Complete Genome of Rose Myrtle, Rhodomyrtus tomentosa, and Its Population Genetics in Thai Peninsula
Abstract
1. Introduction
2. Results
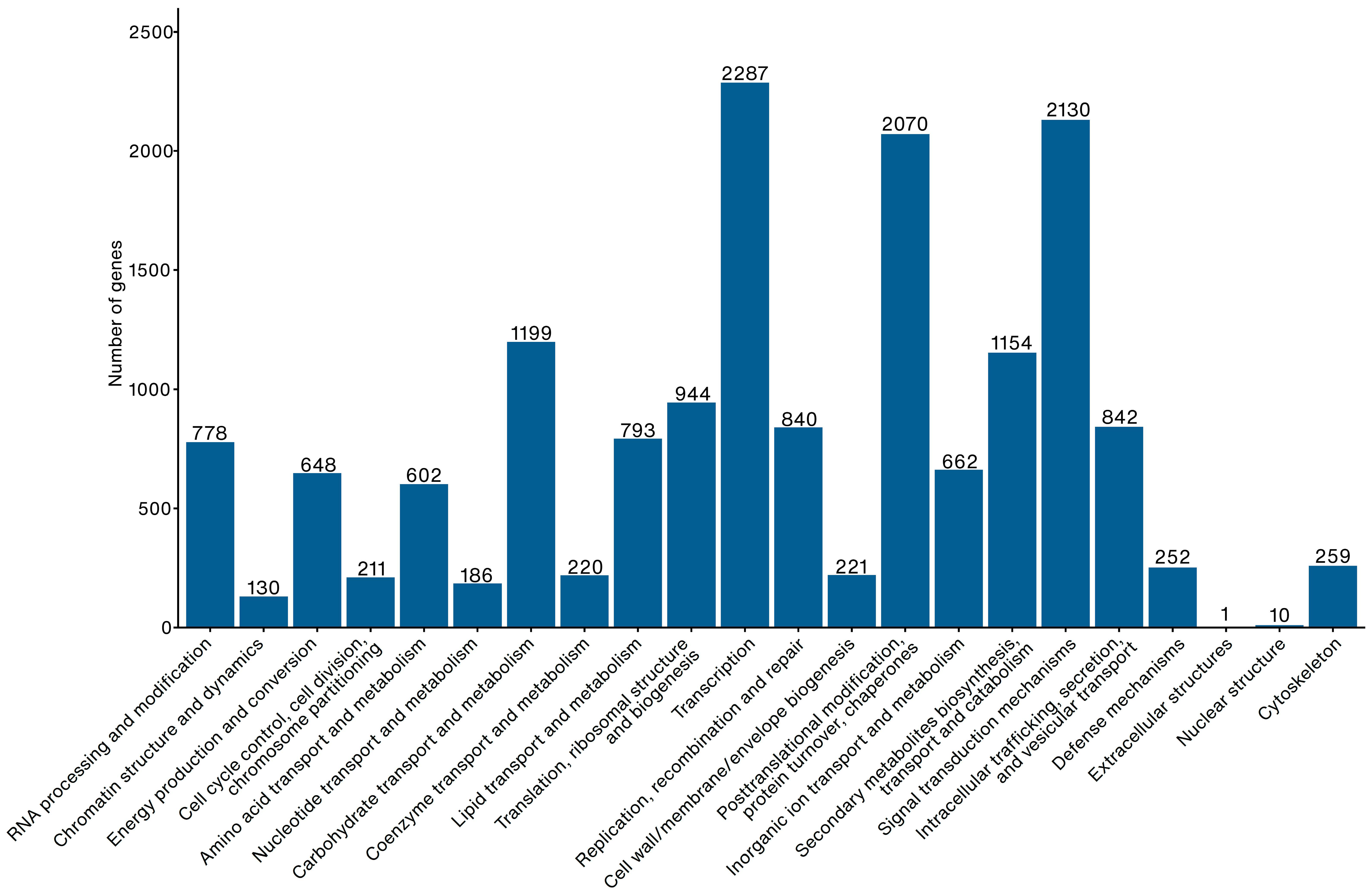
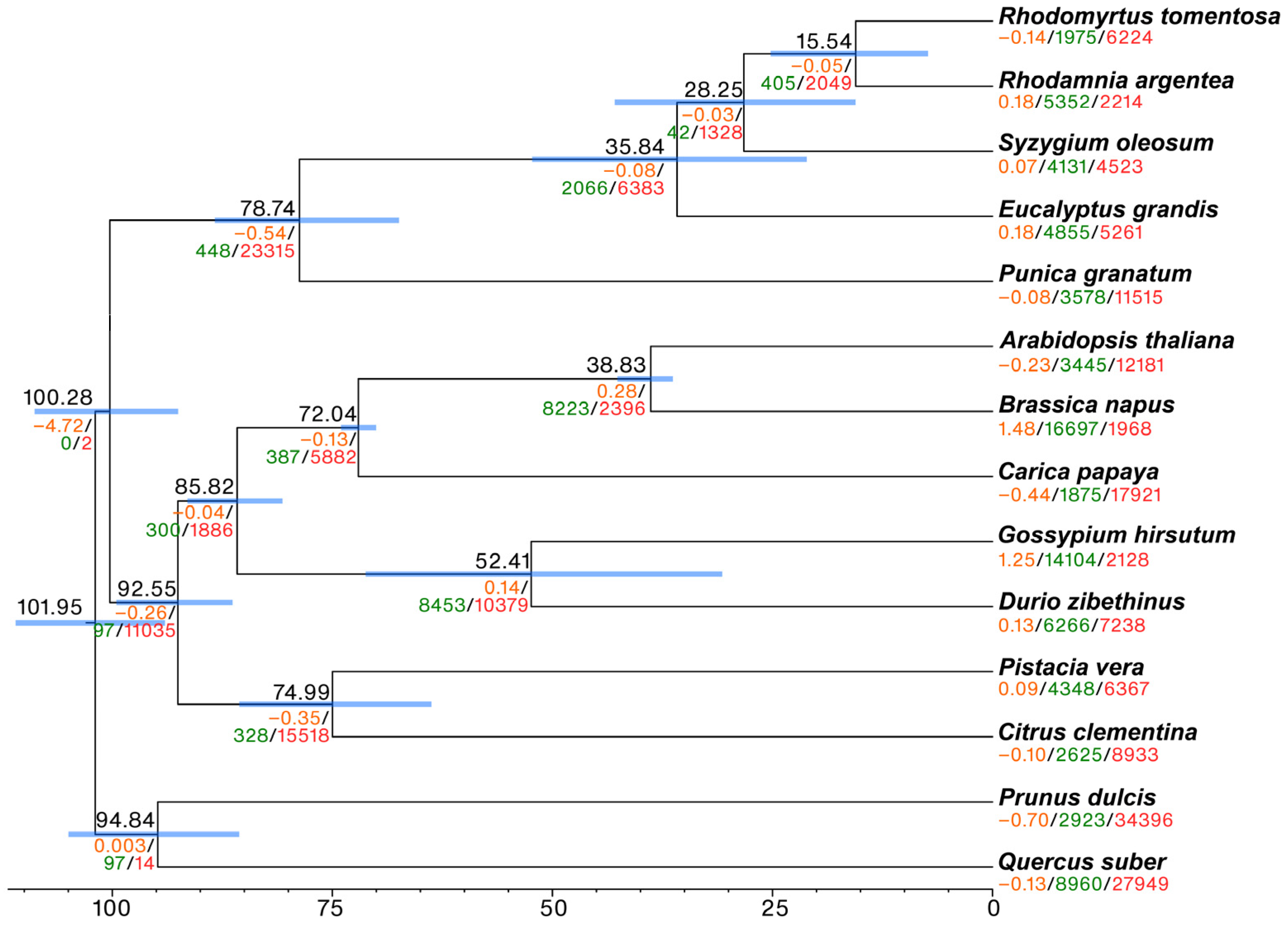
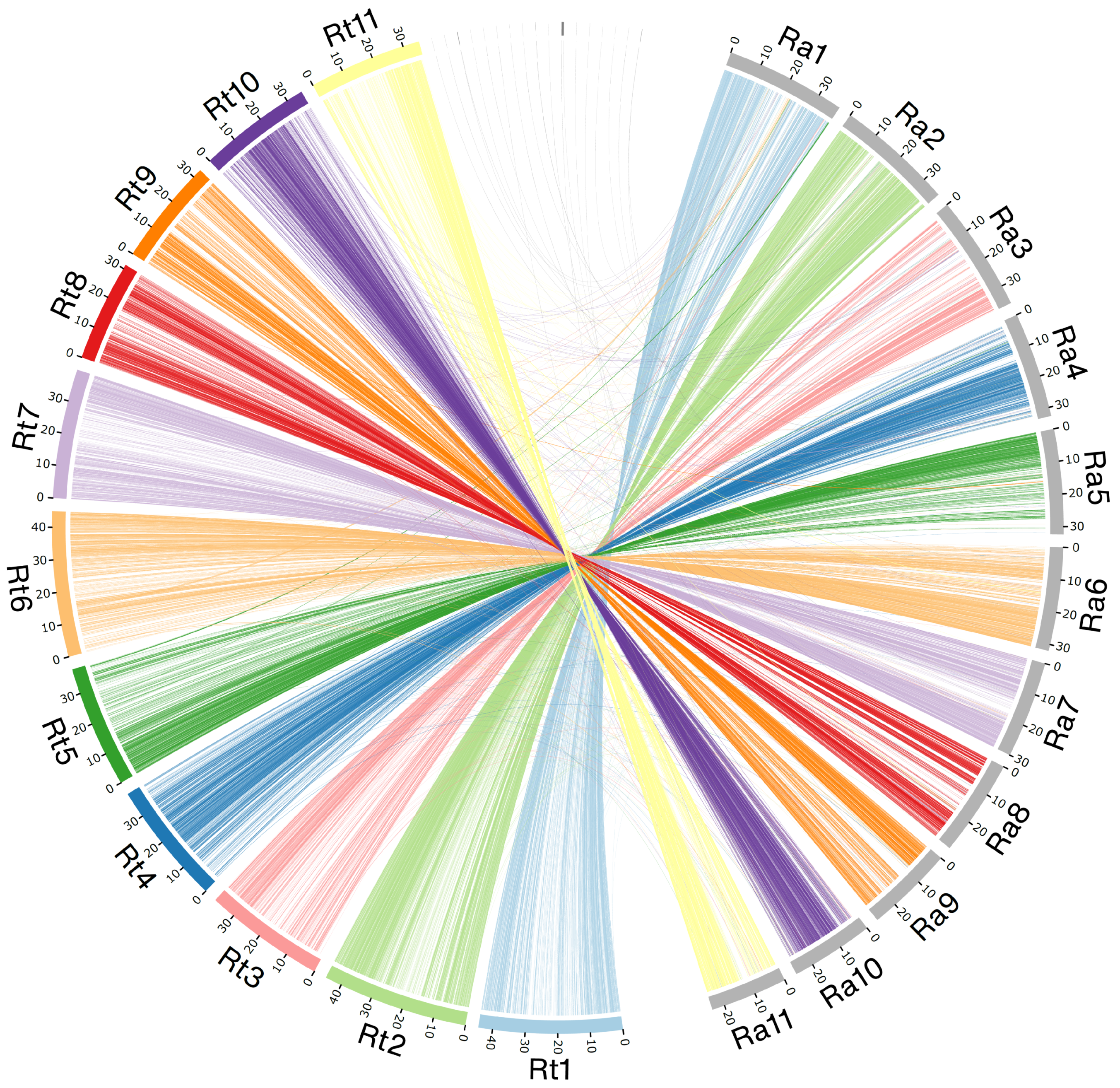
2.1. Genome Assembly
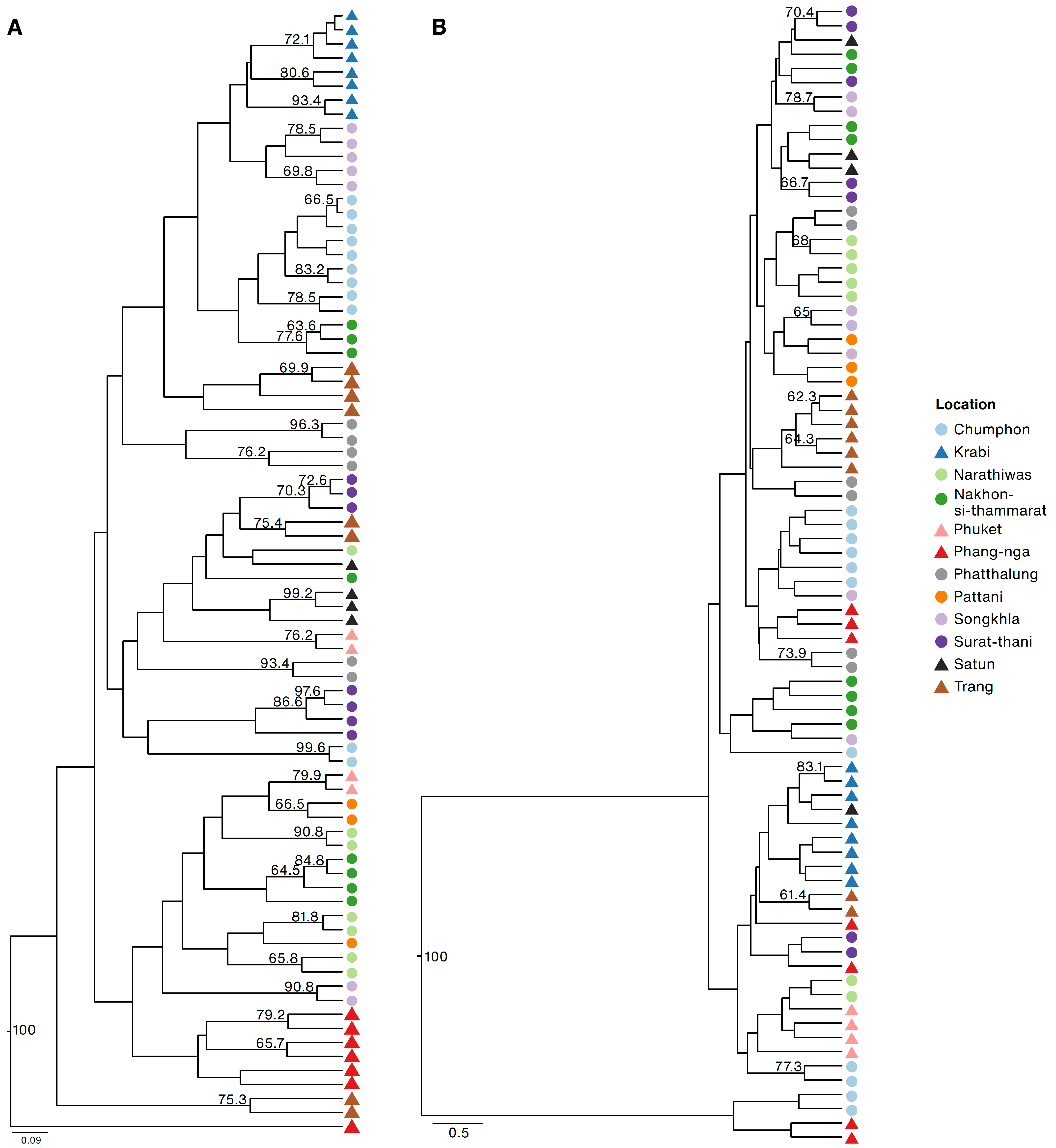
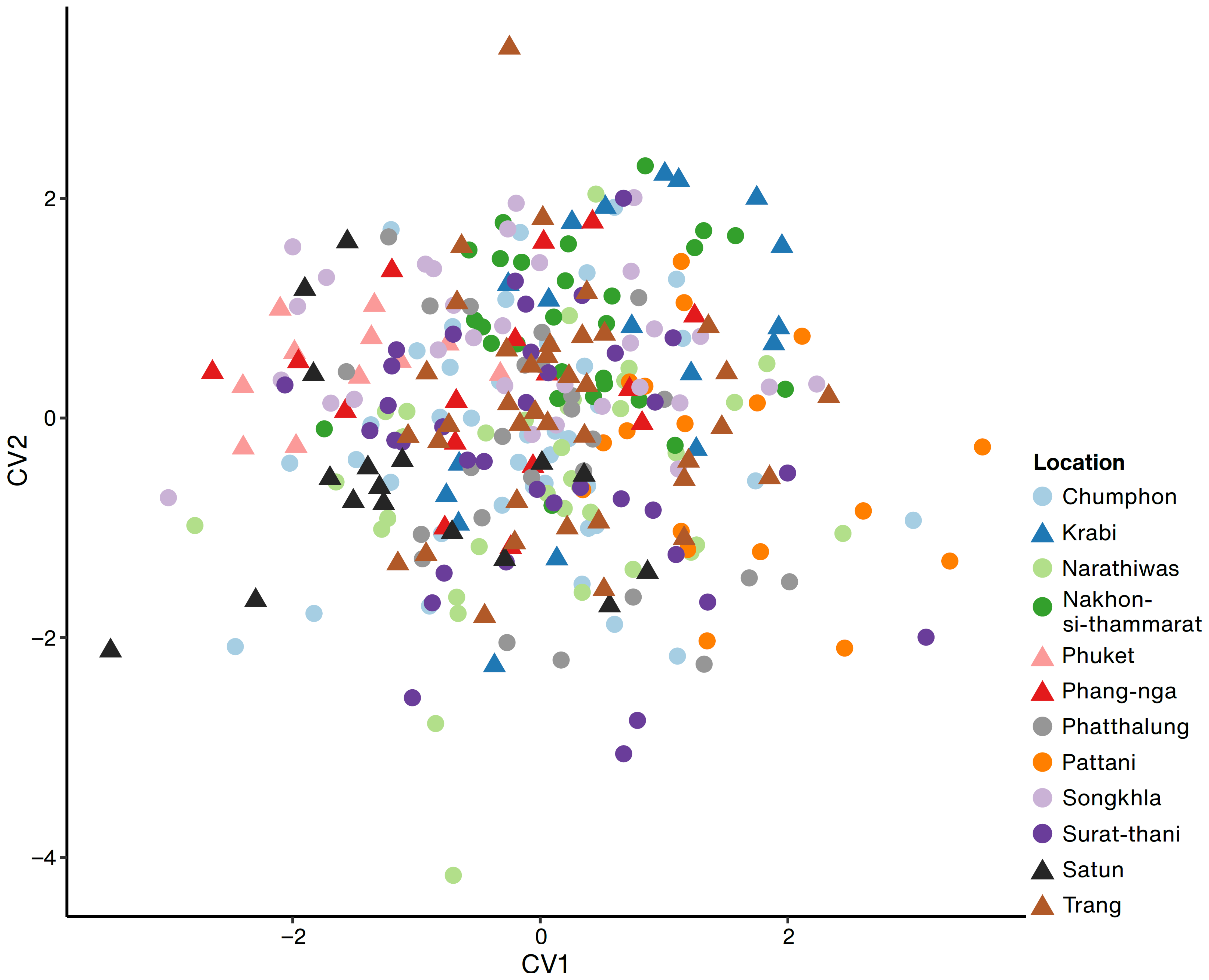
2.2. Population Genetics and Leaf Morphometrics of R. tomentosa on Thai Peninsula
3. Discussion
3.1. R. tomentosa Genome
3.2. Population Genetics and Geometric Morphometrics of R. tomentosa in Thai Peninsula
4. Materials and Methods
4.1. Genome Assembly
4.2. Population Genetic Analysis
4.3. Geometric Morphometrics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, T. (Ed.) Check List of Hong Kong Plants. Agriculture, Fisheries and Conservation Department Bulletin 1 (Revised Edn); Hong Kong Herbarium and the South China Institute of Botany: Hong Kong, China, 2002. [Google Scholar]
- Winotai, A.; Wright, T.; Goolsby, J.A. Herbivores in Thailand on Rhodomyrtus tomentosa (Myrtaceae), an Invasive Weed in Florida. Fla. Entomol. 2005, 88, 104–105. [Google Scholar] [CrossRef]
- Ren, H.; Peng, S.; Sun, G.; Yu, Z. The Ecological Comparison of Psychotria rubra and Rhodomyrtus tomentosa in South China. Chin. J. Plant Ecol. 1997, 21, 386–392. [Google Scholar]
- Langeland, K.A.; Craddock Burks, K. Identification and Biology of Non-Native Plants in Florida’s Natural Areas; University of Florida: Gainesville, FL, USA, 1998. [Google Scholar]
- Wei, M.S.; Chen, Z.H.; Ren, H.; Yin, Z.Y. Reproductive Ecology of Rhodomyrtus tomentosa (Myrtaceae). Nord. J. Bot. 2009, 27, 154–160. [Google Scholar] [CrossRef]
- Ong, H.C.; Nordiana, M. Malay Ethno-Medico Botany in Machang, Kelantan, Malaysia. Fitoterapia 1999, 70, 502–513. [Google Scholar] [CrossRef]
- Wei, F. Manufacture of Oral Liquid Contaning Traditional Chinese Medicine Extract for Treating Gynecopathy; Guangxi Huahong Pharmaceutical Co., Ltd.: Liuzhou, China, 2006. [Google Scholar]
- Zhao, Z.; Wu, L.; Xie, J.; Feng, Y.; Tian, J.; He, X.; Li, B.; Wang, L.; Wang, X.; Zhang, Y.; et al. Rhodomyrtus tomentosa (Aiton.): A Review of Phytochemistry, Pharmacology and Industrial Applications Research Progress. Food Chem. 2020, 309, 125715. [Google Scholar] [CrossRef]
- Limsuwan, S.; Trip, E.N.; Kouwen, T.R.H.M.; Piersma, S.; Hiranrat, A.; Mahabusarakam, W.; Voravuthikunchai, S.P.; van Dijl, J.M.; Kayser, O. Rhodomyrtone: A New Candidate as Natural Antibacterial Drug from Rhodomyrtus tomentosa. Phytomedicine 2009, 16, 645–651. [Google Scholar] [CrossRef]
