Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants
Abstract
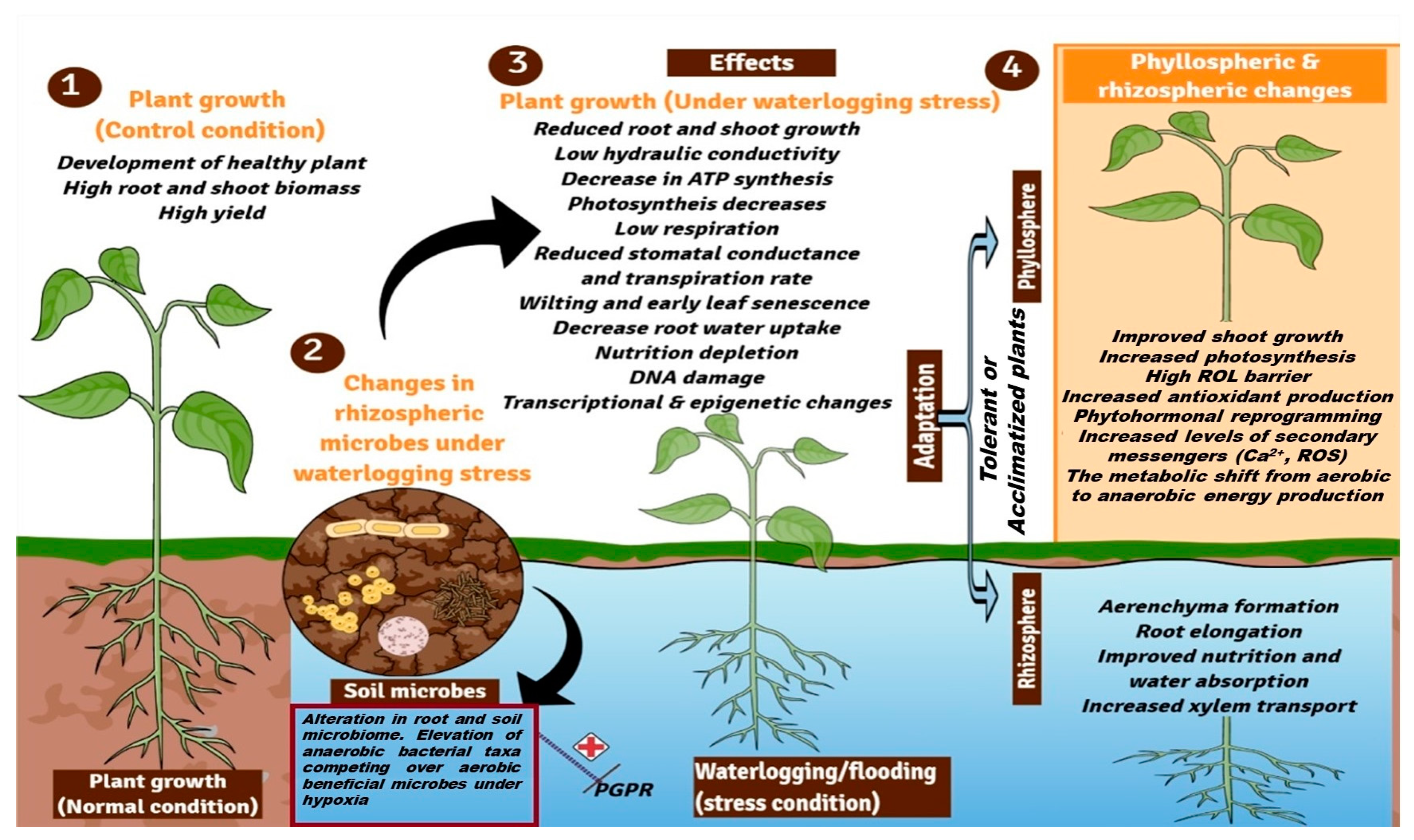
1. Introduction
2. Flooding Affects Diverse Crop Traits
3. Adaptive Responses of Plants against Waterlogging Stress
4. Waterlogging-Mediated Signaling Mechanism in Plants
5. Strategies for Improving Waterlogging Tolerance in Plants: Past, Present, and Future
5.1. Past: Classical Breeding and Genetic Engineering Approaches Used for Waterlogging Tolerance in Plants
5.2. Present: Omics Approaches for Understanding Waterlogging Tolerance in Plants
5.3. Transcriptional, Metabolic, and Translational Profiling under Waterlogging Stress in Plants
5.4. Future: Integrated Omics and Panomics for Waterlogging Tolerance in Plants
6. Role of High-Throughput Phenotyping Tool in Waterlogging Stress
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flooding Tolerance: O2 Sensing and Survival Strategies. Curr. Opin. Plant Biol. 2013, 16, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Westra, S.; Fowler, H.J.; Evans, J.P.; Alexander, L.V.; Berg, P.; Johnson, F.; Kendon, E.J.; Lenderink, G.; Roberts, N. Future Changes to the Intensity and Frequency of Short-duration Extreme Rainfall. Rev. Geophys. 2014, 52, 522–555. [Google Scholar] [CrossRef]
- Masson-Delmotte, V.; Zhai, P.; Pörtner, H.-O.; Roberts, D.; Skea, J.; Shukla, P.R.; Pirani, A.; Moufouma-Okia, W.; Péan, C.; Pidcock, R. Global Warming of 1.5 °C; Cambridge University Press: Cambridge, UK, 2018; Volume 1. [Google Scholar]
- FAO. The Future of Food and Agriculture: Trends and Challenges; Food and Agriculture Organization of the United Nations Rome: Rome, Italy, 2017; Volume 4. [Google Scholar]
- Shabala, S. Physiological and Cellular Aspects of Phytotoxicity Tolerance in Plants: The Role of Membrane Transporters and Implications for Crop Breeding for Waterlogging Tolerance. New Phytol. 2011, 190, 289–298. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Sasidharan, R. Ethylene-and Oxygen Signalling-Drive Plant Survival during Flooding. Plant Biol. 2013, 15, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, J.; Salhab, M.; Jafino, B.A. Flood Exposure and Poverty in 188 Countries. Nat. Commun. 2022, 13, 3527. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making Sense of Low Oxygen Sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Submergence and Waterlogging Stress in Plants: A Review Highlighting Research Opportunities and Understudied Aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef]
- Yohan, Y. Influence of Waterlogging on Certain Biochemical and Yield Parameters of Pigeonpea (Cajanus cajan (L.) Millsp). Int. J. Pure Appl. Biosci. 2017, 5, 1862–1868. [Google Scholar] [CrossRef]
- Talukdar, D.; Sinjushin, A. Cytogenomics and Mutagenomics in Plant Functional Biology and Breeding. In PlantOmics: The Omics of Plant Science; Springer: New Delhi, India, 2015; pp. 113–156. [Google Scholar]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vuong, T.; Valliyodan, B.; Nguyen, H.T. Integrating Omic Approaches for Abiotic Stress Tolerance in Soybean. Front. Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef]
- Shah, T.; Xu, J.; Zou, X.; Cheng, Y.; Nasir, M.; Zhang, X. Omics Approaches for Engineering Wheat Production under Abiotic Stresses. Int. J. Mol. Sci. 2018, 19, 2390. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Bae, H. Ionomic Approaches for Discovery of Novel Stress-resilient Genes in Plants. Int. J. Mol. Sci. 2021, 22, 7182. [Google Scholar] [CrossRef]
- Yadav, C.B.; Pandey, G.; Muthamilarasan, M.; Prasad, M. Epigenetics and Epigenomics of Plants. Plant Genet. Mol. Biol. 2018, 164, 237–261. [Google Scholar]
- Jogaiah, S.; Govind, S.R.; Tran, L.S.P. Systems Biology-Based Approaches toward Understanding Drought Tolerance in Food Crops. Crit. Rev. Biotechnol. 2013, 33, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Luo, J. Metabolite-Based Genome-Wide Association Studies in Plants. Curr. Opin. Plant Biol. 2015, 24, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Nakabayashi, R.; Yang, Z.; Okazaki, Y.; Yonemaru, J.I.; Ebana, K.; Yano, M.; Saito, K. Metabolome-Genome-Wide Association Study Dissects Genetic Architecture for Generating Natural Variation in Rice Secondary Metabolism. Plant J. 2015, 81, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS Meets Germplasm. Plant Biotechnol. J. 2020, 18, 1507–1525. [Google Scholar] [CrossRef]
- Wen, X. Bayesian Model Selection in Complex Linear Systems, as Illustrated in Genetic Association Studies. Biometrics 2014, 70, 73–83. [Google Scholar] [CrossRef]
- Dhanapal, A.P.; Ray, J.D.; Singh, S.K.; Hoyos-Villegas, V.; Smith, J.R.; Purcell, L.C.; King, C.A.; Fritschi, F.B. Genome-Wide Association Analysis of Diverse Soybean Genotypes Reveals Novel Markers for Nitrogen Traits. Plant Genome 2015, 8, 1–15. [Google Scholar] [CrossRef]
- Weckwerth, W. Green Systems Biology-From Single Genomes, Proteomes and Metabolomes to Ecosystems Research and Biotechnology. J. Proteom. 2011, 75, 284–305. [Google Scholar] [CrossRef]
- Weckwerth, W. Toward a Unification of System-Theoretical Principles in Biology and Ecology—The Stochastic Lyapunov Matrix Equation and Its Inverse Application. Front. Appl. Math. Stat. 2019, 5, 29. [Google Scholar] [CrossRef]
- Xu, X.; Wang, H.; Qi, X.; Xu, Q.; Chen, X. Waterlogging-Induced Increase in Fermentation and Related Gene Expression in the Root of Cucumber (Cucumis sativus L.). Sci. Hortic. 2014, 179, 388–395. [Google Scholar] [CrossRef]
- Zhang, P.; Lyu, D.; Jia, L.; He, J.; Qin, S. Physiological and de Novo Transcriptome Analysis of the Fermentation Mechanism of Cerasus Sachalinensis Roots in Response to Short-Term Waterlogging. BMC Genom. 2017, 18, 181. [Google Scholar] [CrossRef] [PubMed]
