Integrative Omic Analysis Reveals the Dynamic Change in Phenylpropanoid Metabolism in Morus alba under Different Stress
Abstract
:1. Introduction
2. Results
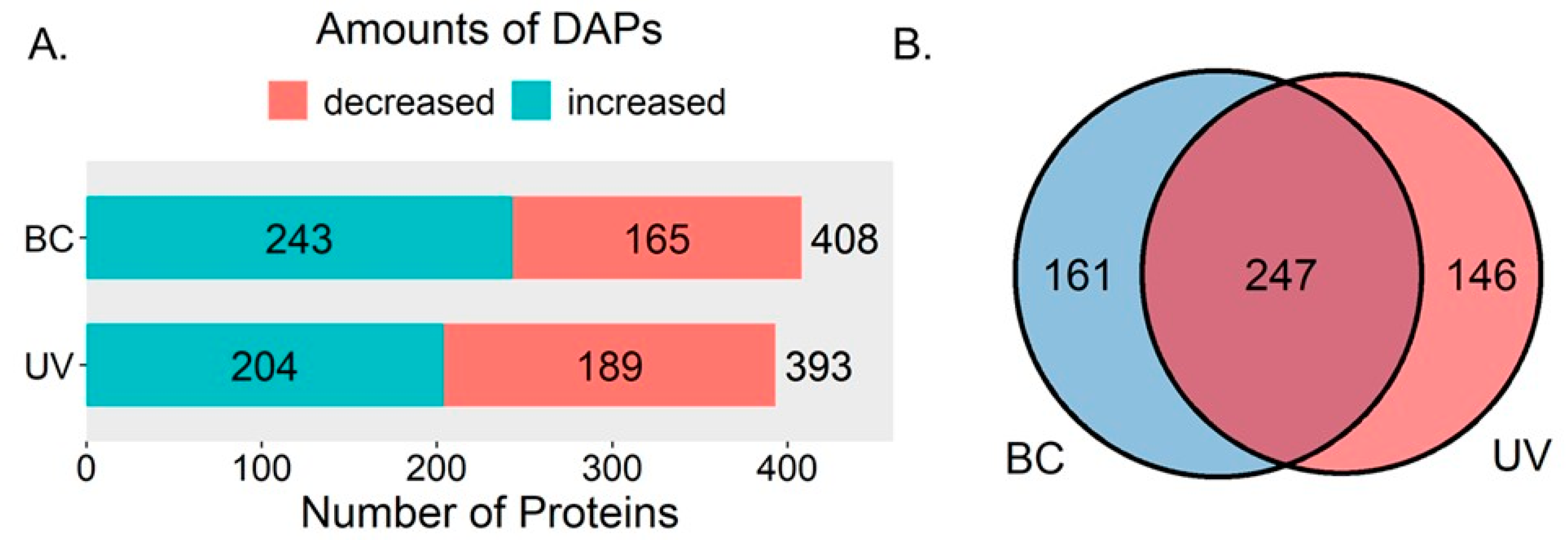
2.1. Overview of Differentially Expressed Proteins in the BC Group and UVB Group