- Leejae, S.; Hasap, L.; Voravuthikunchai, S.P. Inhibition of Staphyloxanthin Biosynthesis in Staphylococcus aureus by Rhodomyrtone, a Novel Antibiotic Candidate. J. Med. Microbiol. 2013, 62, 421–428. [Google Scholar] [CrossRef]
- Chorachoo, J.; Lambert, S.; Furnholm, T.; Roberts, L.; Reingold, L.; Auepemkiate, S.; Voravuthikunchai, S.P.; Johnston, A. The Small Molecule Rhodomyrtone Suppresses TNF-∞ and IL-17A-Induced Keratinocyte Inflammatory Responses: A Potential New Therapeutic for Psoriasis. PLoS ONE 2018, 13, e0205340. [Google Scholar] [CrossRef]
- Saising, J.; Nguyen, M.T.; Härtner, T.; Ebner, P.; Al Mamun Bhuyan, A.; Berscheid, A.; Muehlenkamp, M.; Schäkermann, S.; Kumari, N.; Maier, M.E.; et al. Rhodomyrtone (Rom) Is a Membrane-Active Compound. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1114–1124. [Google Scholar] [CrossRef]
- Wunnoo, S.; Bilhman, S.; Amnuaikit, T.; Ontong, J.C.; Singh, S.; Auepemkiate, S.; Voravuthikunchai, S.P. Rhodomyrtone as a New Natural Antibiotic Isolated from Rhodomyrtus tomentosa Leaf Extract: A Clinical Application in the Management of Acne Vulgaris. Antibiotics 2021, 10, 108. [Google Scholar] [CrossRef]
- Lai, T.N.H.; Herent, M.F.; Quetin-Leclercq, J.; Nguyen, T.B.T.; Rogez, H.; Larondelle, Y.; André, C.M. Piceatannol, a Potent Bioactive Stilbene, as Major Phenolic Component in Rhodomyrtus tomentosa. Food Chem. 2013, 138, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Grattapaglia, D.; Vaillancourt, R.E.; Shepherd, M.; Thumma, B.R.; Foley, W.; Külheim, C.; Potts, B.M.; Myburg, A.A. Progress in Myrtaceae Genetics and Genomics: Eucalyptus as the Pivotal Genus. Tree Genet. Genomes 2012, 8, 463–508. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, Z.; Huang, S.; An, W.; Li, J.; Zheng, X. Comprehensive Analysis of Rhodomyrtus tomentosa Chloroplast Genome. Plants 2019, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xu, S.; Xiao, Z.; Wang, J.; Mei, Y.; Hu, H.; Li, J.; Liu, J.; Hou, Z.; Zhao, J.; et al. Gap-Free Genome Assembly and Comparative Analysis Reveal the Evolution and Anthocyanin Accumulation Mechanism of Rhodomyrtus tomentosa. Hortic. Res. 2023, 10, uhad005. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, N.; Subramanian, D. Cytotaxonomical Studies in South Indian Myrtaceae. Cytologia 1985, 50, 513–520. [Google Scholar] [CrossRef]
- Hue, T.S.; Abdullah, T.L.; Abdullah, N.A.P.; Sinniah, U.R. Genetic Variation in Rhodomyrtus tomentosa (Kemunting) Populations from Malaysia as Revealed by Inter-Simple Sequence Repeat Markers. Genet. Mol. Res. 2015, 14, 16827–16839. [Google Scholar] [CrossRef]
