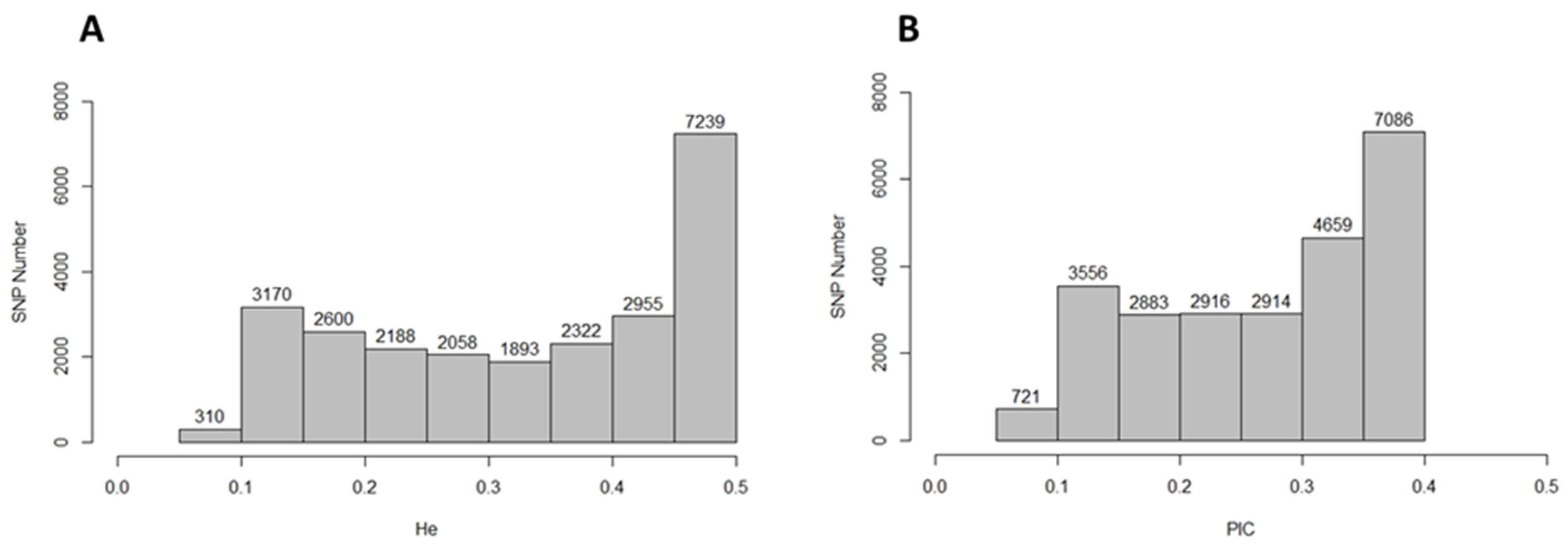
2.1. Single-Nucleotide Polymorphism Markers Coverage and Polymorphism Analyses
2.2. Analysis of Population Structure
2.3. Analysis of Molecular Variance (AMOVA) and Genetic Diversity Indices
2.4. Linkage Disequilibrium
2.5. Applications of High-Throughput Genotyping of Sesame Accessions
3. Materials and Methods
3.1. Plant Materials
3.2. DNA Extraction and Genotyping-by-Sequencing (GBS)
3.3. GBS Sequencing and Genotyping Pipeline Analyses
3.4. Population Genetic Analyses
3.4.1. Marker Polymorphism Analyses
3.4.2. Analysis of Population Structure
3.5. Analysis of Molecular Variance (AMOVA) and Genetic Diversity Indices
3.6. Linkage Disequilibrium (LD)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miao, H.; Langham, D.R.; Zhang, H. Botanical Descriptions of Sesame. In The Sesame Genome; Miao, H., Zhang, H., Kole, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 19–57. [Google Scholar]
- Wei, P.; Zhao, F.; Wang, Z.; Wang, Q.; Chai, X.; Hou, G.; Meng, Q. Sesame (Sesamum indicum L.): A Comprehensive Review of Nutritional Value, Phytochemical Composition, Health Benefits, Development of Food, and Industrial Applications. Nutrients 2022, 14, 4079. [Google Scholar] [CrossRef] [PubMed]
- Pathak, N.; Rai, A.K.; Kumari, R.; Bhat, K.V. Value addition in sesame: A perspective on bioactive components for enhancing utility and profitability. Pharmacogn. Rev. 2014, 8, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Shayanowako, A.I.T. Analyses of genetic diversity and population structure of sesame (Sesamum indicum L.) germplasm collections through seed oil and fatty acid compositions and SSR markers. J. Food Compos. Anal. 2022, 110, 104545. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, F.; Tang, X.; Yang, Y.; Zhou, T.; Liu, H. Morphology and SSR Markers-Based Genetic Diversity Analysis of Sesame (Sesamum indicum L.) Cultivars Released in China. Agriculture 2023, 13, 1885. [Google Scholar] [CrossRef]
- Pham, T.; Nguyen, T.-D.; Carlsson, A.; Bui, T. Morphological evaluation of sesame (Sesamum indicum L.) varieties from different origins. Aust. J. Crop Sci. 2010, 4, 498–504. [Google Scholar]
- Frary, A.; Tekin, P.; Celik, I.; Furat, S.; Uzun, B.; Doganlar, S. Morphological and Molecular Diversity in Turkish Sesame Germplasm and Core Set Selection. Crop Sci. 2015, 55, 702–711. [Google Scholar] [CrossRef]
- Gedifew, S.; Demelash, H.; Abate, A.; Abebe, T.D. Association of quantitative traits and genetic diversity in Ethiopian sesame (Sesamum indicum L.) genotypes. Heliyon 2024, 10, e26676. [Google Scholar] [CrossRef] [PubMed]
- Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Mashilo, J. Genetic diversity and association of yield-related traits in sesame. Plant Breed. 2021, 140, 331–341. [Google Scholar] [CrossRef]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef]
- Kim, D.H.; Zur, G.; Danin-Poleg, Y.; Lee, S.W.; Shim, K.B.; Kang, C.W.; Kashi, Y. Genetic relationships of sesame germplasm collection as revealed by inter-simple sequence repeats. Plant Breed. 2002, 121, 259–262. [Google Scholar] [CrossRef]
