Plant Immunity: At the Crossroads of Pathogen Perception and Defense Response
Abstract
1. Introduction
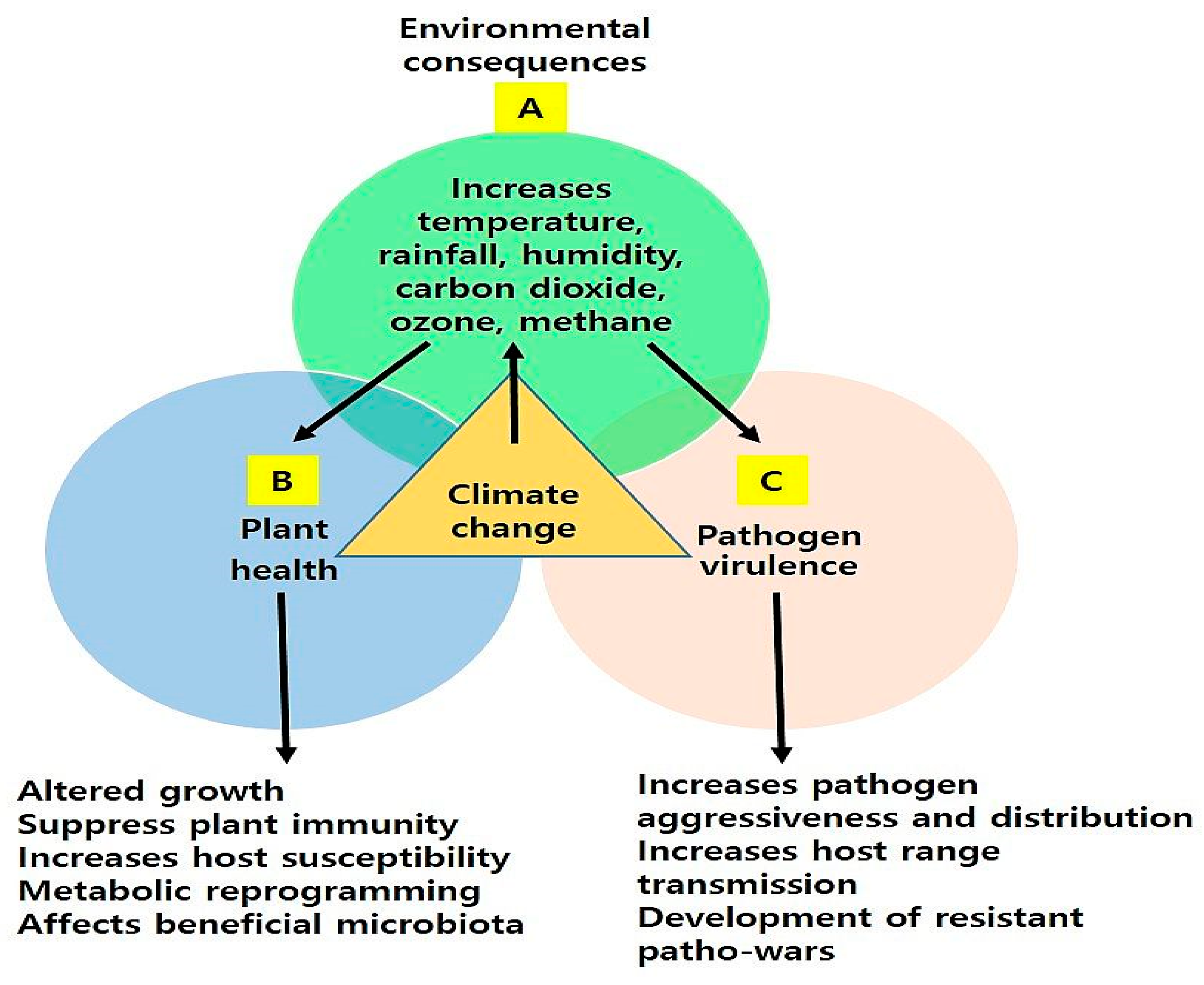
2. Impact of Climate Change or Environmental Factors on Plant–Pathogen Interaction
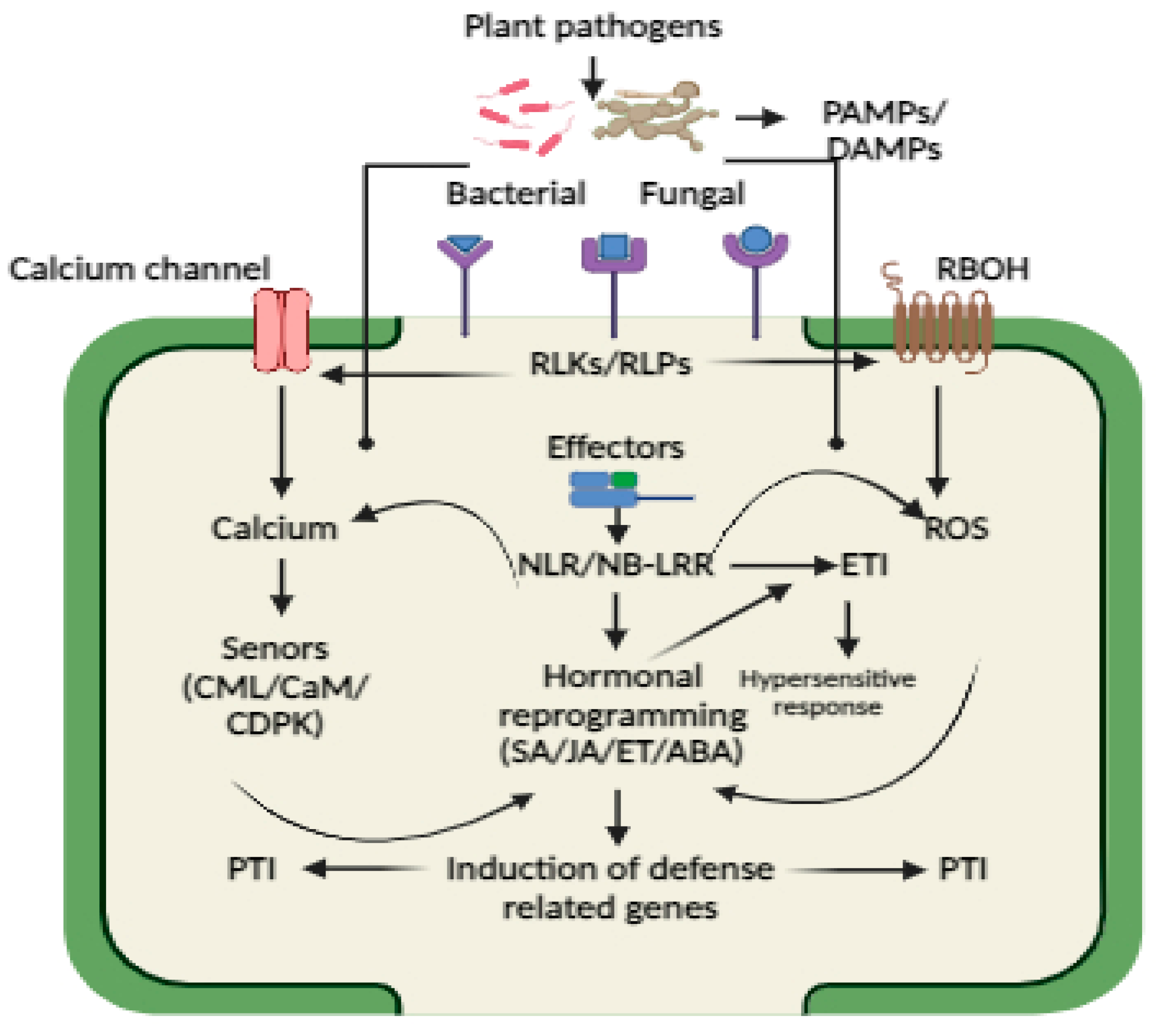
3. Pathogen Perception and Plant Immunity
4. Role of Calcium and ROS in Plant Immunity
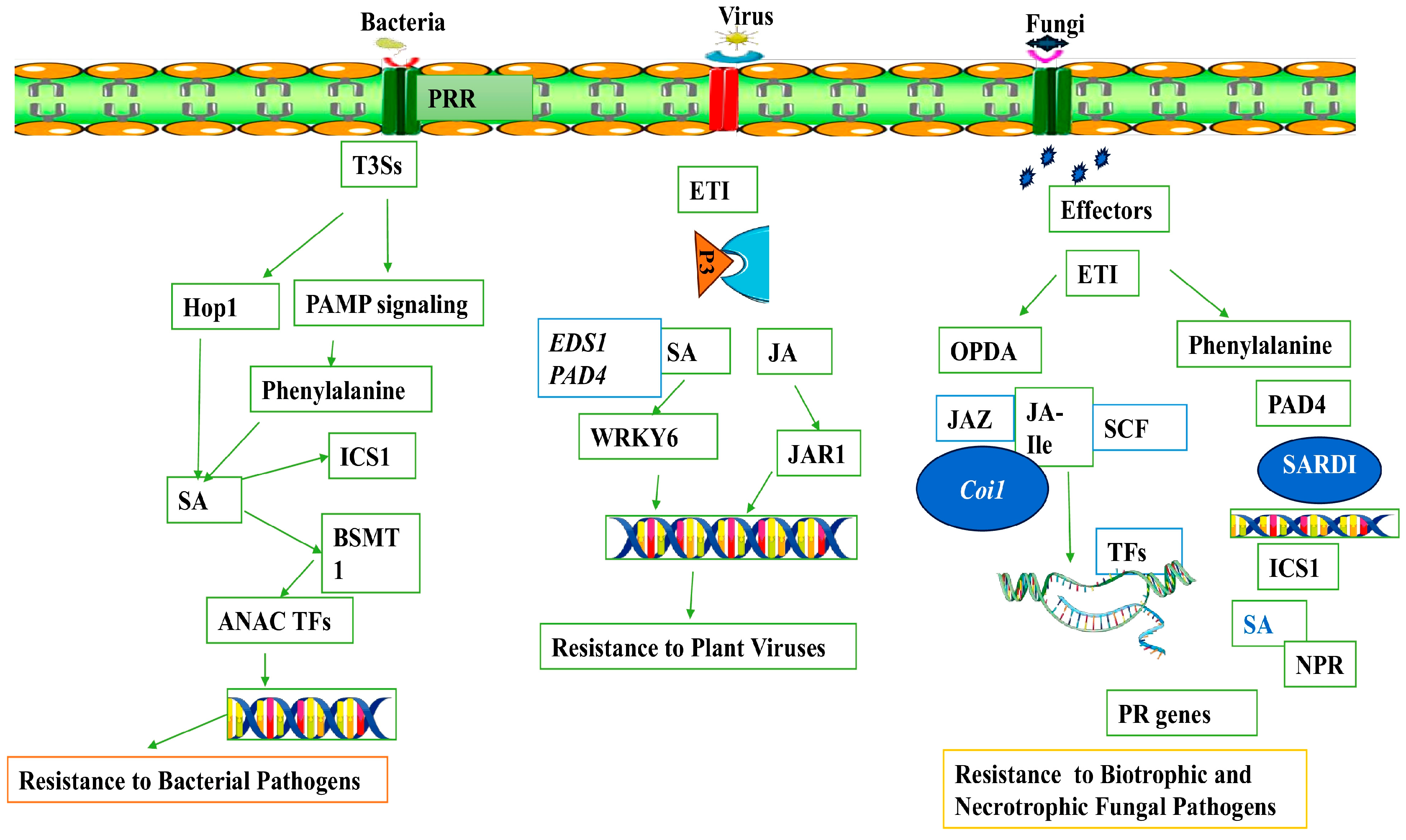
5. Revisiting the Role of Hormones in Plant Defense Response
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ali, S.; Tyagi, A.; Bae, H. Plant microbiome: An ocean of possibilities for improving disease resistance in plants. Microorganisms 2023, 11, 392. [Google Scholar] [CrossRef] [PubMed]
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Tyagi, A.; Rajarammohan, S.; Mir, Z.A.; Bae, H. Revisiting Alternaria-host interactions: New insights on its pathogenesis, defense mechanisms and control strategies. Sci. Hortic. 2023, 322, 112424. [Google Scholar] [CrossRef]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Petre, B.; Kamoun, S. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 2014, 12, e1001801. [Google Scholar] [CrossRef] [PubMed]
- Doehlemann, G.; Van Der Linde, K.; Aßmann, D.; Schwammbach, D.; Hof, A.; Mohanty, A.; Jackson, D.; Kahmann, R. Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 2009, 5, e1000290. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyan, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef]
- Cornelis, G.R. The type III secretion injectisome, a complex nanomachine for intracellular “toxin” delivery. Biol. Chem. 2010, 391, 745–751. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Bourdais, G.; He, S.Y.; Robatzek, S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014, 202, 259–269. [Google Scholar] [CrossRef]
- Nazarov, T.; Chen, X.; Carter, A.; See, D. Fine mapping of high-temperature adult-plant resistance to stripe rust in wheat cultivar Louise. J. Plant Prot. Res. 2020, 60, 126–133. [Google Scholar]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.; Latunde-Dada, A.O.; Woods-Tör, A.; Lynn, J.; Lucas, J.A.; Crute, I.R.; Holub, E.B. Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad-spectrum suppression of innate immunity. Mol. Plant-Microbe Interact. 2008, 21, 745–756. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.P.; Brown, R.; Kazan, K.; et al. NPR1 modulates cross-talk between salicylate-and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Notz, R.; Maurhofer, M.; Dubach, H.; Haas, D.; Défago, G. Fusaric acid-producing strains of Fusarium oxysporum alter 2, 4-diacetylphloroglucinol biosynthetic gene expression in Pseudomonas fluorescens CHA0 in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 2002, 68, 2229–2235. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhang, C.; Zheng, H.; Sun, M.; Zhang, F.; Zhang, M.; Cui, F.; Lv, D.; Liu, L.; Guo, S.; et al. Antagonistic interaction between auxin and SA signaling pathways regulates bacterial infection through lateral root in Arabidopsis. Cell Rep. 2020, 32, 108060. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; De Veylder, L.; Holsters, M.; Vereecke, D. Eternal youth, the fate of developing Arabidopsis leaves upon Rhodococcus fascians infection. Plant Physiol. 2009, 149, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Dolezal, K.; Van Lijsebettens, M.; Moritz, T.; Holsters, M.; Vereecke, D. Modulation of the hormone setting by Rhodococcus fascians results in ectopic KNOX activation in Arabidopsis. Plant Physiol. 2008, 146, 1267–1281. [Google Scholar] [CrossRef] [PubMed]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 44526. [Google Scholar] [CrossRef]
- Garcia-Brugger, A.; Lamotte, O.; Vandelle, E.; Bourque, S.; Lecourieux, D.; Poinssot, B.; Wendehenne, D.; Pugin, A. Early signaling events induced by elicitors of plant defenses. Mol. Plant-Microbe Interact. 2006, 19, 711–724. [Google Scholar] [CrossRef]