- Sun, L.; Li, J.; Sun, K.; Wang, H.; Yang, K.; Chen, Q.; Lin, M. Development and Characterization of EST-SSR Markers in Rhodomyrtus tomentosa Based on Transcriptome. Genet. Resour. Crop Evol. 2022. [Google Scholar] [CrossRef]
- Snow, N.; McFadden, J.; Evans, T.M.; Salywon, A.M.; Wojciechowski, M.F.; Wilson, P.G. Morphological and Molecular Evidence of Polyphyly in Rhodomyrtus (Myrtaceae: Myrteae). Syst. Bot. 2011, 36, 390–404. [Google Scholar] [CrossRef]
- Carvalho, G.M.A.; Carvalho, C.R.; Soares, F.A.F. Flow Cytometry and Cytogenetic Tools in Eucalypts: Genome Size Variation × Karyotype Stability. Tree Genet. Genomes 2017, 13, 106. [Google Scholar] [CrossRef]
- Wang, W.; Das, A.; Kainer, D.; Schalamun, M.; Morales-Suarez, A.; Schwessinger, B.; Lanfear, R. The Draft Nuclear Genome Assembly of Eucalyptus pauciflora: A Pipeline for Comparing de novo Assemblies. Gigascience 2020, 9, giz160. [Google Scholar] [CrossRef]
- Lai, T.N.H.; André, C.; Rogez, H.; Mignolet, E.; Nguyen, T.B.T.; Larondelle, Y. Nutritional Composition and Antioxidant Properties of the Sim Fruit (Rhodomyrtus tomentosa). Food Chem. 2015, 168, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, M.; Kubo, Y.; Hatakeyama, K.; Imamura, J.; Ezura, H.; Nanasato, Y.; Tabei, Y.; Takano, Y.; Shirasu, K.; Narusaka, Y. Interfamily Transfer of Dual NB-LRR Genes Confers Resistance to Multiple Pathogens. PLoS ONE 2013, 8, e55954. [Google Scholar] [CrossRef]
- Dang, P.M.; Lamb, M.C.; Chen, C.Y. Association of Differentially Expressed R-Gene Candidates with Leaf Spot Resistance in Peanut (Arachis hypogaea L.). Mol. Biol. Rep. 2021, 48, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Goggin, F.L.; Milligan, S.B.; Kaloshian, I.; Ullman, D.E.; Williamson, V.M. The Nematode Resistance Gene Mi of Tomato Confers Resistance against the Potato Aphid. Proc. Natl. Acad. Sci. USA 1998, 95, 9750–9754. [Google Scholar] [CrossRef] [PubMed]
- Kubaa, R.A.; Giampetruzzi, A.; Altamura, G.; Saponari, M.; Saldarelli, P. Infections of the Xylella fastidiosa Subsp. pauca Strain “De Donno” in Alfalfa (Medicago sativa) Elicits an Overactive Immune Response. Plants 2019, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Li, X.; Zheng, J. Transcriptome Profiling Using Illumina- and SMRT-Based RNA-Seq of Hot Pepper for in-Depth Understanding of Genes Involved in CMV Infection. Gene 2018, 666, 123–133. [Google Scholar] [CrossRef]