- Basak, M.; Uzun, B.; Yol, E. Genetic diversity and population structure of the Mediterranean sesame core collection with use of genome-wide SNPs developed by double digest RAD-Seq. PLoS ONE 2019, 14, e0223757. [Google Scholar] [CrossRef] [PubMed]
- Teklu, D.H.; Shimelis, H.; Abady, S. Genetic Improvement in Sesame (Sesamum indicum L.): Progress and Outlook: A Review. Agronomy 2022, 12, 2144. [Google Scholar] [CrossRef]
- Kefale, H.; Wang, L. Discovering favorable genes, QTLs, and genotypes as a genetic resource for sesame (Sesamum indicum L.) improvement. Front. Genet. 2022, 13, 1002182. [Google Scholar] [CrossRef] [PubMed]
- Mesfer, A.S.; Safhi, F.A.; Alshaya, D.S.; Ibrahim, A.A.; Mansour, H.; Abd El Moneim, D. Genetic diversity using biochemical, physiological, karyological and molecular markers of Sesamum indicum L. Front. Genet. 2022, 13, 1035977. [Google Scholar] [CrossRef]
- Salazar, B.; Laurentín, H.; Davila, M.; Castillo, A. Reliability of the RAPD technique for germplasm analysis of sesame (Sesamum indicum L.) from Venezuela. Interciencia 2006, 31, 456–460. [Google Scholar]
- Ercan, A.; Taskin, K.; Turgut, K. Analysis of genetic diversity in Turkish sesame (Sesamum indicum L.) populations using RAPD markers. Genet. Resour. Crop Evol. 2004, 51, 599–607. [Google Scholar] [CrossRef]
- Bhat, K.; Babrekar, P.; Lakhanpaul, S. Study of genetic diversity in Indian and exotic sesame (Sesamum indicum L.) germplasm using random amplified polymorphic DNA (RAPD) markers. Euphytica 1999, 110, 21–34. [Google Scholar] [CrossRef]
- Asekova, S.; Kulkarni, K.; Oh, K.W.; Lee, M.-H.; Oh, E.; Kim, J.-I.; Yeo, U.-S.; Pae, S.-B.; Ha, T.J.; Kim, S.U. Analysis of Molecular Variance and Population Structure of Sesame (Sesamum indicum L.) Genotypes Using Simple Sequence Repeat Markers. Plant Breed. Biotech. 2018, 6, 321–336. [Google Scholar] [CrossRef]
- Sasipriya, S.; Balram, M.; Kamireddy, P.; Eswari, K. Assessment of molecular divergence in sesame (Sesamum indicum L.) genotypes using microsatellite (SSR) markers. Int. J. Ecol. Environ. Sci. 2020, 2, 182–187. [Google Scholar]
- Anggraeni, T.D.A.; Fadilah, S.N.; Kusnadi, J.; Basuki, S. The Use of ISSR markers for clustering sesame genotypes based on geographical origin. IOP Conf. Ser. Earth Environ. Sci. 2022, 974, 012031. [Google Scholar] [CrossRef]
- Laurentin, H.E.; Karlovsky, P. Genetic relationship and diversity in a sesame (Sesamum indicum L.) germplasm collection using amplified fragment length polymorphism (AFLP). BMC Genet. 2006, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Ali Al-Somain, B.H.; Migdadi, H.M.; Al-Faifi, S.A.; Alghamdi, S.S.; Muharram, A.A.; Mohammed, N.A.; Refay, Y.A. Assessment of genetic diversity of sesame accessions collected from different ecological regions using sequence-related amplified polymorphism markers. 3 Biotech 2017, 7, 82. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Gupta, S.; Bandhiwal, N.; Kumar, T.; Bharadwaj, C.; Bhatia, S. High-density linkage map construction and mapping of seed trait QTLs in chickpea (Cicer arietinum L.) using genotyping-by-sequencing (GBS). Sci. Rep. 2015, 5, 17512. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, T.; Tesfaye, K.; Keneni, G.; Ziyomo, C.; Alemu, T. Genetic diversity of Sesame (Sesamum indicum L) using high throughput diversity array technology. J. Crop Sci. Biotechnol. 2022, 25, 359–371. [Google Scholar] [CrossRef]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; Larose, S.; Jean, M.; et al. An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef] [PubMed]