- Camisón, Á.; Ángela Martín, M.; Dorado, F.J.; Moreno, G.; Solla, A. Changes in Carbohydrates Induced by Drought and Waterlogging in Castanea sativa. Trees-Struct. Funct. 2020, 34, 579–591. [Google Scholar] [CrossRef]
- Jaeger, C.; Gessler, A.; Biller, S.; Rennenberg, H.; Kreuzwieser, J. Differences in C Metabolism of Ash Species and Provenances as a Consequence of Root Oxygen Deprivation by Waterlogging. J. Exp. Bot. 2009, 60, 4335–4345. [Google Scholar] [CrossRef]
- Li, M.; López, R.; Venturas, M.; Pita, P.; Gordaliza, G.G.; Gil, L.; Rodríguez-Calcerrada, J. Greater Resistance to Flooding of Seedlings of Ulmus Laevis than Ulmus Minor Is Related to the Maintenance of a More Positive Carbon Balance. Trees 2015, 29, 835–848. [Google Scholar] [CrossRef]
- Kozlowski, T.T. Responses of Woody Plants to Flooding and Salinity. Tree Physiol. 1997, 17, 490. [Google Scholar] [CrossRef]
- Shukla, V.; Lombardi, L.; Iacopino, S.; Pencik, A.; Novak, O.; Perata, P.; Giuntoli, B.; Licausi, F. Endogenous Hypoxia in Lateral Root Primordia Controls Root Architecture by Antagonizing Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 538–551. [Google Scholar] [CrossRef]
- Jackson, M.B. Long-Distance Signalling from Roots to Shoots Assessed: The Flooding Story. J. Exp. Bot. 2002, 53, 175–181. [Google Scholar] [CrossRef]
- Limami, A.M. Adaptations of Nitrogen Metabolism to Oxygen Deprivation in Plants. Plant Cell Monogr. 2014, 21, 209–221. [Google Scholar] [CrossRef]
- Emdadul Haque, M.; Kawaguchi, K.; Komatsu, S. Analysis of Proteins in Aerenchymatous Seminal Roots of Wheat Grown in Hypoxic Soils under Waterlogged Conditions. Protein Pept. Lett. 2011, 18, 912–924. [Google Scholar] [CrossRef]
- Dawood, T.; Yang, X.; Visser, E.J.W.; Te Beek, T.A.H.; Kensche, P.R.; Cristescu, S.M.; Lee, S.; Floková, K.; Nguyen, D.; Mariani, C.; et al. A Co-Opted Hormonal Cascade Activates Dormant Adventitious Root Primordia upon Flooding in Solanum Dulcamara. Plant Physiol. 2016, 170, 2351–2364. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Qiao, X.; Tian, Z.; Zhang, X.; Zou, X.; Cheng, Y.; Lu, G.; Zeng, L.; Fu, G.; Ding, X.; et al. Proteomic Analysis of Rapeseed Root Response to Waterlogging Stress. Plants 2018, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- Tamang, B.G.; Magliozzi, J.O.; Maroof, M.A.S.; Fukao, T. Physiological and Transcriptomic Characterization of Submergence and Reoxygenation Responses in Soybean Seedlings. Plant Cell Environ. 2014, 37, 2350–2365. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Hwang, S.J.; Waqas, M.; Khan, A.L.; Lee, J.H.; Lee, J.D.; Nguyen, H.T.; Lee, I.J. Comparative Analysis of Endogenous Hormones Level in Two Soybean (Glycine max L.) Lines Differing in Waterlogging Tolerance. Front. Plant Sci. 2015, 6, 714. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Zhang, J.; Li, X.; Fan, X.; Dong, S.; Liu, P.; Zhao, B. Effects of Waterlogging on the Yield and Growth of Summer Maize under Field Conditions. Can. J. Plant Sci. 2014, 94, 23–31. [Google Scholar] [CrossRef]
- Shen, C.; Yuan, J.; Qiao, H.; Wang, Z.; Liu, Y.; Ren, X.; Wang, F.; Liu, X.; Zhang, Y.; Chen, X.; et al. Transcriptomic and Anatomic Profiling Reveal the Germination Process of Different Wheat Varieties in Response to Waterlogging Stress. BMC Genet. 2020, 21, 93. [Google Scholar] [CrossRef]
- Luan, H.; Li, H.; Li, Y.; Chen, C.; Li, S.; Wang, Y.; Yang, J.; Xu, M.; Shen, H.; Qiao, H. Transcriptome Analysis of Barley (Hordeum vulgare L.) under Waterlogging Stress, and Overexpression of the HvADH4 Gene Confers Waterlogging Tolerance in Transgenic Arabidopsis. BMC Plant Biol. 2023, 23, 62. [Google Scholar] [CrossRef]
- Fry, E.L.; Zhu, F.; Greenwood, B. Adapting to Environmental Change. In Microbiomes of Soils, Plants and Animals; Cambridge University Press: Cambridge, UK, 2020; pp. 154–181. [Google Scholar]
- Ali, S.; Tyagi, A.; Mushtaq, M.; Al-Mahmoudi, H.; Bae, H. Harnessing Plant Microbiome for Mitigating Arsenic Toxicity in Sustainable Agriculture. Environ. Pollut. 2022, 300, 118940. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Park, S.; Mir, R.A.; Mushtaq, M.; Bhat, B.; Al-Mahmoudi, H.; Bae, H. Deciphering the Plant Microbiome to Improve Drought Tolerance: Mechanisms and Perspectives. Environ. Exp. Bot. 2022, 201, 104933. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Bae, H. Plant Microbiome: An Ocean of Possibilities for Improving Disease Resistance in Plants. Microorganisms 2023, 11, 392. [Google Scholar] [CrossRef]
- Francioli, D.; Cid, G.; Kanukollu, S.; Ulrich, A.; Hajirezaei, M.R.; Kolb, S. Flooding Causes Dramatic Compositional Shifts and Depletion of Putative Beneficial Bacteria on the Spring Wheat Microbiota. Front. Microbiol. 2021, 12, 773116. [Google Scholar] [CrossRef]
- Hamonts, K.; Clough, T.J.; Stewart, A.; Clinton, P.W.; Richardson, A.E.; Wakelin, S.A.; O’Callaghan, M.; Condron, L.M. Effect of Nitrogen and Waterlogging on Denitrifier Gene Abundance, Community Structure and Activity in the Rhizosphere of Wheat. FEMS Microbiol. Ecol. 2013, 83, 568–584. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhu, D.; Lin, X. Effects of Water Management and Organic Fertilization with SRI Crop Practices on Hybrid Rice Performance and Rhizosphere Dynamics. Paddy Water Environ. 2011, 9, 33–39. [Google Scholar] [CrossRef]
- Li, H.Y.; Zhao, C.A.; Liu, C.J.; Xu, X.F. Endophytic Fungi Diversity of Aquatic/Riparian Plants and Their Antifungal Activity in Vitro. J. Microbiol. 2010, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.Y.; Cao, Y.; Zhang, K.Q. Metagenomic Insights into Communities, Functions of Endophytes, and Their Associates with Infection by Root-Knot Nematode, Meloidogyne Incognita, in Tomato Roots. Sci. Rep. 2015, 5, 17087. [Google Scholar] [CrossRef] [PubMed]