- Brophy, J.J.; Goldsack, R.J.; Forster, P.I. The Leaf Essential Oils of the Australian Species of Rhodamnia (Myrtaceae). Flavour Fragr. J. 1997, 12, 345–354. [Google Scholar] [CrossRef]
- Vasconcelos, T.N.C.; Proença, C.E.B.; Ahmad, B.; Aguilar, D.S.; Aguilar, R.; Amorim, B.S.; Campbell, K.; Costa, I.R.; De-Carvalho, P.S.; Faria, J.E.Q.; et al. Myrteae Phylogeny, Calibration, Biogeography and Diversification Patterns: Increased Understanding in the Most Species Rich Tribe of Myrtaceae. Mol. Phylogenetics Evol. 2017, 109, 113–137. [Google Scholar] [CrossRef]
- Feng, C.; Feng, C.; Lin, X.; Liu, S.; Li, Y.; Kang, M. A Chromosome-Level Genome Assembly Provides Insights into Ascorbic Acid Accumulation and Fruit Softening in Guava (Psidium guajava). Plant Biotechnol. J. 2021, 19, 717–730. [Google Scholar] [CrossRef]
- Healey, A.L.; Shepherd, M.; King, G.J.; Butler, J.B.; Freeman, J.S.; Lee, D.J.; Potts, B.M.; Silva-Junior, O.B.; Baten, A.; Jenkins, J.; et al. Pests, Diseases, and Aridity Have Shaped the Genome of Corymbia citriodora. Commun. Biol. 2021, 4, 537. [Google Scholar] [CrossRef]
- Izuno, A.; Wicker, T.; Hatakeyama, M.; Copetti, D.; Shimizu, K.K. Updated Genome Assembly and Annotation for Metrosideros polymorpha, an Emerging Model Tree Species of Ecological Divergence. G3 Genes Genomes Genet. 2019, 9, 3513. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, A.H.; Ho, S.Y.W.; Külheim, C.; Crisp, M.D. Interpreting the Modern Distribution of Myrtaceae Using a Dated Molecular Phylogeny. Mol. Phylogenetics Evol. 2015, 93, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Balbinott, N.; Rodrigues, N.F.; Guzman, F.L.; Turchetto-Zolet, A.C.; Margis, R. Perspectives in Myrtaceae Evolution from Plastomes and Nuclear Phylogenies. Genet. Mol. Biol. 2022, 45, e20210191. [Google Scholar] [CrossRef]
- Padovan, A.; Keszei, A.; Külheim, C.; Foley, W.J. The Evolution of Foliar Terpene Diversity in Myrtaceae. Phytochem. Rev. 2014, 13, 695–716. [Google Scholar] [CrossRef]
- Calvert, J.; Baten, A.; Butler, J.; Barkla, B.; Shepherd, M. Terpene Synthase Genes in Melaleuca alternifolia: Comparative Analysis of Lineage-Specific Subfamily Variation within Myrtaceae. Plant Syst. Evol. 2018, 304, 111–121. [Google Scholar] [CrossRef]
- Külheim, C.; Padovan, A.; Hefer, C.; Krause, S.T.; Köllner, T.G.; Myburg, A.A.; Degenhardt, J.; Foley, W.J. The Eucalyptus Terpene Synthase Gene Family. BMC Genom. 2015, 16, 450. [Google Scholar] [CrossRef]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of Lineage-Specific Expansion of Plant Tandem Duplicates in the Adaptive Response to Environmental Stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef]