- Desaint, H.; Aoun, N.; Deslandes, L.; Vailleau, F.; Roux, F.; Berthomé, R. Fight hard or die trying: When plants face pathogens under heat stress. New Phytol. 2021, 229, 712–734. [Google Scholar] [CrossRef]
- Roussin-Léveillée, C.; Rossi, C.A.; Castroverde, C.D.; Moffett, P. The plant disease triangle facing climate change: A molecular perspective. Trends Plant Sci. 2024. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, A.C.; Castroverde, C.D.; He, S.Y. Plant–pathogen warfare under changing climate conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef]
- Zarattini, M.; Farjad, M.; Launay, A.; Cannella, D.; Soulié, M.C.; Bernacchia, G.; Fagard, M. Every cloud has a silver lining: How abiotic stresses affect gene expression in plant-pathogen interactions. J. Exp. Bot. 2021, 72, 1020–1033. [Google Scholar] [CrossRef]
- Bidzinski, P.; Ballini, E.; Ducasse, A.; Michel, C.; Zuluaga, P.; Genga, A.; Chiozzotto, R.; Morel, J.B. Transcriptional basis of drought-induced susceptibility to the rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 2016, 7, 1558. [Google Scholar] [CrossRef]
- Wakelin, S.A.; Gomez-Gallego, M.; Jones, E.; Smaill, S.; Lear, G.; Lambie, S. Climate change induced drought impacts on plant diseases in New Zealand. Australas. Plant Pathol. 2018, 47, 101–114. [Google Scholar] [CrossRef]
- Shakya, S.K.; Goss, E.M.; Dufault, N.S.; Van Bruggen, A.H. Potential effects of diurnal temperature oscillations on potato late blight with special reference to climate change. Phytopathology 2015, 105, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Milus, E.A.; Kristensen, K.; Hovmøller, M.S. Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 2009, 99, 89–94. [Google Scholar]
- Gustafson, E.J.; Miranda, B.R.; Dreaden, T.J.; Pinchot, C.C.; Jacobs, D.F. Beyond blight: Phytophthora root rot under climate change limits populations of reintroduced American chestnut. Ecosphere 2022, 13, e3917. [Google Scholar] [CrossRef]
- Váry, Z.; Mullins, E.; McElwain, J.C.; Doohan, F.M. The severity of wheat diseases increases when plants and pathogens are acclimatized to elevated carbon dioxide. Glob. Change Biol. 2015, 21, 2661–2669. [Google Scholar] [CrossRef]
- Serrano, M.S.; Romero, M.Á.; Homet, P.; Gómez-Aparicio, L. Climate change impact on the population dynamics of exotic pathogens: The case of the worldwide pathogen Phytophthora cinnamomi. Agric. For. Meteorol. 2022, 322, 109002. [Google Scholar] [CrossRef]
- Hanson, M.C.; Petch, G.M.; Ottosen, T.B.; Skjøth, C.A. Climate change impact on fungi in the atmospheric microbiome. Sci. Total Environ. 2022, 830, 154491. [Google Scholar] [CrossRef] [PubMed]
- Lehsten, V.; Wiik, L.; Hannukkala, A.; Andreasson, E.; Chen, D.; Ou, T.; Liljeroth, E.; Lankinen, Å.; Grenville-Briggs, L. Earlier occurrence and increased explanatory power of climate for the first incidence of potato late blight caused by Phytophthora infestans in Fennoscandia. PLoS ONE 2017, 12, e0177580. [Google Scholar] [CrossRef] [PubMed]
- Francisco, C.S.; Ma, X.; Zwyssig, M.M.; McDonald, B.A.; Palma-Guerrero, J. Morphological changes in response to environmental stresses in the fungal plant pathogen Zymoseptoria tritici. Sci. Rep. 2019, 9, 9642. [Google Scholar] [CrossRef] [PubMed]
- Reina-Pinto, J.J.; Yephremov, A. Surface lipids and plant defenses. Plant Physiol. Biochem. 2009, 47, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Jedrzejczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Chandrashekar, N.; Yadav, P.; Rawat, S.; Sultana, M.; Grover, A. Isolation and characterization of systemic acquired resistance marker gene PR1 and its promoter from Brassica juncea. 3 Biotech 2018, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Chandrashekar, N.; Rawat, S.; Nayanakantha, N.M.C.; Mir, Z.A.; Manoharan, A.; Sultana, M.; Grover, A. Isolation and molecular characterization of pathogenesis related PR2 gene and its promoter from Brassica juncea. Biol. Plant 2017, 61, 763–773. [Google Scholar] [CrossRef]