- Vishwanathan, K.; Zienkiewicz, K.; Liu, Y.; Janz, D.; Feussner, I.; Polle, A.; Haney, C.H. Ectomycorrhizal Fungi Induce Systemic Resistance against Insects on a Nonmycorrhizal Plant in a CERK1-Dependent Manner. New Phytol. 2020, 228, 728–740. [Google Scholar] [CrossRef]
- Xu, X.; Ji, J.; Ma, X.; Xu, Q.; Qi, X.; Chen, X. Comparative Proteomic Analysis Provides Insight into the Key Proteins Involved in Cucumber (Cucumis sativus L.) Adventitious Root Emergence under Waterlogging Stress. Front. Plant Sci. 2016, 7, 1515. [Google Scholar] [CrossRef]
- Eysholdt-Derzsó, E.; Sauter, M. Hypoxia and the Group VII Ethylene Response Transcription Factor HRE2 Promote Adventitious Root Elongation in Arabidopsis. Plant Biol. 2019, 21, 103–108. [Google Scholar] [CrossRef]
- Visser, E.J.W.; Voesenek, L.A.C.J. Acclimation to Soil Flooding-Sensing and Signal-Transduction. Plant Soil 2005, 274, 197–214. [Google Scholar] [CrossRef]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-Gibberellin Signaling Underlies Adaptation of Rice to Periodic Flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef]
- Caruso, P.; Baldoni, E.; Mattana, M.; Pietro Paolo, D.; Genga, A.; Coraggio, I.; Russo, G.; Picchi, V.; Reforgiato Recupero, G.; Locatelli, F. Ectopic Expression of a Rice Transcription Factor, Mybleu, Enhances Tolerance of Transgenic Plants of Carrizo Citrange to Low Oxygen Stress. Plant Cell Tissue Organ Cult. 2012, 109, 327–339. [Google Scholar] [CrossRef]
- Borella, J.; Becker, R.; Lima, M.C.; de Oliveira, D.D.S.C.; Braga, E.J.B.; de Oliveira, A.C.B.; Do Amarante, L. Nitrogen Source Influences the Antioxidative System of Soybean Plants under Hypoxia and Re-Oxygenation. Sci. Agric. 2019, 76, 51–62. [Google Scholar] [CrossRef]
- Doupis, G.; Kavroulakis, N.; Psarras, G.; Papadakis, I.E. Growth, Photosynthetic Performance and Antioxidative Response of ‘Hass’ and ‘Fuerte’ Avocado (Persea americana Mill.) Plants Grown under High Soil Moisture. Photosynthetica 2017, 55, 655–663. [Google Scholar] [CrossRef]
- Tyagi, A.; Sharma, S.; Ali, S.; Gaikwad, K. Crosstalk between H2S and NO: An emerging signalling pathway during waterlogging stress in legume crops. Plant Biol. 2022, 24(4), 576–586. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Sharma, S.; Srivastava, H.; Singh, N.K.; Gaikwad, K. In silico characterization and homology modeling of cytosolic APX gene predicts novel glycine residue modulating waterlogging stress response in pigeon pea. PeerJ 2021, 9, e10888. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Shen, H.; Pan, Y.; Guo, B.; Lv, C.; Xu, R. Elucidating the Hypoxic Stress Response in Barley (Hordeum vulgare L.) during Waterlogging: A Proteomics Approach. Sci. Rep. 2018, 8, 9655. [Google Scholar] [CrossRef]
- Alpuerto, J.B.; Hussain, R.M.F.; Fukao, T. The Key Regulator of Submergence Tolerance, SUB1A, Promotes Photosynthetic and Metabolic Recovery from Submergence Damage in Rice Leaves. Plant Cell Environ. 2016, 39, 672–684. [Google Scholar] [CrossRef]
- Hartman, S.; Sasidharan, R.; Voesenek, L.A.C.J. The Role of Ethylene in Metabolic Acclimations to Low Oxygen. New Phytol. 2021, 229, 64–70. [Google Scholar] [CrossRef]
- Hess, N.; Klode, M.; Anders, M.; Sauter, M. The Hypoxia Responsive Transcription Factor Genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis Are Differentially Regulated by Ethylene. Physiol. Plant. 2011, 143, 41–49. [Google Scholar] [CrossRef]
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustropha, A. Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved Cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef]
- Giuntoli, B.; Perata, P. Group VII Ethylene Response Factors in Arabidopsis: Regulation and Physiological Roles. Plant Physiol. 2018, 176, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A Group VII Ethylene Response Factor Gene, ZmEREB180, Coordinates Waterlogging Tolerance in Maize Seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, S.; Nishimura, T.; Koshiba, T.; Yamamoto, R.; Hiraga, S.; Nakamura, T.; Komatsu, S. Effects of Anti-Auxins on Secondary Aerenchyma Formation in Flooded Soybean Hypocotyls. Plant Prod. Sci. 2016, 19, 154–160. [Google Scholar] [CrossRef]
- Liu, P.; Sun, F.; Gao, R.; Dong, H. RAP2.6L Overexpression Delays Waterlogging Induced Premature Senescence by Increasing Stomatal Closure More than Antioxidant Enzyme Activity. Plant Mol. Biol. 2012, 79, 609–622. [Google Scholar] [CrossRef]
- McDonald, M.P.; Visser, E.J.W. A Study of the Interaction between Auxin and Ethylene in Wild Type and Transgenic Ethylene-Insensitive Tobacco during Adventitious Root Formation Induced by Stagnant Root Zone Conditions. Plant Biol. 2003, 5, 550–556. [Google Scholar] [CrossRef]
- Qi, X.; Li, Q.; Ma, X.; Qian, C.; Wang, H.; Ren, N.; Shen, C.; Huang, S.; Xu, X.; Xu, Q.; et al. Waterlogging-Induced Adventitious Root Formation in Cucumber Is Regulated by Ethylene and Auxin through Reactive Oxygen Species Signalling. Plant Cell Environ. 2019, 42, 1458–1470. [Google Scholar] [CrossRef]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal Interplay during Adventitious Root Formation in Flooded Tomato Plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef]
- Wang, G.; Fan, W.; Peng, F. Physiological Responses of the Young Peach Tree to Water-Logging and Spraying SA at Different Timing. J. Fruit Sci. 2015, 32, 872–878. [Google Scholar]
- Kamal, A.H.M.; Komatsu, S. Jasmonic Acid Induced Protein Response to Biophoton Emissions and Flooding Stress in Soybean. J. Proteom. 2016, 133, 33–47. [Google Scholar] [CrossRef]