- Ravi Kiran, S.; Sita Devi, P. Evaluation of Mosquitocidal Activity of Essential Oil and Sesquiterpenes from Leaves of Chloroxylon swietenia DC. Parasitol. Res. 2007, 101, 413–418. [Google Scholar] [CrossRef]
- Rai, M.K.; Phulwaria, M.; Harish; Gupta, A.K.; Shekhawat, N.S.; Jaiswal, U. Genetic Homogeneity of Guava Plants Derived from Somatic Embryogenesis Using SSR and ISSR Markers. Plant Cell Tissue Organ Cult. 2012, 111, 259–264. [Google Scholar] [CrossRef]
- Quezada, M.; Pastina, M.M.; Ravest, G.; Silva, P.; Vignale, B.; Cabrera, D.; Hinrichsen, P.; Garcia, A.A.F.; Pritsch, C. A First Genetic Map of Acca sellowiana Based on ISSR, AFLP and SSR Markers. Sci. Hortic. 2014, 169, 138–146. [Google Scholar] [CrossRef]
- Liao, P.C.; Chiang, Y.C.; Huang, S.; Wang, J.C. Gene Flow of Ceriops tagal (Rhizophoraceae) across the Kra Isthmus in the Thai Malay Peninsula. Bot. Stud. 2009, 50, 193–204. [Google Scholar]
- Nguyen, V.X.; Detcharoen, M.; Tuntiprapas, P.; Soe-Htun, U.; Sidik, J.B.; Harah, M.Z.; Prathep, A.; Papenbrock, J. Genetic Species Identification and Population Structure of Halophila (Hydrocharitaceae) from the Western Pacific to the Eastern Indian Ocean. BMC Evol. Biol. 2014, 14, 92. [Google Scholar] [CrossRef]
- Wee, A.K.S.; Noreen, A.M.E.; Ono, J.; Takayama, K.; Kumar, P.P.; Tan, H.T.W.; Saleh, M.N.; Kajita, T.; Webb, E.L. Genetic Structures across a Biogeographical Barrier Reflect Dispersal Potential of Four Southeast Asian Mangrove Plant Species. J. Biogeogr. 2020, 47, 1258–1271. [Google Scholar] [CrossRef]
- Hargreaves, S.; Maxted, N.; Hirano, R.; Abberton, M.; Skøt, L.; Ford-Lloyd, B.V. Islands as Refugia of Trifolium repens Genetic Diversity. Conserv. Genet. 2010, 11, 1317–1326. [Google Scholar] [CrossRef]
- Dias, E.F.; Moura, M.; Schaefer, H.; Silva, L. Geographical Distance and Barriers Explain Population Genetic Patterns in an Endangered Island Perennial. AoB Plants 2016, 8, plw072. [Google Scholar] [CrossRef]
- Burton, G.P.; Campbell, K.C.S.E.; Lucas, E.J. Morphometric Analysis as a Tool to Resolve a Taxonomic Complex in Myrcia Sect. Calyptranthes (Myrtaceae, Myrteae). Syst. Bot. 2022, 46, 1016–1025. [Google Scholar] [CrossRef]
- Dalastra, C.; Sausen, T.L.; Capellesso, E.S.; Fornel, R. Variations in Leaf Size and Leaf Shape in Four Species of Eugenia (Myrtaceae) Using Geometric Morphometrics Approach. Pesqui. Botânica 2021, 75, 143–154. [Google Scholar]