- Staskawicz, B.; Dahlbeck, D.; Keen, N.T. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc. Natl. Acad. Sci. USA 1984, 81, 6024–6028. [Google Scholar] [CrossRef] [PubMed]
- de Wit, P.J.G.M.; Hofman AEv Velthuis, G.C.M.; Kuc, J.A. Isolation and characterization of an elicitor of necrosis isolated from intercellular fluids of compatible interactions of Cladosporium-Fulvum (Syn Fulvia-Fulva) and tomato. Plant Physiol. 1985, 77, 642–647. [Google Scholar] [CrossRef]
- Martin, G.B.; Brommonschenkel, S.H.; Chunwongse, J.; Frary, A.; Ganal, M.W.; Spivey, R.; Wu, T.; Earle, E.D.; Tanksley, S.D. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 1993, 262, 1432–1436. [Google Scholar] [CrossRef]
- Bent, A.F.; Kunkel, B.N.; Dahlbeck, D.; Brown, K.L.; Schmidt, R.; Giraudat, J.; Leung, J.; Staskawicz, B.J. RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.A.; Thomas, C.M.; Hammondkosack, K.E.; Balintkurti, P.J.; Jones, J.D.G. Isolation of the tomato Cf-9 gene for resistance to Cladosporium-Fulvum by transposon tagging. Science 1994, 266, 789–793. [Google Scholar] [CrossRef]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef]
- Dievart, A.; Gottin, C.; Périn, C.; Ranwez, V.; Chantret, N. Origin and diversity of plant receptor-like kinases. Ann. Rev. Plant Biol. 2020, 71, 131–156. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, T.A.; Zipfel, C. Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 2021, 81, 3449–3467. [Google Scholar] [CrossRef]
- Bauer, Z.; Gomez-Gomez, L.; Boller, T.; Felix, G. Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 2001, 276, 45669–45676. [Google Scholar] [CrossRef]
- Shen, Q.; Bourdais, G.; Pan, H.; Robatzek, S.; Tang, D. Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc. Natl. Acad. Sci. USA 2017, 114, 5749–5754. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef]
- Willmann, R.; Lajunen, H.M.; Erbs, G.; Newman, M.A.; Kolb, D.; Tsuda, K.; Katagiri, F.; Fliegmann, J.; Bono, J.J.; Cullimore, J.V.; et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829. [Google Scholar] [CrossRef]
- Liu, B.; Li, J.F.; Ao, Y.; Qu, J.; Li, Z.; Su, J.; Zhang, Y.; Liu, J.; Feng, D.; Qi, K.; et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 2012, 24, 3406–3419. [Google Scholar] [CrossRef]
- Ron, M.; Avni, A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef]
- Jehle, A.K.; Lipschis, M.; Albert, M.; Fallahzadeh-Mamaghani, V.; Furst, U.; Mueller, K.; Felix, G. The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 2013, 25, 2330–2340. [Google Scholar] [CrossRef]
- Huffaker, A.; Ryan, C.A. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 2007, 104, 10732–10736. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Huffaker, A.; Bryan, A.C.; Tax, F.E.; Ryan, C.A. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 2010, 22, 508–522. [Google Scholar] [CrossRef]
- Krol, E.; Mentzel, T.; Chinchilla, D.; Boller, T.; Felix, G.; Kemmerling, B.; Postel, S.; Arents, M.; Jeworutzki, E.; Al-Rasheid, K.A.; et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 2010, 285, 13471–13479. [Google Scholar] [CrossRef]
- de Jonge, R.; van Esse, H.P.; Maruthachalam, K.; Bolton, M.D.; Santhanam, P.; Saber, M.K.; Zhang, Z.; Usami, T.; Lievens, B.; Subbarao, K.V.; et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 5110–5115. [Google Scholar] [CrossRef] [PubMed]
- Joosten, M.H.; Vogelsang, R.; Cozijnsen, T.J.; Verberne, M.C.; De Wit, P.J. The biotrophic fungus Cladosporium fulvum circumvents Cf-4-mediated resistance by producing unstable AVR4 elicitors. Plant Cell 1997, 9, 367–379. [Google Scholar]
- Dixon, M.S.; Jones, D.A.; Keddie, J.S.; Thomas, C.M.; Harrison, K.; Jones, J.D. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 1996, 84, 451–459. [Google Scholar] [CrossRef]
- Kruger, J.; Thomas, C.M.; Golstein, C.; Dixon, M.S.; Smoker, M.; Tang, S.; Mulder, L.; Jones, J.D. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 2002, 296, 744–747. [Google Scholar] [CrossRef]