- Bird, K.A.; An, H.; Gazave, E.; Gore, M.A.; Pires, J.C.; Robertson, L.D.; Labate, J.A. Population structure and phylogenetic relationships in a diverse panel of Brassica rapa L. Front. Plant Sci. 2017, 8, 321. [Google Scholar] [CrossRef] [PubMed]
- Khedikar, Y.; Clarke, W.E.; Chen, L.; Higgins, E.E.; Kagale, S.; Koh, C.S.; Bennett, R.; Parkin, I.A.P. Narrow genetic base shapes population structure and linkage disequilibrium in an industrial oilseed crop, Brassica carinata A. Braun. Sci. Rep. 2020, 10, 12629. [Google Scholar] [CrossRef]
- Abdel-Haleem, H.; Luo, Z.; Szczepanek, A. Genetic diversity and population structure of the USDA collection of Brassica juncea L. Ind. Crop. Prod. 2022, 187, 115379. [Google Scholar] [CrossRef]
- Luo, Z.; Brock, J.; Dyer, J.M.; Kutchan, T.; Schachtman, D.; Augustin, M.; Ge, Y.; Fahlgren, N.; Abdel-Haleem, H. Genetic Diversity and Population Structure of a Camelina sativa Spring Panel. Front. Plant Sci. 2019, 10, 184. [Google Scholar] [CrossRef]
- Islam, A.; Sanders, D.; Mishra, A.K.; Joshi, V. Genetic Diversity and Population Structure Analysis of the USDA Olive Germplasm Using Genotyping-By-Sequencing (GBS). Genes 2021, 12, 2007. [Google Scholar] [CrossRef]
- Fu, Y.B.; Cober, E.R.; Morrison, M.J.; Marsolais, F.; Peterson, G.W.; Horbach, C. Patterns of Genetic Variation in a Soybean Germplasm Collection as Characterized with Genotyping-by-Sequencing. Plants 2021, 10, 1611. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.J.; Lee, J.R.; Sebastin, R.; Shin, M.J.; Kim, S.H.; Cho, G.T.; Hyun, D.Y. Genetic Diversity Assessed by Genotyping by Sequencing (GBS) in Watermelon Germplasm. Genes 2019, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tan, B.; Liu, H.; Zhu, W.; Xu, L.; Wang, Y.; Fan, X.; Sha, L.; Zhang, H.; Zeng, J.; et al. Genetic Diversity and Population Structure of Asian and European Common Wheat Accessions Based on Genotyping-By-Sequencing. Front. Genet. 2020, 11, 580782. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.; Geleta, M.; Bui, T.M.; Bui, T.C.; Merker, A.; Carlsson, A.S. Comparative analysis of genetic diversity of sesame (Sesamum indicum L.) from Vietnam and Cambodia using agro-morphological and molecular markers. Hereditas 2011, 148, 28–35. [Google Scholar] [CrossRef]
- Yates, H.E.; Frary, A.; Doganlar, S.; Frampton, A.; Eannetta, N.T.; Uhlig, J.; Tanksley, S.D. Comparative fine mapping of fruit quality QTLs on chromosome 4 introgressions derived from two wild tomato species. Euphytica 2004, 135, 283–296. [Google Scholar] [CrossRef]
- Cui, C.; Mei, H.; Liu, Y.; Zhang, H.; Zheng, Y. Genetic Diversity, Population Structure, and Linkage Disequilibrium of an Association-Mapping Panel Revealed by Genome-Wide SNP Markers in Sesame. Front. Plant Sci. 2017, 8, 1189. [Google Scholar] [CrossRef]