- Jones, R.C.; Steane, D.A.; Potts, B.M.; Vaillancourt, R.E. Microsatellite and Morphological Analysis of Eucalyptus globulus Populations. Can. J. For. Res. 2002, 32, 59–66. [Google Scholar] [CrossRef]
- Jensen, R.J.; Schwoyer, M.; Crawford, D.J.; Stuessy, T.F.; Anderson, G.J.; Baeza, C.M.; Mario Silva, O.; Ruiz, E. Patterns of Morphological and Genetic Variation among Populations of Myrceugenia fernandeziana (Myrtaceae) on Masatierra Island: Implications for Conservation. Syst. Bot. 2002, 27, 534–547. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure from Small Quantities of Fresh Leaf Material. Phytochem Bull 1987, 19, 11–15. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A Fast, Lock-Free Approach for Efficient Parallel Counting of Occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for Reference-Free Profiling of Polyploid Genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and Accurate de novo Genome Assembly from Long Uncorrected Reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Guan, D.; McCarthy, S.A.; Wood, J.; Howe, K.; Wang, Y.; Durbin, R.; Durbin, R. Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics 2020, 36, 2896–2898. [Google Scholar] [CrossRef]
- Laetsch, D.R.; Blaxter, M.L. BlobTools: Interrogation of Genome Assemblies. F1000Res 2017, 6, 1287. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for Automated Genomic Discovery of Transposable Element Families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0 2015. Available online: https://www.repeatmasker.org/ (accessed on 9 February 2023).
- Luo, X.; Chen, S.; Zhang, Y. PlantRep: A Database of Plant Repetitive Elements. Plant Cell Rep. 2022, 41, 1163–1166. [Google Scholar] [CrossRef] [PubMed]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. RnaSPAdes: A de novo Transcriptome Assembler and Its Application to RNA-Seq Data. Gigascience 2019, 8, giz100. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio Prediction of Alternative Transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Lomsadze, A.; Ter-Hovhannisyan, V.; Chernoff, Y.O.; Borodovsky, M. Gene Identification in Novel Eukaryotic Genomes by Self-Training Algorithm. Nucleic Acids Res. 2005, 33, 6494–6506. [Google Scholar] [CrossRef]
- Brůna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: Automatic Eukaryotic Genome Annotation with GeneMark-EP+ and AUGUSTUS Supported by a Protein Database. NAR Genom. Bioinform. 2021, 3, lqaa108. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Beilstein, M.A.; Nagalingum, N.S.; Clements, M.D.; Manchester, S.R.; Mathews, S. Dated Molecular Phylogenies Indicate a Miocene Origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 18724–18728. [Google Scholar] [CrossRef]