- Dixon, M.S.; Hatzixanthis, K.; Jones, D.A.; Harrison, K.; Jones, J.D. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 1998, 10, 1915–1925. [Google Scholar] [CrossRef]
- Takken, F.L.; Thomas, C.M.; Joosten, M.H.; Golstein, C.; Westerink, N.; Hille, J.; Nijkamp, H.J.; De Wit, P.J.; Jones, J.D. A second gene at the tomato Cf-4 locus confers resistance to cladosporium fulvum through recognition of a novel avirulence determinant. Plant J. 1999, 20, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Westerink, N.; Brandwagt, B.F.; de Wit, P.J.; Joosten, M.H. Cladosporium fulvum circumvents the second functional resistance gene homologue at the Cf-4 locus (Hcr9-4E) by secretion of a stable avr4E isoform. Mol. Microbiol. 2004, 54, 533–545. [Google Scholar] [CrossRef]
- Panter, S.N.; Hammond-Kosack, K.E.; Harrison, K.; Jones, J.D.; Jones, D.A. Developmental control of promoter activity is not responsible for mature onset of Cf-9B-mediated resistance to leaf mold in tomato. Mol. Plant Microbe Interact. 2002, 15, 1099–1107. [Google Scholar] [CrossRef]
- Mosher, S.; Seybold, H.; Rodriguez, P.; Stahl, M.; Davies, K.A.; Dayaratne, S.; Morillo, S.A.; Wierzba, M.; Favery, B.; Keller, H.; et al. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 2013, 73, 469–482. [Google Scholar] [CrossRef]
- Llorente, F.; Alonso-Blanco, C.; Sanchez-Rodriguez, C.; Jorda, L.; Molina, A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 2005, 43, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Alcazar, R.; Garcia, A.V.; Kronholm, I.; de Meaux, J.; Koornneef, M.; Parker, J.E.; Reymond, M. Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat. Genet. 2010, 42, 1135–1139. [Google Scholar] [CrossRef]
- Kawahigashi, H.; Kasuga, S.; Ando, T.; Kanamori, H.; Wu, J.; Yonemaru, J.; Sazuka, T.; Matsumoto, T. Positional cloning of ds1, the target leaf spot resistance gene against Bipolaris sorghicola in sorghum. Theor. Appl. Genet. 2011, 123, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Mantelin, S.; Peng, H.C.; Li, B.; Atamian, H.S.; Takken, F.L.; Kaloshian, I. The receptor-like kinase SlSERK1 is required for Mi-1-mediated resistance to potato aphids in tomato. Plant J. 2011, 67, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Tanaka, K.; Zhang, X.C.; Son, G.H.; Brechenmacher, L.; Nguyen, T.H.; Stacey, G. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012, 160, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Velasquez, A.C.; Munkvold, K.R.; Zhang, J.; Martin, G.B. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 2012, 69, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Hematy, K.; Sado, P.E.; Van Tuinen, A.; Rochange, S.; Desnos, T.; Balzergue, S.; Pelletier, S.; Renou, J.P.; Hofte, H. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 2007, 17, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.A.; Shimosato-Asano, H.; Keinath, N.F.; Wuest, S.E.; Ingram, G.; Panstruga, R.; Grossniklaus, U. Conserved molecular components for pollen tube reception and fungal invasion. Science 2010, 330, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef]
- Li, H.; Zhou, S.Y.; Zhao, W.S.; Su, S.C.; Peng, Y.L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar] [CrossRef]
- Zhou, H.; Li, S.; Deng, Z.; Wang, X.; Chen, T.; Zhang, J.; Chen, S.; Ling, H.; Zhang, A.; Wang, D.; et al. Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. Plant J. 2007, 52, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Cheng, Y.T.; Li, X.; Zhang, Y. Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol. 2010, 153, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Schachermayr, G.; Keller, B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. 1997, 11, 4552. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nurnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef]