- Hudgins, J.W.; Franceschi, V.R. Methyl Jasmonate-Induced Ethylene Production Is Responsible for Conifer Phloem Defense Responses and Reprogramming of Stem Cambial Zone for Traumatic Resin Duct Formation. Plant Physiol. 2004, 135, 2134–2149. [Google Scholar] [CrossRef]
- Huang, H.; Liu, D.; Li, L.; Wu, J.; Wang, S.; Li, X. Effects of Spraying Plant Growth Regulators on Peanut Growth and Yield & Quality under Waterlogging Stress. J. Hum. Agric. 2018, 44, 129. [Google Scholar]
- Kang, Y.Y.; Guo, S.R.; Li, J.; Duan, J.J. Effect of Root Applied 24-Epibrassinolide on Carbohydrate Status and Fermentative Enzyme Activities in Cucumber (Cucumis sativus L.) Seedlings under Hypoxia. Plant Growth Regul. 2009, 57, 259–269. [Google Scholar] [CrossRef]
- Ma, Y.H.; Guo, S.R. 24-Epibrassinolide Improves Cucumber Photosynthesis under Hypoxia by Increasing CO2 Assimilation and Photosystem II Efficiency. Photosynthetica 2014, 52, 96–104. [Google Scholar] [CrossRef]
- Li, H.; Vaillancourt, R.; Mendham, N.; Zhou, M. Comparative Mapping of Quantitative Trait Loci Associated with Waterlogging Tolerance in Barley (Hordeum vulgare L.). BMC Genom. 2008, 9, 401. [Google Scholar] [CrossRef]
- Mano, Y.; Muraki, M.; Komatsu, T.; Fujimori, M.; Akiyama, F.; Takamizo, T. Varietal Difference in Pre-Germination Flooding Tolerance and Waterlogging Tolerance at the Seedling Stage in Maize Inbred Lines. Jpn. J. Crop Sci. 2002, 71, 361–367. [Google Scholar] [CrossRef]
- Boru, G.; Van Ginkel, M.; Kronstad, W.E.; Boersma, L. Expression and Inheritance of Tolerance to Waterlogging Stress in Wheat. Euphytica 2001, 117, 91–98. [Google Scholar] [CrossRef]
- Cai, S.; Cao, Y.; Fang, X. Studies on the Variability and Combining Ability of Waterlogging Tolerance in Common Wheat. Jiangsu J. Agric. Sci. 1996, 12, 1–5. [Google Scholar]
- Yang, C.; Shibin, C.; Zhaosu, W. Studies on Genetic Features of Waterlogging Tolerance in Wheat. Jiangsu J. Agric. Sci. 1995, 11, 11–15. [Google Scholar]
- Ikeda, T. Studies on the Wet-Injury Resistance of Wheat and Barley Varieties. (II) Varietal Difference of Wet-Injury Resistance of Wheat and Barley. Bull. Div. Plant Breed. Cultiv. Tokai-Kinki Natl. Agric. Exp. Stn. 1955, 2, 11–16. [Google Scholar]
- Reyna, N.; Cornelious, B.; Shannon, J.G.; Sneller, C.H. Evaluation of a QTL for Waterlogging Tolerance in Southern Soybean Germplasm. Crop. Sci. 2003, 43, 2077–2082. [Google Scholar] [CrossRef]
- Hamaci, Y.; Yoshino, M.; Furusho, M.; Yoshida, T. Index of Screening for Wet Endurance in Malting Barley. Jpn. J. Breed. 1990, 40, 361–366. [Google Scholar] [CrossRef]
- Fang, X.W.; Cao, Y.; Cai, S.B.; Xiong, E.H.; Zhu, W. Genetic Evaluation of Waterlogging Tolerance in Triticum Macha. Jiangsu J. Agric. Sci. 1997, 13, 73–75. [Google Scholar]
- Sachs, M.M. Molecular Genetic Basis of Metabolic Adaptation to Anoxia in Maize and Its Possible Utility for Improving Tolerance of Crops to Soil Waterlogging. In Interacting Stresses on Plants in a Changing Climate; Springer: Berlin/Heidelberg, Germany, 1993; pp. 375–393. [Google Scholar]
- Hamachi, Y.; Furusho, M.; Yoshida, T. Heritability of Wet Endurance in Malting Barley. Jpn. J. Breed. 1989, 39, 195–202. [Google Scholar] [CrossRef]
- Mazur, B.J.; Tingey, S.V. Genetic Mapping and Introgression of Genes of Agronomic Importance. Curr. Opin. Biotechnol. 1995, 6, 175–182. [Google Scholar] [CrossRef]
- Campbell, M.T.; Proctor, C.A.; Dou, Y.; Schmitz, A.J.; Phansak, P.; Kruger, G.R.; Zhang, C.; Walia, H. Genetic and Molecular Characterization of Submergence Response Identifies Subtol6 as a Major Submergence Tolerance Locus in Maize. PLoS ONE 2015, 10, e0120385. [Google Scholar] [CrossRef]
- Mano, Y.; Omori, F.; Muraki, M.; Takamizo, T. QTL Mapping of Adventitious Root Formation under Flooding Conditions in Tropical Maize (Zea mays L.) Seedlings. Breed. Sci. 2005, 55, 343–347. [Google Scholar] [CrossRef]
- Mano, Y.; Muraki, M.; Fujimori, M.; Takamizo, T.; Kindiger, B. Identification of QTL Controlling Adventitious Root Formation during Flooding Conditions in Teosinte (Zea mays Ssp. Huehuetenangensis) Seedlings. Euphytica 2005, 142, 33–42. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, B.; Yu, F.; Li, L.; Wang, M.; Xue, Y.; Zhang, Z.; Yan, J.; Yue, B.; Zheng, Y.; et al. Identification of Major QTL for Waterlogging Tolerance Using Genome-Wide Association and Linkage Mapping of Maize Seedlings. Plant Mol. Biol. Report. 2013, 31, 594–606. [Google Scholar] [CrossRef]
- Qiu, F.; Zheng, Y.; Zhang, Z.; Xu, S. Mapping of QTL Associated with Waterlogging Tolerance during the Seedling Stage in Maize. Ann. Bot. 2007, 99, 1067–1081. [Google Scholar] [CrossRef]
- Hattori, Y.; Miura, K.; Asano, K.; Yamamoto, E.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. A Major QTL Confers Rapid Internode Elongation in Response to Water Rise in Deepwater Rice. Breed. Sci. 2007, 57, 305–314. [Google Scholar] [CrossRef]
- Ballesteros, D.C.; Mason, R.E.; Addison, C.K.; Andrea Acuña, M.; Nelly Arguello, M.; Subramanian, N.; Miller, R.G.; Sater, H.; Gbur, E.E.; Miller, D.; et al. Tolerance of Wheat to Vegetative Stage Soil Waterlogging Is Conditioned by Both Constitutive and Adaptive QTL. Euphytica 2015, 201, 329–343. [Google Scholar] [CrossRef]
- Ma, Y.U.; Mao, S.L.; Chen, G.Y.; Liu, Y.X.; Wei, L.I.; Wei, Y.M.; Liu, C.J.; Zheng, Y.L. QTLs for Waterlogging Tolerance at Germination and Seedling Stages in Population of Recombinant Inbred Lines Derived from a Cross Between Synthetic and Cultivated Wheat Genotypes. J. Integr. Agric. 2014, 13, 31–39. [Google Scholar] [CrossRef]
- Xue, D.W.; Zhou, M.X.; Zhang, X.Q.; Chen, S.; Wei, K.; Zeng, F.R.; Mao, Y.; Wu, F.B.; Zhang, G.P. Identification of QTLs for Yield and Yield Components of Barley under Different Growth Conditions. J. Zhejiang Univ. Sci. B 2010, 11, 169–176. [Google Scholar] [CrossRef]