- Wikström, N.; Savolainen, V.; Chase, M.W. Evolution of the Angiosperms: Calibrating the Family Tree. Proc. R. Soc. B Biol. Sci. 2001, 268, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The Pomegranate (Punica granatum L.) Genome and the Genomics of Punicalagin Biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef] [PubMed]
- Han, M.V.; Thomas, G.W.C.; Lugo-Martinez, J.; Hahn, M.W. Estimating Gene Gain and Loss Rates in the Presence of Error in Genome Assembly and Annotation Using CAFE 3. Mol. Biol. Evol. 2013, 30, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Alonge, M.; Soyk, S.; Ramakrishnan, S.; Wang, X.; Goodwin, S.; Sedlazeck, F.J.; Lippman, Z.B.; Schatz, M.C. RaGOO: Fast and Accurate Reference-Guided Scaffolding of Draft Genomes. Genome Biol. 2019, 20, 224. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and Open Software for Comparing Large Genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, gkw955. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Martins, W.S.; Lucas, D.C.S.; Neves, K.F.S.; Bertioli, D.J. WebSat—A Web Software for Microsatellite Marker Development. Bioinformation 2009, 3, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias-Pazaran, G.; Diaz-Garcia, L.; Schlautman, B.; Salazar, W.; Zalapa, J. Fragman: An R Package for Fragment Analysis. BMC Genet. 2016, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R. A Language and Environment for Statistical Computing; R Core Team R: Vienna, Austria, 2020. [Google Scholar]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2014, e281. [Google Scholar] [CrossRef]
- Winter, D.J. MMOD: An R Library for the Calculation of Population Differentiation Statistics. Mol. Ecol. Resour. 2012, 12, 1158–1160. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IMEC: Online Marker Efficiency Calculator. Appl. Plant Sci. 2018, 6, e01159. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New Tools for the Analysis of Genome-Wide SNP Data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Klingenberg, C.P. MorphoJ: An Integrated Software Package for Geometric Morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P. Vegan: Community Ecology Package. R Package Version 2.5-6. 2019. Available online: https://cran.r-project.org/package=vegan (accessed on 9 February 2023).