- Schulze, B.; Mentzel, T.; Jehle, A.K.; Mueller, K.; Beeler, S.; Boller, T.; Felix, G.; Chinchilla, D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 2010, 285, 9444–9451. [Google Scholar] [CrossRef]
- Postel, S.; Kufner, I.; Beuter, C.; Mazzotta, S.; Schwedt, A.; Borlotti, A.; Halter, T.; Kemmerling, B.; Nurnberger, T. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol. 2010, 89, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tor, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Bar, M.; Sharfman, M.; Ron, M.; Avni, A. BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 2010, 63, 791–800. [Google Scholar] [CrossRef]
- Liebrand, T.W.; van den Berg, G.C.; Zhang, Z.; Smit, P.; Cordewener, J.H.; America, A.H.; Sklenar, J.; Jones, A.M.; Tameling, W.I.; Robatzek, S.; et al. Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA 2013, 110, 10010–10015. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Wang, B.; Chen, J.Q. Tracking ancestral lineages and recent expansions of NBS-LRR genes in angiosperms. Plant Signal. Behav. 2016, 11, 2095–2109. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, Q.; Gao, S.; Yu, N.; Zhao, L.; Wang, J.; Zhao, J.; Huang, P.; Yao, L.; Wang, M.; et al. Disruption of the primary salicylic acid hydroxylases in rice enhances broad-spectrum resistance against pathogens. Plant Cell Environ. 2022, 45, 2211–2225. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Bruce, T.J.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Cao, L.; Yoo, H.; Chen, T.; Mwimba, M.; Zhang, X.; Dong, X. H2O2 sulfenylates CHE linking local infection to establishment of systemic acquired resistance. bioRxiv 2023. [Google Scholar] [CrossRef]
- Leong, J.X.; Langin, G.; Üstün, S. Selective autophagy: Adding precision in plant immunity. Essays Biochem. 2022, 66, 189–206. [Google Scholar]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Czymmek, K.; Tallóczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef]
- Lenz, H.D.; Haller, E.; Melzer, E.; Kober, K.; Wurster, K.; Stahl, M.; Bassham, D.C.; Vierstra, R.D.; Parker, J.E.; Bautor, J.; et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011, 66, 818–830. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Dinesh-Kumar, S.P. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 2008, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, I.V.; Shein, M.Y.; Burkhanova, G.F. RNA Interference in Plant Defense Systems. Russ. J. Plant Physiol. 2021, 68, 613–625. [Google Scholar] [CrossRef]
- Vance, V.; Vaucheret, H. RNA silencing in plants--defense and counterdefense. Science 2001, 292, 2277–2280. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gomollon, S.; Baulcombe, D.C. Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat. Rev. Mol. Cell Biol. 2022, 23, 645–662. [Google Scholar] [CrossRef] [PubMed]
- Marcec, M.J.; Gilroy, S.; Poovaiah, B.W.; Tanaka, K. Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 2019, 283, 343–354. [Google Scholar] [CrossRef]
- Seybold, H.; Trempel, F.; Ranf, S.; Scheel, D.; Romeis, T.; Lee, J. Ca2+ signalling in plant immune response: From pattern recognition receptors to Ca2+ decoding mechanisms. New Phytol. 2014, 204, 782–790. [Google Scholar] [CrossRef]
- Moeder, W.; Phan, V.; Yoshioka, K. Ca2+ to the rescue– Ca2+ channels and signaling in plant immunity. Plant Sci. 2019, 279, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef]
- Edel, K.H.; Marchadier, E.; Brownlee, C.; Kudla, J.; Hetherington, A.M. The evolution of calcium-based signalling in plants. Curr. Biol. 2017, 27, R667–R679. [Google Scholar] [CrossRef]
- Costa, A.; Navazio, L.; Szabo, I. The contribution of organelles to plant intracellular calcium signalling. J. Exp. Bot. 2018, 69, 4175–4193. [Google Scholar] [CrossRef]
- Saand, M.A.; Xu, Y.P.; Munyampundu, J.P.; Li, W.; Zhang, X.R.; Cai, X.Z. Phylogeny and evolution of plant cyclic nucleotide-gated ion channel (CNGC) gene family and functional analyses of tomato CNGCs. DNA Res. 2015, 22, 471–483. [Google Scholar] [CrossRef]