- Osman, K.A.; Tang, B.; Wang, Y.; Chen, J.; Yu, F.; Li, L.; Han, X.; Zhang, Z.; Yan, J.; Zheng, Y.; et al. Dynamic QTL Analysis and Candidate Gene Mapping for Waterlogging Tolerance at Maize Seedling Stage. PLoS ONE 2013, 8, e79305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, G.; Shabala, S.; Koutoulis, A.; Shabala, L.; Johnson, P.; Li, C.; Zhou, M. Identification of Aerenchyma Formation-Related QTL in Barley That Can Be Effective in Breeding for Waterlogging Tolerance. Theor. Appl. Genet. 2016, 129, 1167–1177. [Google Scholar] [CrossRef]
- Mano, Y.; Omori, F.; Takamizo, T.; Kindiger, B.; Bird, R.M.; Loaisiga, C.H.; Takahashi, H. QTL Mapping of Root Aerenchyma Formation in Seedlings of a Maize × Rare Teosinte “Zea nicaraguensis” cross. Plant Soil 2007, 295, 103–113. [Google Scholar] [CrossRef]
- Zhou, M.; Johnson, P.; Zhou, G.; Li, C.; Lance, R. Quantitative Trait Loci for Waterlogging Tolerance in a Barley Cross of Franklin × YuYaoXiangTian Erleng and the Relationship between Waterlogging and Salinity Tolerance. Crop. Sci. 2012, 52, 2082–2088. [Google Scholar] [CrossRef]
- Cavanagh, C.; Morell, M.; Mackay, I.; Powell, W. From Mutations to MAGIC: Resources for Gene Discovery, Validation and Delivery in Crop Plants. Curr. Opin. Plant Biol. 2008, 11, 215–221. [Google Scholar] [CrossRef]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.L.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.W.; McCouch, S.; Thomson, M.; Mauleon, R.; et al. Multi-Parent Advanced Generation Inter-Cross (MAGIC) Populations in Rice: Progress and Potential for Genetics Research and Breeding. Rice 2013, 6, 11. [Google Scholar] [CrossRef]
- Dell’Acqua, M.; Gatti, D.M.; Pea, G.; Cattonaro, F.; Coppens, F.; Magris, G.; Hlaing, A.L.; Aung, H.H.; Nelissen, H.; Baute, J.; et al. Genetic Properties of the MAGIC Maize Population: A New Platform for High Definition QTL Mapping in Zea mays. Genome Biol. 2015, 16, 167. [Google Scholar] [CrossRef]
- Sannemann, W.; Huang, B.E.; Mathew, B.; Léon, J. Multi-Parent Advanced Generation Inter-Cross in Barley: High-Resolution Quantitative Trait Locus Mapping for Flowering Time as a Proof of Concept. Mol. Breed. 2015, 35, 86. [Google Scholar] [CrossRef]
- Du, H.; Shen, X.; Huang, Y.; Huang, M.; Zhang, Z. Overexpression of Vitreoscilla Hemoglobin Increases Waterlogging Tolerance in Arabidopsis and Maize. BMC Plant Biol. 2016, 16, 35. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Peng, R.H.; Fan, H.Q.; Xiong, A.S.; Yao, Q.H.; Cheng, Z.M.; Li, Y. Vitreoscilla Hemoglobin Overexpression Increases Submergence Tolerance in Cabbage. Plant Cell Rep. 2005, 23, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.C.; Hu, Y.L.; Zhong, J.; Wang, L.X.; Guo, J.Y.; Lin, Z.P. Improvement of the Hydroponic Growth and Waterlogging Tolerance of Petunias by the Introduction of Vhb Gene. Acta Bot. Sin. 2003, 45, 205–210. [Google Scholar]
- Raineri, J.; Caraballo, L.; Rigalli, N.; Portapila, M.; Otegui, M.E.; Chan, R.L. HaHB11 Transformed Maize Has Improved Yield under Waterlogging and Defoliation in Control and Field Conditions. bioRxiv 2021, 2021-10. [Google Scholar]
- Rauf, M.; Arif, M.; Fisahn, J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. NAC Transcription Factor speedy hyponastic growth Regulates Flooding-Induced Leaf Movement in Arabidopsis. Plant Cell 2013, 25, 4941–4955. [Google Scholar] [CrossRef]
- Xu, X.; Ji, J.; Xu, Q.; Qi, X.; Weng, Y.; Chen, X. The major-effect quantitative trait locus csarn6.1 encodes an aaa atpase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. Plant J. 2018, 93, 917–930. [Google Scholar] [CrossRef]
- Yin, D.; Sun, D.; Han, Z.; Ni, D.; Norris, A.; Jiang, C.Z. PhERF2, an Ethylene-Responsive Element Binding Factor, Plays an Essential Role in Waterlogging Tolerance of Petunia. Hortic. Res. 2019, 6, 83. [Google Scholar] [CrossRef]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive Expression of a Stabilized Transcription Factor Group VII Ethylene Response Factor Enhances Waterlogging Tolerance in Wheat without Penalizing Grain Yield. Plant Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef]
- Luan, H.; Guo, B.; Shen, H.; Pan, Y.; Hong, Y.; Lv, C.; Xu, R. Overexpression of Barley Transcription Factor HvERF2.11 in Arabidopsis Enhances Plant Waterlogging Tolerance. Int. J. Mol. Sci. 2020, 21, 1982. [Google Scholar] [CrossRef]
- Cabello, J.V.; Giacomelli, J.I.; Piattoni, C.V.; Iglesias, A.A.; Chan, R.L. The Sunflower Transcription Factor HaHB11 Improves Yield, Biomass and Tolerance to Flooding in Transgenic Arabidopsis Plants. J. Biotechnol. 2016, 222, 73–83. [Google Scholar] [CrossRef]
- Xuan, L.; Hua, J.; Zhang, F.; Wang, Z.; Pei, X.; Yang, Y.; Yin, Y.; Creech, D.L. Identification and Functional Analysis of Thadh1 and Thadh4 Genes Involved in Tolerance to Waterlogging Stress in Taxodium Hybrid ‘Zhongshanshan 406’. Genes 2021, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Quimio, C.A.; Torrizo, L.B.; Setter, T.L.; Ellis, M.; Grover, A.; Abrigo, E.M.; Oliva, N.P.; Ella, E.S.; Carpena, A.L.; Ito, O.; et al. Enhancement of Submergence Tolerance in Transgenic Rice Overproducing Pyruvate Decarboxylase. J. Plant Physiol. 2000, 156, 516–521. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A Is an Ethylene-Response-Factor-like Gene That Confers Submergence Tolerance to Rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Submergence Tolerance Conferred by Sub1A Is Mediated by SLR1 and SLRL1 Restriction of Gibberellin Responses in Rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The Ethylene Response Factors SNORKEL1 and SNORKEL2 Allow Rice to Adapt to Deep Water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Ismond, K.P.; Dolferus, R.; De Pauw, M.; Dennis, E.S.; Good, A.G. Enhanced Low Oxygen Survival in Arabidopsis through Increased Metabolic Flux in the Fermentative Pathway. Plant Physiol. 2003, 132, 1292–1302. [Google Scholar] [CrossRef]