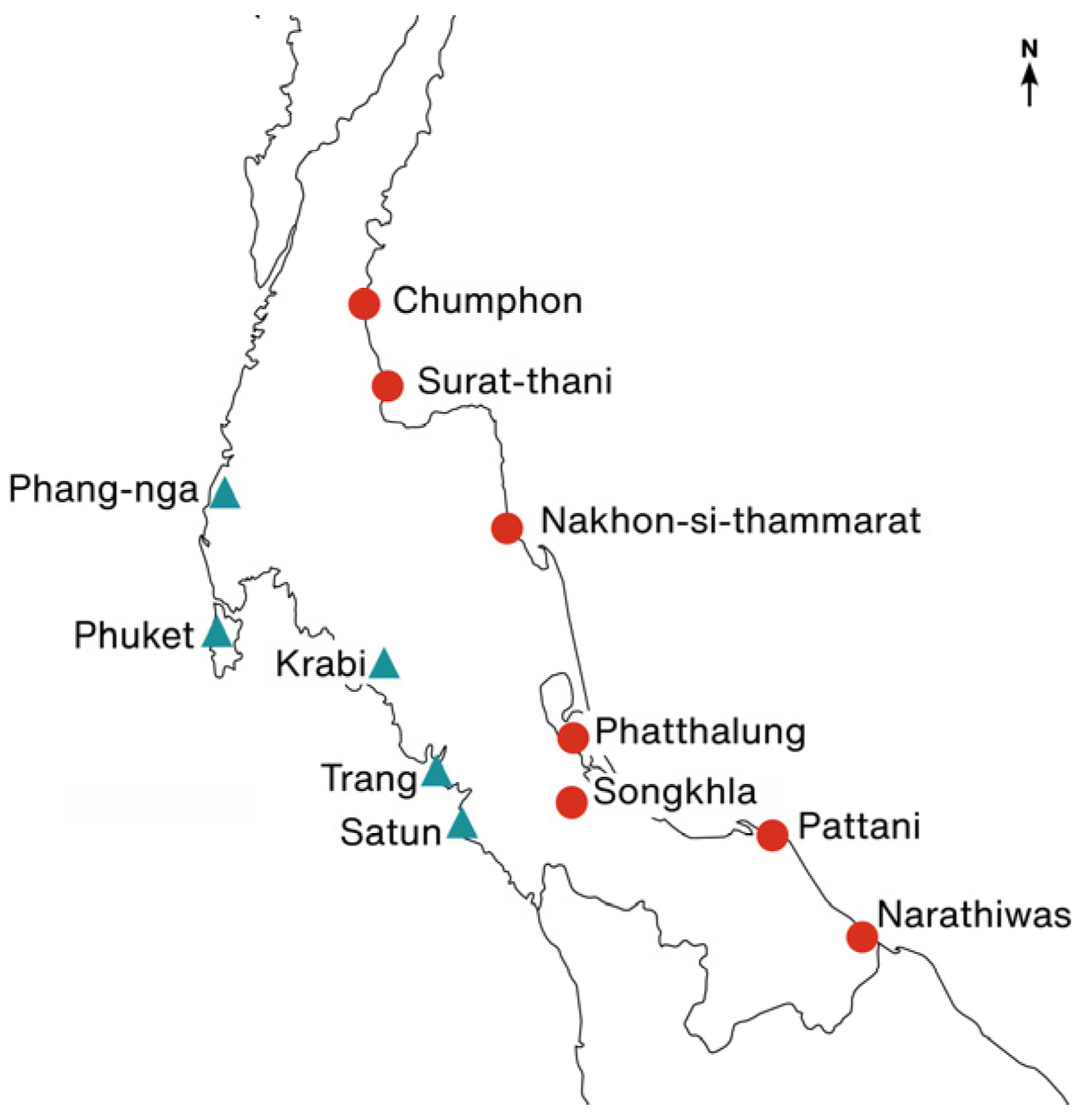
| Location | Coordinate | Side | n Samples |
|---|---|---|---|
| Chumphon | 9°53′49.8″ N 99°08′16.2″ E | East | 11 |
| Krabi | 7°51′43.4″ N 99°10′08.4″ E | West | 8 |
| Narathiwas | 6°17′30.0″ N 101°58′41.3″ E | East | 7 |
| Nakhon-si-thammarat | 8°37′09.7″ N 99°57′07.2″ E | East | 8 |
| Phuket | 8°02′09.1″ N 98°17′51.5″ E | West | 4 |
| Phang-nga | 8°49′36.1″ N 98°20′33.0″ E | West | 7 |
| Phatthalung | 7°09′46.3″ N 100°04′58.4″ E | East | 6 |
| Pattani | 6°52′10.4″ N 101°27′58.8″ E | East | 3 |
| Songkhla | 7°13′45.0″ N 100°22′21.5″ E | East | 7 |
| Surat-thani | 9°25′49.2″ N 99°16′16.2″ E | East | 7 |
| Satun | 6°56′46.2″ N 99°41′44.5″ E | West | 4 |
| Trang | 7°14′35.9″ N 99°33′01.4″ E | West | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Detcharoen, M.; Bumrungsri, S.; Voravuthikunchai, S.P. Complete Genome of Rose Myrtle, Rhodomyrtus tomentosa, and Its Population Genetics in Thai Peninsula. Plants 2023, 12, 1582. https://doi.org/10.3390/plants12081582
Detcharoen M, Bumrungsri S, Voravuthikunchai SP. Complete Genome of Rose Myrtle, Rhodomyrtus tomentosa, and Its Population Genetics in Thai Peninsula. Plants. 2023; 12(8):1582. https://doi.org/10.3390/plants12081582
Chicago/Turabian StyleDetcharoen, Matsapume, Sara Bumrungsri, and Supayang Piyawan Voravuthikunchai. 2023. "Complete Genome of Rose Myrtle, Rhodomyrtus tomentosa, and Its Population Genetics in Thai Peninsula" Plants 12, no. 8: 1582. https://doi.org/10.3390/plants12081582
APA StyleDetcharoen, M., Bumrungsri, S., & Voravuthikunchai, S. P. (2023). Complete Genome of Rose Myrtle, Rhodomyrtus tomentosa, and Its Population Genetics in Thai Peninsula. Plants, 12(8), 1582. https://doi.org/10.3390/plants12081582