- Toyota, M. Conservation of Long-Range Signaling in Land Plants via Glutamate Receptor–Like Channels. Plant Cell Physiol. 2024, 65, 657–659. [Google Scholar] [CrossRef]
- Thor, K.; Jiang, S.; Michard, E.; George, J.; Scherzer, S.; Huang, S.; Dindas, J.; Derbyshire, P.; Leitão, N.; DeFalco, T.A.; et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 2020, 585, 569–573. [Google Scholar] [CrossRef]
- Ali, R.; Ma, W.; Lemtiri-chlieh, F.; Tsaltas, D.; Leng, Q.; Bodman, S.; Berkowitz, G. Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 2007, 19, 1081–1095. [Google Scholar] [CrossRef]
- Ma, Y.; Walker, R.; Zhao, Y.; Berkowitz, G. Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 19852–19857. [Google Scholar] [CrossRef]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Zhang, A.; Ren, Y.; Wu, F.; Wang, G.; Xu, Y.; Lei, C.; Zhu, S.; Pan, T.; et al. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019, 29, 820–831. [Google Scholar] [CrossRef]
- Meena, M.; Prajapati, R.; Krishna, D.; Divakaran, K.; Pandey, Y.; Reichelt, M.; Mathew, M.; Boland, W.; Mithöfer, A.; Vadassery, J. The Ca2+ channel CNGC19 regulates Arabidopsis defense against Spodoptera herbivory. Plant Cell 2019, 31, 1539–1562. [Google Scholar] [CrossRef]
- Yu, X.; Xie, Y.; Luo, D.; Liu, H.; de Oliveira, M.; Qi, P.; Kim, S.; Ortiz-Morea, F.; Liu, J.; Chen, Y.; et al. A phospho-switch constrains BTL2-mediated phytocytokine signaling in plant immunity. Cell 2013, 186, 2329–2344. [Google Scholar] [CrossRef]
- Espinoza, C.; Liang, Y.; Stacey, G. Chitin receptor CERK1 links salt stress and chitin-triggered innate immunity in Arabidopsis. Plant J. 2017, 89, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Bjornson, M.; Pimprikar, P.; Nürnberger, T.; Zipfel, C. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat. Plants 2021, 7, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Berrios, L.; Rentsch, J.D. Linking reactive oxygen species (ROS) to abiotic and biotic feedbacks in plant microbiomes: The dose makes the poison. Inter. J. Mol. Sci. 2022, 23, 4402. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S.; Eschen-Lippold, L.; Fröhlich, K.; Westphal, L.; Scheel, D.; Lee, J. Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 2014, 14, 374. [Google Scholar] [CrossRef]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal 2009, 2, ra45. [Google Scholar] [CrossRef]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, calcium, and electric signals: Key mediators of rapid systemic signaling in plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- López, M.A.; Bannenberg, G.; Castresana, C. Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr. Opin. Plant Biol. 2008, 11, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Kobayashi, M.; Mitsuhara, I.; Seo, S. Jasmonic acid negatively regulates resistance to Tobacco mosaic virus in tobacco. Plant Cell Physiol. 2013, 54, 1999–2010. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Ann. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mir, Z.A.; Tyagi, A.; Mehari, H.; Meena, R.P.; Bhat, J.A.; Yadav, P.; Papalou, P.; Rawat, S.; Grover, A. Overexpression of NPR1 in Brassica juncea confers broad-spectrum resistance to fungal pathogens. Front. Plant Sci. 2017, 8, 1693. [Google Scholar] [CrossRef] [PubMed]
- Zavaliev, R.; Mohan, R.; Chen, T.; Dong, X. Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 2020, 182, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic acid biosynthesis in plants. Front. Plant Sci. 2020, 11, 521987. [Google Scholar] [CrossRef]
- Tandon, G.; Jaiswal, S.; Iquebal, M.A.; Kumar, S.; Kaur, S.; Rai, A.; Kumar, D. Evidence of salicylic acid pathway with EDS1 and PAD4 proteins by molecular dynamics simulation for grape improvement. J. Biomol. Struct. Dyn. 2015, 33, 2180–2191. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.E.; Munné-Bosch, S. Salicylic acid deficiency in NahG transgenic lines and sid2 mutant’s increases seed yield in the annual plant Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Lefert, P.; Robatzek, S. Plant pathogens trick guard cells into opening the gates. Cell 2006, 126, 831–834. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. The missing link in jasmonic acid biosynthesis. Nat. Plants 2019, 5, 776–777. [Google Scholar] [CrossRef]