- Dolferus, R.; Wolansky, M.; Carroll, R.; Miyashita, Y.; Ismond, K.; Good, A. Functional Analysis of Lactate Dehydrogenase during Hypoxic Stress in Arabidopsis. Funct. Plant Biol. 2008, 35, 131–140. [Google Scholar] [CrossRef]
- Hunt, P.W.; Klok, E.J.; Trevaskis, B.; Watts, R.A.; Ellis, M.H.; Peacock, W.J.; Dennis, E.S. Increased Level of Hemoglobin 1 Enhances Survival of Hypoxic Stress and Promotes Early Growth in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2002, 99, 17197–17202. [Google Scholar] [CrossRef]
- Grichko, V.P.; Glick, B.R. Flooding Tolerance of Transgenic Tomato Plants Expressing the Bacterial Enzyme ACC Deaminase Controlled by the 35S, RolD or PRB-1b Promoter. Plant Physiol. Biochem. 2001, 39, 19–25. [Google Scholar] [CrossRef]
- Zhang, J.; Van Toai, T.; Huynh, L.; Preiszner, J. Development of Flooding-Tolerant Arabidopsis Thaliana by Autoregulated Cytokinin Production. Mol. Breed. 2000, 6, 135–144. [Google Scholar] [CrossRef]
- Huynh, L.N.; VanToai, T.; Streeter, J.; Banowetz, G. Regulation of Flooding Tolerance of SAG12:Ipt Arabidopsis Plants by Cytokinin. J. Exp. Bot. 2005, 56, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.L.; Wang, G.; Wang, T.; Jia, Z.H.; Guo, Z.R.; Zhang, J.Y. Adrap2.3, a Novel Ethylene Response Factor VII from Actinidia Deliciosa, Enhanceswaterlogging Resistance in Transgenic Tobacco through Improving Expression Levels of Pdc and Adh Genes. Int. J. Mol. Sci. 2019, 20, 1189. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of Abiotic Stress on Plants: A Systems Biology Perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, Two Hypoxia-Inducible Ethylene Response Factors, Affect Anaerobic Responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Hoeren, F.U.; Dolferus, R.; Wu, Y.; Peacock, W.J.; Dennis, E.S. Evidence for a Role for AtMYB2 in the Induction of the Arabidopsis Alcohol Dehydrogenase Gene (ADH1) by Low Oxygen. Genetics 1998, 149, 479–490. [Google Scholar] [CrossRef]
- Bond, D.M.; Wilson, I.W.; Dennis, E.S.; Pogson, B.J.; Jean Finnegan, E. Vernalization Insensitive 3 (VIN3) Is Required for the Response of Arabidopsis thaliana Seedlings Exposed to Low Oxygen Conditions. Plant J. 2009, 59, 576–587. [Google Scholar] [CrossRef]
- Christianson, J.A.; Wilson, I.W.; Llewellyn, D.J.; Dennis, E.S. The Low-Oxygen-Induced NAC Domain Transcription Factor ANAC102 Affects Viability of Arabidopsis Seeds Following Low-Oxygen Treatment. Plant Physiol. 2009, 149, 1724–1738. [Google Scholar] [CrossRef]
- Tournaire-Roux, C.; Sutka, M.; Javot, H.; Gout, E.; Gerbeau, P.; Luu, D.T.; Bligny, R.; Maurel, C. Cytosolic PH Regulates Root Water Transport during Anoxic Stress through Gating of Aquaporins. Nature 2003, 425, 393–397. [Google Scholar] [CrossRef]
- Aroca, R.; Porcel, R.; Ruiz-Lozano, J.M. Regulation of Root Water Uptake under Abiotic Stress Conditions. J. Exp. Bot. 2012, 63, 43–57. [Google Scholar] [CrossRef]
- Steffens, B.; Geske, T.; Sauter, M. Aerenchyma Formation in the Rice Stem and Its Promotion by H2O2. New Phytol. 2011, 190, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Zheng, L.; Li, J.; Mao, Y.; Zhang, R.; Niu, X.; Geng, M.; Zhang, X.; Huang, W.; Luo, K.; et al. Transcriptomic Profiling Suggests Candidate Molecular Responses to Waterlogging in Cassava. PLoS ONE 2022, 17, e0261086. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Sharma, S.; Srivastava, H.; Singh, A.; Kaila, T.; Ali, S.; Gaikwad, A.B.; Singh, N.K.; Gaikwad, D. Transcriptome Profiling of Two Contrasting Pigeon Pea (Cajanus cajan) Genotypes in Response to Waterloging Stress. Front. Genet. 2023, 13, 3757. [Google Scholar] [CrossRef] [PubMed]
- Butsayawarapat, P.; Juntawong, P.; Khamsuk, O.; Somta, P. Comparative Transcriptome Analysis of Waterlogging-Sensitive and Tolerant Zombi Pea (Vigna vexillata) Reveals Energy Conservation and Root Plasticity Controlling Waterlogging Tolerance. Plants 2019, 8, 264. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, X.; Zhang, Z.; Liu, N.; Li, D.; Hu, L. Melatonin Improved Waterlogging Tolerance in Alfalfa (Medicago sativa) by Reprogramming Polyamine and Ethylene Metabolism. Front. Plant Sci. 2019, 10, 4. [Google Scholar] [CrossRef]
- Casarotto, G.; Kaspary, T.E.; Cutti, L.; Thomas, A.L.; Neto, J.F.B. Expression of Genes Related to Soil Flooding Tolerance in Soybeans. Acta Sci. Agron. 2019, 41, e42709. [Google Scholar] [CrossRef]
- Oh, M.W.; Nanjo, Y.; Komatsu, S. Gel-Free Proteomic Analysis of Soybean Root Proteins Affected by Calcium under Flooding Stress. Front. Plant Sci. 2014, 5, 559. [Google Scholar] [CrossRef]
- Wang, X.; Deng, Z.; Zhang, W.; Meng, Z.; Chang, X.; Lv, M. Effect of Waterlogging Duration at Different Growth Stages on the Growth, Yield and Quality of Cotton. PLoS ONE 2017, 12, e0169029. [Google Scholar] [CrossRef]
- Jung, H.J.; Roy, S.K.; Cho, S.W.; Kwon, S.J.; Kun, C.; Chun, H.C.; Woo, S.H. Proteome Analysis of Sesame Leaves in Response to Waterlogging Stress at Vegetative and Flowering Stages. Biol. Plant. 2019, 63, 733–749. [Google Scholar] [CrossRef]