- Dossa, K.; Wei, X.; Zhang, Y.; Fonceka, D.; Yang, W.; Diouf, D.; Liao, B.; Cissé, N.; Zhang, X. Analysis of Genetic Diversity and Population Structure of Sesame Accessions from Africa and Asia as Major Centers of Its Cultivation. Genes 2016, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.S.; Zhang, J.; Tollenaere, R.; Teuber, P.V.; Dalton-Morgan, J.; Hu, L.Y.; Yan, G.J.; Edwards, D.; Redden, R.; Batley, J. High-throughput genotyping for species identification and diversity assessment in germplasm collections. Mol. Ecol. Resour. 2015, 15, 1091–1101. [Google Scholar] [CrossRef]
- Mohd Saad, N.S.; Severn-Ellis, A.A.; Pradhan, A.; Edwards, D.; Batley, J. Genomics armed with diversity leads the way in brassica improvement in a changing global environment. Front. Genet. 2021, 12, 600789. [Google Scholar] [CrossRef]
- Ashri, A. Sesame (Sesamum indicum L.). In Genetic Resources, Chromosome Engineering, and Crop Improvement: Oilseed Crops, Volume 4; Singh, R.J., Ed.; CRC Press: Boca Raton, FL, USA, 2006; pp. 231–289. [Google Scholar]
- Wang, M.; Huang, J.; Liu, S.; Liu, X.; Li, R.; Luo, J.; Fu, Z. Improved assembly and annotation of the sesame genome. DNA Res. 2022, 29, dsac041. [Google Scholar] [CrossRef]
- Bancroft, I.; Morgan, C.; Fraser, F.; Higgins, J.; Wells, R.; Clissold, L.; Baker, D.; Long, Y.; Meng, J.; Wang, X.; et al. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 2011, 29, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Schnable, J.C.; Springer, N.M.; Freeling, M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 2011, 108, 4069–4074. [Google Scholar] [CrossRef] [PubMed]
- Delourme, R.; Falentin, C.; Fomeju, B.F.; Boillot, M.; Lassalle, G.; André, I.; Duarte, J.; Gauthier, V.; Lucante, N.; Marty, A.J.B.g. High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genom. 2013, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Yu, H.-J.; Mun, J.-H.; Lee, S.-C. Genome-wide discovery of DNA polymorphism in Brassica rapa. Mol. Genet. Genom. 2009, 283, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Park, J.-S.; Shin, Y.-H.; Park, Y.-D. Identification and validation of genetic variations in transgenic Chinese cabbage plants (Brassica rapa ssp. pekinensis) by next-generation sequencing. Genes 2021, 12, 621. [Google Scholar] [CrossRef] [PubMed]
- Bus, A.; Hecht, J.; Huettel, B.; Reinhardt, R.; Stich, B. High-throughput polymorphism detection and genotyping in Brassica napus using next-generation RAD sequencing. BMC Genom. 2012, 13, 281. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Iaffaldano, B.J.; Zhuang, X.; Fresnedo-Ramírez, J.; Cornish, K. Analysis of the first Taraxacum kok-saghyz transcriptome reveals potential rubber yield related SNPs. Sci. Rep. 2017, 7, 9939. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Harris, A.M.; DeGiorgio, M. An unbiased estimator of gene diversity with improved variance for samples containing related and inbred individuals of any ploidy. G3 Gene. Genom. Genet. 2017, 7, 671–691. [Google Scholar] [CrossRef]