- Van Moerkercke, A.; Duncan, O.; Zander, M.; Šimura, J.; Broda, M.; Vanden Bossche, R.; Lewsey, M.G.; Lama, S.; Singh, K.B.; Ljung, K.; et al. A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. USA 2019, 116, 23345–23356. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.K.; Gao, X.; Jones, A.D.; Howe, G.A. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009, 59, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Ruiz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes. Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef]
- Taylor, J.E.; Hatcher, P.E.; Paul, N.D. Crosstalk between plant responses to pathogens and herbivores: A view from the outside in. J. Exp. Bot. 2004, 55, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Hilleary, R.; Swanson, S.J.; Kim, S.H.; Gilroy, S. Rapid, long-distance electrical and calcium signaling in plants. Annu. Rev. Plant Biol. 2016, 67, 287–310. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Raikhi, G.; Kumar, D. Chemical elicitors of systemic acquired resistance—Salicylic acid and its functional analogs. Curr. Plant Biol. 2019, 17, 48–59. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant immunity: From signaling to epigenetic control of defense. Trends Plant Sci. 2018, 23, 833–844. [Google Scholar] [CrossRef]
- Palukaitis, P.; Yoon, J.Y. R gene mediated defense against viruses. Curr. Opin. Virol. 2020, 45, 1–7. [Google Scholar] [CrossRef]
- Campos, L.; Granell, P.; Tárraga, S.; López-Gresa, P.; Conejero, V.; Bellés, J.M.; Rodrigo, I.; Lisón, P. Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiol. Biochem. 2014, 77, 35–43. [Google Scholar] [CrossRef]
- Han, K.; Huang, H.; Zheng, H.; Ji, M.; Yuan, Q.; Cui, W.; Zhang, H.; Peng, J.; Lu, Y.; Rao, S.; et al. Rice stripe virus coat protein induces the accumulation of jasmonic acid, activating plant defence against the virus while also attracting its vector to feed. Mol. Plant Pathol. 2020, 21, 1647–1653. [Google Scholar] [CrossRef]
- Ji, M.; Zhao, J.; Han, K.; Cui, W.; Wu, X.; Chen, B.; Lu, Y.; Peng, J.; Zheng, H.; Rao, S.; et al. Turnip mosaic virus P1 suppresses JA biosynthesis by degrading cpSRP54 that delivers AOCs onto the thylakoid membrane to facilitate viral infection. PLoS Pathog. 2021, 17, e1010108. [Google Scholar] [CrossRef] [PubMed]
- Syller, J.; Grupa, A. Antagonistic within-host interactions between plant viruses: Molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 2016, 17, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kanayama, Y.; Zheng, M.S.; Kusano, T.; Hase, S.; Ikegami, M.; Shah, J. Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol. 2004, 45, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Baldwin, I.T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.X.; Feng, Q.; Liu, P.Q.; He, X.R.; Zhao, J.H.; Xu, Y.J.; Zhang, L.L.; Huang, Y.Y.; Zhao, J.Q.; Fan, J.; et al. RPW8.1 enhances the ethylene-signaling pathway to feedback-attenuate its mediated cell death and disease resistance in Arabidopsis. New Phytol. 2021, 229, 516. [Google Scholar] [CrossRef] [PubMed]
- Pré, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef]
- Mur, L.A.; Lloyd, A.J.; Cristescu, S.M.; Harren, F.J.; Hall, M.; Smith, A. Biphasic ethylene production during the hypersensitive response in Arabidopsis: A window into defence priming mechanisms? Plant Signal Behav. 2009, 4, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; D’Agostino, N.; Tytgat, T.O.G.; Sun, P.; Lortzing, T.; Visser, E.J.W.; Cristescu, S.M.; Steppuhn, A.; Mariani, C.; van Dam, N.M.; et al. Drought and flooding have distinct effects on herbivore-induced responses and resistance in Solanum dulcamara. Plant Cell Environ. 2016, 39, 1485–1499. [Google Scholar] [CrossRef]