- Ahsan, N.; Lee, D.G.; Lee, S.H.; Kang, K.Y.; Bahk, J.D.; Choi, M.S.; Lee, I.J.; Renaut, J.; Lee, B.H. A Comparative Proteomic Analysis of Tomato Leaves in Response to Waterlogging Stress. Physiol. Plant. 2007, 131, 555–570. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A.C.J. Flooding Stress: Acclimations and Genetic Diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Zabalza, A.; Van Dongen, J.T. Regulation of Respiration When the Oxygen Availability Changes. Physiol. Plant. 2009, 137, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.T.; Schurr, U.; Pfister, M.; Geigenberger, P. Phloem Metabolism and Function Have to Cope with Low Internal Oxygen. Plant Physiol. 2003, 131, 1529–1543. [Google Scholar] [CrossRef] [PubMed]
- Benamar, A.; Rolletschek, H.; Borisjuk, L.; Avelange-Macherel, M.H.; Curien, G.; Mostefai, H.A.; Andriantsitohaina, R.; Macherel, D. Nitrite-Nitric Oxide Control of Mitochondrial Respiration at the Frontier of Anoxia. Biochim. Biophys. Acta-Bioenerg. 2008, 1777, 1268–1275. [Google Scholar] [CrossRef]
- Lothier, J.; Diab, H.; Cukier, C.; Limami, A.M.; Tcherkez, G. Metabolic Responses to Waterlogging Differ between Roots and Shoots and Reflect Phloem Transport Alteration in Medicago truncatula. Plants 2020, 9, 1373. [Google Scholar] [CrossRef]
- Komatsu, S.; Nakamura, T.; Sugimoto, Y.; Sakamoto, K. Proteomic and Metabolomic Analyses of Soybean Root Tips Under Flooding Stress. Protein Pept. Lett. 2014, 21, 865–884. [Google Scholar] [CrossRef]
- Cui, J.; Davanture, M.; Zivy, M.; Lamade, E.; Tcherkez, G. Metabolic Responses to Potassium Availability and Waterlogging Reshape Respiration and Carbon Use Efficiency in Oil Palm. New Phytol. 2019, 223, 310–322. [Google Scholar] [CrossRef]
- Zhao, N.; Li, C.; Yan, Y.; Cao, W.; Song, A.; Wang, H.; Chen, S.; Jiang, J.; Chen, F. Comparative Transcriptome Analysis of Waterlogging-Sensitive and Waterlogging-Tolerant Chrysanthemum morifolium Cultivars under Waterlogging Stress and Reoxygenation Conditions. Int. J. Mol. Sci. 2018, 19, 1455. [Google Scholar] [CrossRef]
- Li, Z.; Bai, D.; Zhong, Y.; Lin, M.; Sun, L.; Qi, X.; Hu, C.; Fang, J. Full-Length Transcriptome and RNA-Seq Analyses Reveal the Mechanisms Underlying Waterlogging Tolerance in Kiwifruit (Actinidia valvata). Int. J. Mol. Sci. 2022, 23, 3237. [Google Scholar] [CrossRef]
- Cui, J.; Abadie, C.; Carroll, A.; Lamade, E.; Tcherkez, G. Responses to K Deficiency and Waterlogging Interact via Respiratory and Nitrogen Metabolism. Plant Cell Env. 2019, 42, 647–658. [Google Scholar] [CrossRef]
- Cid, G.A.; Francioli, D.; Kolb, S.; Tandron-Moya, Y.A.; von Wiren, N.; Hajirezaei, M.-R. Elucidating the Systemic Response of Wheat Plants under Waterlogging Based on Transcriptomic and Metabolic Approaches. bioRxiv 2022, 2022-08. [Google Scholar] [CrossRef]
- Pazhamala, L.T.; Kudapa, H.; Weckwerth, W.; Millar, A.H.; Varshney, R.K. Systems Biology for Crop Improvement. Plant Genome 2021, 14, e20098. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of Integrating Data to Uncover Genotype-Phenotype Interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Chen, L.; Hao, L.; Parry, M.A.J.; Phillips, A.L.; Hu, Y.G. Progress in TILLING as a Tool for Functional Genomics and Improvement of Crops. J. Integr. Plant Biol. 2014, 56, 425–443. [Google Scholar] [CrossRef]
- Negrão, S.; Julkowska, M.M. Plant Phenotyping. eLS 2020, 1–14. [Google Scholar] [CrossRef]


| Species | WS Condition | Affected Traits | References |
|---|---|---|---|
| Triticum aestivum L. | 1 wk | Dry weight of stem and root ↓ Length of root ↓ Ratio of root/shoot ↓ Root aerenchyma ↑ | [33] |
| Solanum dulcamara | 1 wk | Stem region ↓ and adventitious root ↑ (ET↑, ABA↓) | [34] |
| Brassica napus L. | 3 d | Length of root and shoot ↓ Fresh weight ↓ | [35] |
| Glycine max (L.) Merr. cv. “Williams 82” | 10 d | Length of root ↓ Development of lateral root and root hairs ↓ | [36] |
| Glycine max L. (S99-2281) | 10 d | Length of shoot ↓ Fresh weight of shoot and root ↓ Root aerenchyma ↑ Adventitious root ↑ | [37] |
| Zea mays L. (DH605, ZD958) | 3 and 6 d | Height of plant and ear ↓ Leaf area index ↓ Yield ↓ Bald tip ↑ | [38] |
| Triticum aestivum L. (ZM22) | 72 h | Germination ↓ Coleoptile height ↓ Amyloplast ↑ | [39] |
| Hordeum vulgare L. (Franklin) | 21 d | Leaf area ↓ Dry and fresh weight of shoot ↓ Plant height ↓ Total length and number of adventitious root ↑ Leaf aerenchyma ↑ Chlorosis and age of leaf ↑ | [40] |
| Species | Mapping Population Type | QTL | Traits | References |
|---|---|---|---|---|
| Maize | BC3F4, RILs, F2, F2:3 | Subtol6, Qarf7.04–7.05, Qarf8.05, sdw9-4, tdw9-2, tdw9-3 | Leaf chlorosis, mean leaf senescence score, adventitious root formation, shoot dry weight, total dry weight | [90,91,92,93,94] |
| Rice | F2 | qTIL1 C9285, qTIL1 T65, Sub1, qTIL12 C9285, qTIL12 W0120, qNEI12 C9285, qNEI12 W0120, qLEI12 C9285, qLEI12 W0120 | Number and total internode length, green leaf recovery, number of elongated internodes | [95] |
| Wheat | RILs | QRfbio.ua-1B-WGH, QSfbio.ua-1B-WGH, QSpadpost.ua-1B-WF, QSpad.ua-1D.5, GRI-7A | Shoot and root fresh biomass, chlorophyll content, shoot and root dry biomass, seed germination rate | [96,97] |
| Barley | DH lines | KWw2.1, GSw1.1/2.1, tfy1.1-1, QWI.YyFr.2H, tfy1.2-1/2.1-1, tfy1.1-2, QWL.YeFr.4H, QTL-AER, QTL-WL-4H, yfy2.2-3, GYw1.2 | Kernel weight, grains per spike, leaf chlorosis, plant healthiness, yellow leaf percentage, survival rate, aerenchyma formation, waterlogging tolerance, root porosity, grain yield | [52,78,98,99,100,101,102] |
| Gene | Transgenic Plant | Gene Source | Waterlogging Tolerance | References |
|---|---|---|---|---|
| Pdc1 (pyruvate decarboxylase isozyme 1) | O. sativa | O. sativa | Enhanced waterlogging tolerance | [118] |
| OsSub1A (ethylene-response-factor-like submergence tolerance gene) | O. sativa | O. sativa | Enhanced waterlogging tolerance in rice plants by increasing the expression of ADH1 | [119,120,121] |