- Yu, Z.; Fredua-Agyeman, R.; Hwang, S.-F.; Strelkov, S.E. Molecular genetic diversity and population structure analyses of rutabaga accessions from Nordic countries as revealed by single nucleotide polymorphism markers. BMC Genom. 2021, 22, 442. [Google Scholar] [CrossRef]
- Guo, X.; Elston, R.C. Linkage information content of polymorphic genetic markers. Human. Hered. 1999, 49, 112–118. [Google Scholar] [CrossRef]
- Singh, B.K.; Mishra, D.C.; Yadav, S.; Ambawat, S.; Vaidya, E.; Tribhuvan, K.U.; Kumar, A.; Kumar, S.; Kumar, S.; Chaturvedi, K.K.; et al. Identification, characterization, validation and cross-species amplification of genic-SSRs in Indian Mustard (Brassica juncea). J. Plant Biochem. Biotechnol. 2016, 25, 410–420. [Google Scholar] [CrossRef]
- Gupta, R.; Chandrashekar, U.S.; Yadav, J.B.; Chakrabarty, S.K.; Dadlani, M. Assessment of genetic relatedness among Indian mustard (Brassica juncea) genotypes using morphological traits and DNA marker. Ind. J. Agri. Sci. 2012, 82, 746–752. [Google Scholar] [CrossRef]
- Raza, A.; Farooq, A.U.; Khan, W.A.; Iqbal, A.; Celik, S.; Ali, M.; Khan, R.S.A. Polymorphic information and genetic diversity in Brassica species revealed by RAPD markers. Biocell 2020, 44, 769–776. [Google Scholar] [CrossRef]
- Qamar, H.; Shabbir, G.; Ilyas, M.; Arshad, A.; Malik, S.I.; Mahmood, T.; Bin Mustafa, H.S. Studies on genetic divergence of rapeseed genotypes using SSR markers. Pak. J. Bot. 2020, 52, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, M.; Mohammmadi, S.A.; Mohebalipour, N.; Toorchi, M.; Aharizad, S.; Javidfar, F. Assessment of genetic diversity in rapeseed cultivars as revealed by RAPD and microsatellite markers. Afr. J. Biotechnol. 2009, 8, 3160–3167. [Google Scholar]
- Wu, J.; Li, F.; Xu, K.; Gao, G.; Chen, B.; Yan, G.; Wang, N.; Qiao, J.; Li, J.; Li, H.; et al. Assessing and broadening genetic diversity of a rapeseed germplasm collection. Breed. Sci. 2014, 64, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.M.; Poland, J.; Baenziger, P.S. Genetic Diversity and Population Structure of F3:6 Nebraska Winter Wheat Genotypes Using Genotyping-By-Sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]
- Zhao, K.; Aranzana, M.J.; Kim, S.; Lister, C.; Shindo, C.; Tang, C.; Toomajian, C.; Zheng, H.; Dean, C.; Marjoram, P.; et al. An Arabidopsis Example of Association Mapping in Structured Samples. PLoS Genet. 2007, 3, e4. [Google Scholar] [CrossRef]
- Puechmaille, S.J. The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Dossou, S.S.K.; Song, S.; Liu, A.; Li, D.; Zhou, R.; Berhe, M.; Zhang, Y.; Sheng, C.; Wang, Z.; You, J.; et al. Resequencing of 410 Sesame Accessions Identifies SINST1 as the Major Underlying Gene for Lignans Variation. Int. J. Mol. Sci. 2023, 24, 1055. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, S.A.; Prasanna, B.J.C.s. Analysis of genetic diversity in crop plants—Salient statistical tools and considerations. Crop Sci. 2003, 43, 1235–1248. [Google Scholar] [CrossRef]
- Zapata, C.; Rodríguez, S.; Visedo, G.; Sacristán, F. Spectrum of nonrandom associations between microsatellite loci on human chromosome 11p15. Genetics 2001, 158, 1235–1251. [Google Scholar] [CrossRef] [PubMed]
- Lewontin, R.C.; Kojima, K.-i. The evolutionary dynamics of complex polymorphisms. Evolution 1960, 14, 458–472. [Google Scholar] [CrossRef]