- García-Andrade, J.; González, B.; Gonzalez-Guzman, M.; Rodriguez, P.L.; Vera, P. The Role of ABA in Plant Immunity is Mediated through the PYR1 Receptor. Int. J. Mol. Sci. 2020, 21, 5852. [Google Scholar] [CrossRef] [PubMed]
- Mine, A.; Berens, M.L.; Nobori, T.; Anver, S.; Fukumoto, K.; Winkelmüller, T.M.; Takeda, A.; Becker, D.; Tsuda, K. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc. Natl. Acad. Sci. USA 2017, 114, 7456–7461. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, G.; Meng, X.; Liu, Y.; Ji, X.; Li, Y.; Nie, X.; Wang, Y. A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol. Biol. 2013, 82, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
| Receptors | Family | Co-Receptor/Ligand | Host Plant | References |
|---|---|---|---|---|
| FLS2 | LRR RLK/LLG1 | Flg22 | A. thaliana | [49,50] |
| EFR | LRR RLK | Elf18 | A. thaliana | [51] |
| CERK1 | LysM RLK | Chitin | A. thaliana, Oryza sativa | [52,53] |
| CEBiP | LysM RLP | Chitin | O. sativa | [54] |
| LYM1/LYM3 | LysM RLP | PGNs | A. thaliana | [55] |
| LYP4/6 | LysM RLP | PGNs/chitin | O. sativa | [56] |
| LeEix2 | LRR RLP | Eix | Solanum lycopersicum | [57] |
| ReMax | LRR RLP | eMax | A. thaliana | [58] |
| PEPR1/2 | LRR RLK | Peps | A. thaliana | [59,60,61] |
| Ve1 | LRR RLP | Ave1 | S. lycopersicum | [62] |
| Cf-2/4/5/9 | LRR RLP | Avr2, Avr4, Avr9 | S. lycopersicum | [63,64,65,66] |
| Cf-4E | LRR RLP | Avr4E | S. lycopersicum | [67,68] |
| Cf-9B | LRR RLP | Unknown | S. lycopersicum | [69] |
| PSKR1 | LRR RLK | PSKα | A. thaliana | [70] |
| BIR1, SOBIR1, ERECTA, SRF3 | LRR RLK | Unknown | A. thaliana | [71,72] |
| ds1 | LRR RLK | Unknown | Sorghum bicolor | [73] |
| SISERK1 | LRR RLK | Unknown | S. lycopersicum | [74] |
| NbSERK1 | LRR RLK | Unknown | Nicotiana benthamiana | [75] |
| LYK4 | LysM RLK | Unknown | A. thaliana | [76] |
| Bti9, SlLyk13 | LysM RLK | Unknown | S. lycopersicum | [77] |
| THE1m, FER | CrRLK1L RLK | Unknown | A. thaliana | [78,79] |
| Pi-d2 | LecRK | Unknown | O. sativa | [80] |
| OsWAK1 | WAK | Unknown | O. sativa | [81] |
| TaRLK-R1, 2, 3 | Other | Unknown | Triticum aestivum | [82] |
| SNC4 | Other | Unknown | A. thaliana | [83] |
| LRK10 | S-domain | Unknown | T. aestivum | [84] |
| BAK1 | LRR RLK | Flg22, elf18, Peps, Eix | A. thaliana | [85,86,87,88] |
| LeEix1 | LRR RLP | Eix | S. lycopersicum | [89] |
| SOBIR1 | LRR RLK | Avr4, Ve1 | S. lycopersicum | [90] |
| Calcium Channel | Family | Activation | Plants | References |
|---|---|---|---|---|
| CNGC2/4 | CNGC family | Flg22, plant elicitor peptide pep3, or lipopolysaccharides (LPSs) | A. thaliana | [115,116,117] |
| OsCNGC9 | CNGC family | Chitin | O. sativa | [118] |
| CNGC19 | CNGC family | Pep1 | A. thaliana | [119] |
| CNGC20 | CNGC family | BAK-TO LIFE 2 (BTL2) | A. thaliana | [120] |
| OSCA1.3 and OSCA1.7 | OSCA family | BIK1 | A. thaliana | [114] |
| ANNEXIN1 (ANN1) | Annexin gene family | CERK1 | A. thaliana | [121] |
| GLR2.7, GLR2.8, and GLR2.9 | GLR family | Flg22-, elf18-, and pep1 | A. thaliana | [122] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, S.; Tyagi, A.; Mir, Z.A. Plant Immunity: At the Crossroads of Pathogen Perception and Defense Response. Plants 2024, 13, 1434. https://doi.org/10.3390/plants13111434
Ali S, Tyagi A, Mir ZA. Plant Immunity: At the Crossroads of Pathogen Perception and Defense Response. Plants. 2024; 13(11):1434. https://doi.org/10.3390/plants13111434
Chicago/Turabian StyleAli, Sajad, Anshika Tyagi, and Zahoor Ahmad Mir. 2024. "Plant Immunity: At the Crossroads of Pathogen Perception and Defense Response" Plants 13, no. 11: 1434. https://doi.org/10.3390/plants13111434
APA StyleAli, S., Tyagi, A., & Mir, Z. A. (2024). Plant Immunity: At the Crossroads of Pathogen Perception and Defense Response. Plants, 13(11), 1434. https://doi.org/10.3390/plants13111434