| Pdc1 (pyruvate decarboxylase isozyme 1) | A. thaliana | A. thaliana | Confers waterlogging tolerance | [122] |
| Pdc2 (pyruvate decarboxylase isozyme 2) | A. thaliana | A. thaliana | Enhanced waterlogging tolerance | [122] |
| AtACO5 (1-aminocyclopropane-1-carboxylic acid oxidase) and AtACS (acetyl-CoA synthetase) | A. thaliana | A. thaliana | Increased ET levels and waterlogging tolerance | [111] |
| AtLDH (lactate dehydrogenase) | A. thaliana | A. thaliana | Confers hypoxia tolerance by increasing PDC enzyme activity | [123] |
| AtRAP2.6L (member of ERF subfamily) | A. thaliana | A. thaliana | Enhanced the activity of antioxidant enzymes and transcript levels of ABA biosynthesis genes, stomatal closure | [68] |
| GLB1 class I hemoglobin (Hb) | A. thaliana | Parasponia andersonii | Enhanced resistance to hypoxia | [124] |
| Hb (hemoglobin) | Brassica oleracea | Vitreoscilla filiformis | Confers waterlogging tolerance | [109] |
| ACC (1-aminocyclopropane-1-carboxylic acid) deaminase | Solanum lycopersicum | Enterobacter | Confers waterlogging tolerance | [125] |
| ipt (isopentenyl transferase in cytokinin biosynthesis) | A. thaliana | A. thaliana | Confers waterlogging tolerance | [126,127] |
| ZmEREB180 (a group VII ethylene response factor gene) | Zea mays | Zea mays | Confers waterlogging tolerance by stimulating AR formation | [66] |
| AdRAP2.3 (member of ERF subfamily) | Actinidia deliciosa | Nicotiana tabacum | Enhanced ADH and PDC enzyme activities | [128] |
| HvERF2.11 (ethylene responsive factor 2) | Hordeum vulgare | A. thaliana | Stimulates the expression level of ET genes and also increases antioxidant enzyme activity | [115] |
| HaHB11 (homeodomain-leucine zipper I subfamily) | Helianthus annus | A. thaliana | Confers waterlogging tolerance | [116] |
| ThADH1 (alcohol dehydrogenase 1) and ThADH4 (alcohol dehydrogenase 4) | Populus alba | Taxodium mucronatum Tenore × Taxodium distichum (L.). Rich | Confers waterlogging and hypoxia tolerance | [117] |
| HvADH4 (alcohol dehydrogenase 4) | Hordeum vulgare | A. thaliana | Confers waterlogging tolerance by enhancing the antioxidant enzyme activity | [40] |
| Omics Study | Species | WS Condition | Key Genes/Metabolites/Proteins | References |
|---|---|---|---|---|
| Transcriptomics | Chrysanthemum morifolium (Nannongxuefeng) | 12 h | N-end rule pathway (RAP2.3, HRE2, ATE, PCO1, PCO2) ↑ ROS signaling (POD, AOX1a) ↑ Anaerobic respiration and carbohydrate metabolism (ADH, PDC, SUS1, PDC1) ↑ Hsp 83-like, Chaperone protein ClpB1-like, Snakin-2-like isoform X1 ↑ | [153] |
| Actinidia valvata (KR5) | 12, 24, 72 h | ROS scavenging pathway (POD, CAT) ↑, NADH-GOGAT/AlaAT, ERF77 ↑ | [154] | |
| Manihot esculenta Grantz | 6 d | Photosynthesis, RNA transport, RNA degradation, amino metabolism ↑ | [137] | |
| T. aestivum L. (ZM22) | 72 h | Oxidoreductase activity, biological response to ABA and SA ↑ | [39] | |
| Hordeum vulgare L. (Franklin) | 24 h | Metabolic process (biosynthesis of secondary metabolites and phenylpropanoid), transferase activity, catalytic activity ↑ | [40] | |
| 72 h | Oxidation–reduction process, protein binding, catalytic activity ↑ | |||
| Proteomics | B. napus L. (ZS9, tolerant cultivar) | 4, 8, 12 h | Oxidation–reduction process (BnaA09g29780D), response to ethylene (BnaA09g07120D) ↑ Abiotic stress response (BnaC08g02330D), (BnaC02g24210D), response to jasmonic acid (BnaC02g24210D) ↓ | [35] |
| B. napus L. (GH01, sensitive cultivar) | 4, 8, 12 h | Abiotic stress response (BnaC08g02330D), response to ethylene (BnaA09g07120D) ↑ Oxidation–reduction process (BnaA09g29780D), response to jasmonic acid (BnaC02g24210D) ↓ | ||
| G. max L. cultivar Enrei | 2 d | Fermentation and glycolysis-related proteins ↑ Degradation/synthesis/posttranslational modification of proteins, hormone/cell wall metabolisms, and DNA synthesis ↓ | [142] | |
| Sesamum indicum L., cv. Miryang 44 | 2, 3 d | Photosynthesis (OEE1), stress defense (HSPs, Chaperones), energy metabolism (ATPs, GS) ↑ | [144] | |
| Lycopersicon esculentum L. cv. Koma | 24, 48, 72 h | Stress and defense related (Hsp cognate 70, plastidic cysteine synthase1) ↑ Photosynthesis (rubisco large/small subunits, rubisco activase), biosynthesis and metabolism of protein (Cytochrome P450, glycinamide ribonucleotide synthetase) ↓ | [145] | |
| Metabolomics | M. truncatula | 7 and 21 d | Sugars, organic acid, aromatics, glycine, alanine, glutamine, lysine ↑ Nitrogenous compounds, threitol ↓ | [150] |
| H. annuus | 2, 7, 14 d | Alanine, sugars, polyols, aconitate, citrate, phosphate ↑ Aspartate, fumarate ↓ | [155] | |
| Elaeis guineensis | 1, 2, 3, 7 wks | Polyol (myoinositol) ↑ Aconitate, citrate, serine, asparine ↓ | [152] | |
| G. max L. cultivar Enrei | 2 d | Alanine, AMP, cysteine, DHAP, GABA, glycine ↑ 2-oxoglutarate, acetyl-CoA, allantonin, aspartic acid, fumarate, cinnamate, glutamine ↓ | [151] | |
| T. aestivum (Chinese spring) | 12 d | Glycine, alanine, GABA ↑ Asparagine, pyruvate ↓ | [156] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyagi, A.; Ali, S.; Park, S.; Bae, H. Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants. Plants 2023, 12, 1544. https://doi.org/10.3390/plants12071544
Tyagi A, Ali S, Park S, Bae H. Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants. Plants. 2023; 12(7):1544. https://doi.org/10.3390/plants12071544
Chicago/Turabian StyleTyagi, Anshika, Sajad Ali, Suvin Park, and Hanhong Bae. 2023. "Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants" Plants 12, no. 7: 1544. https://doi.org/10.3390/plants12071544
APA StyleTyagi, A., Ali, S., Park, S., & Bae, H. (2023). Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants. Plants, 12(7), 1544. https://doi.org/10.3390/plants12071544