- Ward, R.A.; Kim, K.S.; Diers, B.W. Yield drag associated with the soybean aphid resistance gene Rag2 from PI 200538. Crop Sci. 2017, 57, 3035–3042. [Google Scholar] [CrossRef]
- Andrade, A.C.B.; Viana, J.M.S.; Pereira, H.D.; Pinto, V.B.; Fonseca E Silva, F. Linkage disequilibrium and haplotype block patterns in popcorn populations. PLoS ONE 2019, 14, e0219417. [Google Scholar] [CrossRef] [PubMed]
- Wall, J.D.; Pritchard, J.K. Haplotype blocks and linkage disequilibrium in the human genome. Nat. Rev. Genet. 2003, 4, 587–597. [Google Scholar] [CrossRef]
- Voss-Fels, K.; Snowdon, R.J. Understanding and utilizing crop genome diversity via high-resolution genotyping. Plant Biotechnol. J. 2016, 14, 1086–1094. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S.t. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Linkage disequilibrium-understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Han, X.; Zhang, Y.; Li, D.; Wei, X.; Ding, X.; Zhang, X. Deep resequencing reveals allelic variation in Sesamum indicum. BMC Plant Biol. 2014, 14, 225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Hu, D.; Raman, R.; Guo, S.; Wei, Z.; Shen, X.; Meng, J.; Raman, H.; Zou, J. Investigation of the Genetic Diversity and Quantitative Trait Loci Accounting for Important Agronomic and Seed Quality Traits in Brassica carinata. Front. Plant Sci. 2017, 8, 615. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Qian, W.; Snowdon, R.J. Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. BMC Genom. 2014, 15, 1170. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, A.; Morgante, M. Corn and humans: Recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 2004, 20, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Kelso, J. High-throughput DNA sequencing--concepts and limitations. BioEssays 2010, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Luo, Z.; Fahlgren, N.; Kutchan, T.; Schachtman, D.; Ge, Y.; Gesch, R.; George, S.; Dyer, J.; Abdel-Haleem, H. Discovering candidate genes related to flowering time in the spring panel of Camelina sativa. Ind. Crop. Prod. 2021, 173, 114104. [Google Scholar] [CrossRef]
- Luo, Z.; Szczepanek, A.; Abdel-Haleem, H. Genome-wide association study (GWAS) analysis of camelina seedling germination under salt stress condition. Agronomy 2020, 10, 1444. [Google Scholar] [CrossRef]
- Luo, Z.; Tomasi, P.; Fahlgren, N.; Abdel-Haleem, H. Genome-wide association study (GWAS) of leaf cuticular wax components in Camelina sativa identifies genetic loci related to intracellular wax transport. BMC Plant Biol. 2019, 19, 187. [Google Scholar] [CrossRef] [PubMed]
- Muthulakshmi, C.; Sivaranjani, R.; Selvi, S. Modification of sesame (Sesamum indicum L.) for Triacylglycerol accumulation in plant biomass for biofuel applications. Biotechnol. Rep. 2021, 32, e00668. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lei, R.; Ding, S.-W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A High capacity genotyping by sequencing analysis ppeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Method. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. Pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Weir, B.S. Estimating F-statistics: A historical view. Philos. Sci. 2012, 79, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Del Carpio, D.P.; Basnet, R.K.; De Vos, R.C.; Maliepaard, C.; Visser, R.; Bonnema, G. The patterns of population differentiation in a Brassica napus L. core collection. Theor. Appl. Genet. 2011, 122, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2018, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]




| Chromosomes * | Length (Mbp) * | No. of SNP | Maker Density ** | |
|---|---|---|---|---|
| Kbp | SNP/Mbp | |||
| LG01 | 23.75 | 2389 | 9.94 | 100.60 |
| LG02 | 23.37 | 1240 | 18.85 | 53.05 |
| LG03 | 31.44 | 3118 | 10.08 | 99.18 |
| LG04 | 21.23 | 1466 | 14.48 | 69.07 |
| LG05 | 20.96 | 1434 | 14.62 | 68.40 |
| LG06 | 27.99 | 2467 | 11.35 | 88.13 |
| LG07 | 16.12 | 1259 | 12.80 | 78.11 |
| LG08 | 31.99 | 2342 | 13.66 | 73.22 |
| LG09 | 26.74 | 2336 | 11.45 | 87.36 |
| LG10 | 22.21 | 1705 | 13.03 | 76.76 |
| LG11 | 17.33 | 1716 | 10.10 | 99.01 |
| LG12 | 19.10 | 2065 | 9.25 | 108.11 |
| LG13 | 19.50 | 1198 | 16.28 | 61.44 |
| Total | 301.73 | 24,735 | - | - |
| Average | - | 1903 | 12.76 | 81.73 |
| SNP Type | Transitions | Transversions | ||||
|---|---|---|---|---|---|---|
| A/G | C/T | A/T | A/C | G/T | G/C | |
| Number of SNPs | 7145 | 7292 | 2609 | 2555 | 2529 | 2605 |
| Allele frequency | 0.289 | 0.295 | 0.105 | 0.103 | 0.102 | 0.105 |
| Total (percentage) | 0.584 | 0.416 | ||||
| Source | df | Sum of Squares | Variance Components | Variation % |
|---|---|---|---|---|
| Among subpopulations | 1 | 155,958.45 | 308.77 | 29.50 |
| Among accessions within subpopulations | 499 | 667,180.45 | 599.25 | 57.26 |
| Within accessions | 501 | 69,405.00 | 138.53 | 13.24 |
| Total | 892,543.90 | 1046.56 |
| Chromosome | LD |
|---|---|
| LG01 | 148.77 |
| LG02 | 132.90 |
| LG03 | 148.88 |
| LG04 | 131.75 |
| LG05 | 123.26 |
| LG06 | 151.14 |
| LG07 | 149.29 |
| LG08 | 161.50 |
| LG09 | 167.65 |
| LG10 | 133.18 |
| LG11 | 146.04 |
| LG12 | 159.01 |
| LG13 | 148.82 |
| Whole population | 160.69 |
| Subpopulation 1 | 166.45 |
| Subpopulation 2 | 143.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seay, D.; Szczepanek, A.; De La Fuente, G.N.; Votava, E.; Abdel-Haleem, H. Genetic Diversity and Population Structure of a Large USDA Sesame Collection. Plants 2024, 13, 1765. https://doi.org/10.3390/plants13131765
Seay D, Szczepanek A, De La Fuente GN, Votava E, Abdel-Haleem H. Genetic Diversity and Population Structure of a Large USDA Sesame Collection. Plants. 2024; 13(13):1765. https://doi.org/10.3390/plants13131765
Chicago/Turabian StyleSeay, Damien, Aaron Szczepanek, Gerald N. De La Fuente, Eric Votava, and Hussein Abdel-Haleem. 2024. "Genetic Diversity and Population Structure of a Large USDA Sesame Collection" Plants 13, no. 13: 1765. https://doi.org/10.3390/plants13131765
APA StyleSeay, D., Szczepanek, A., De La Fuente, G. N., Votava, E., & Abdel-Haleem, H. (2024). Genetic Diversity and Population Structure of a Large USDA Sesame Collection. Plants, 13(13), 1765. https://doi.org/10.3390/plants13131765