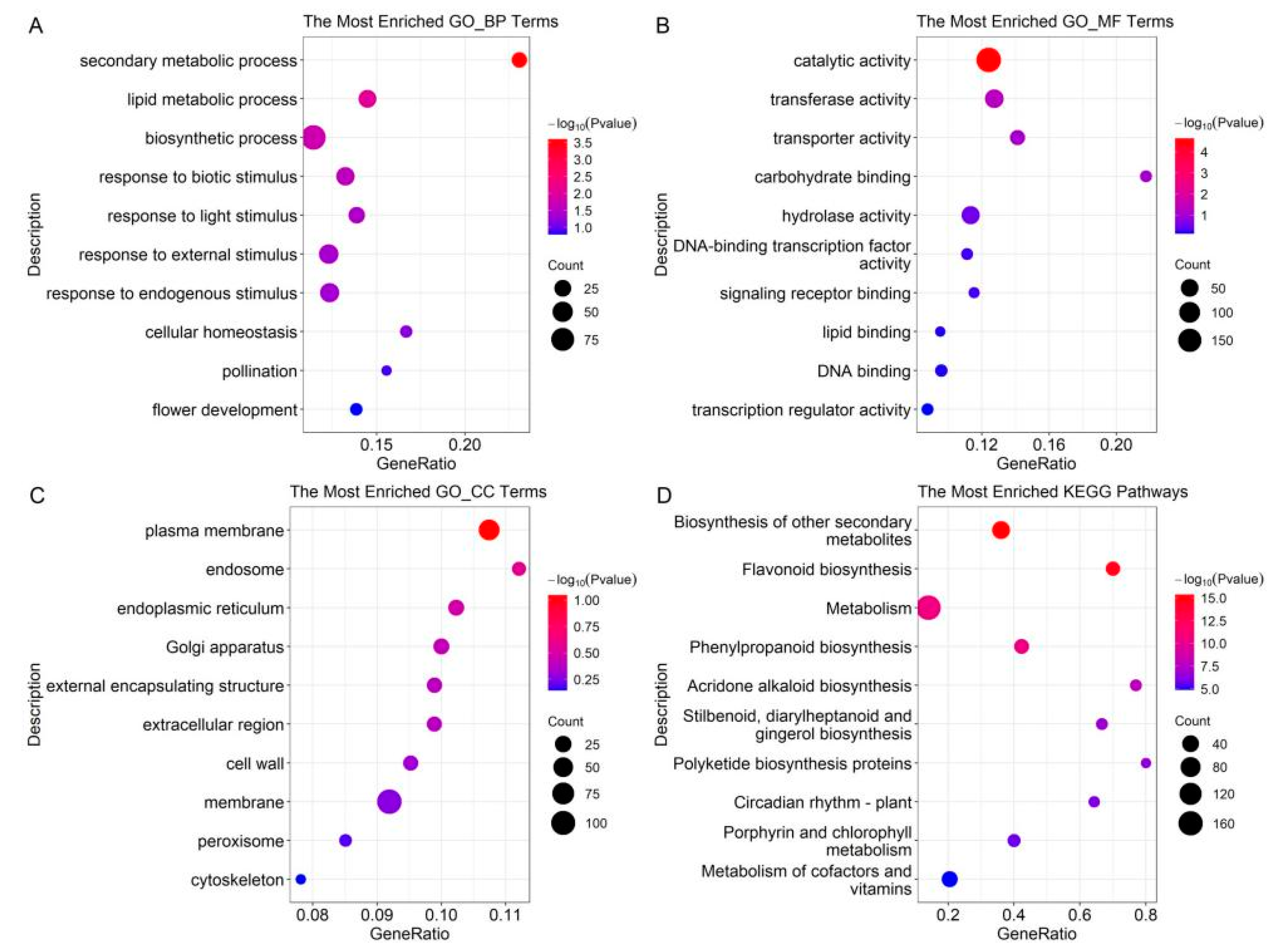
2.2. GO Term and KEGG Pathway Enrichment of DEPs in the BC Group
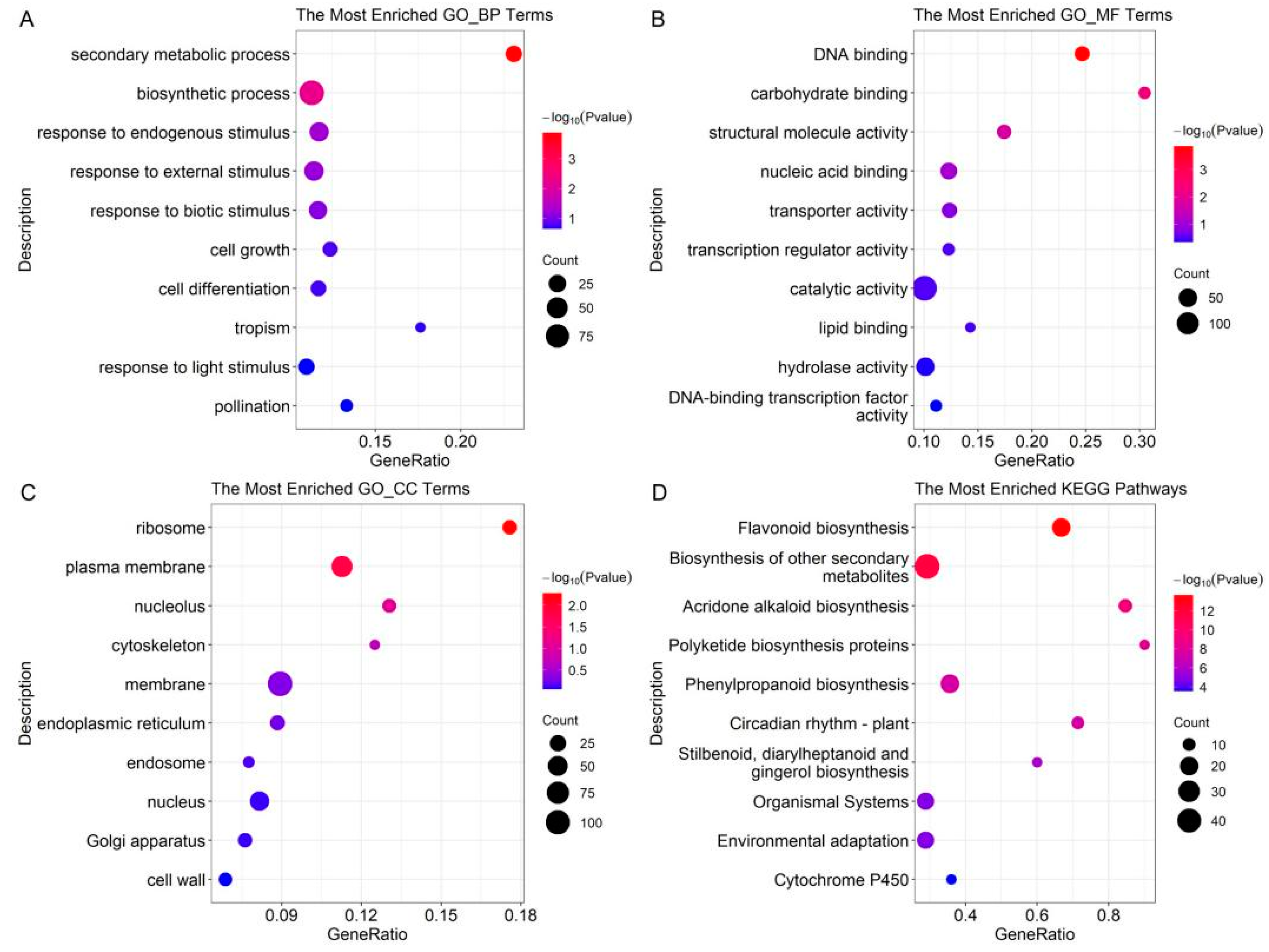
2.3. GO Term and KEGG Pathway Enrichment of DEPs in the UVB Group
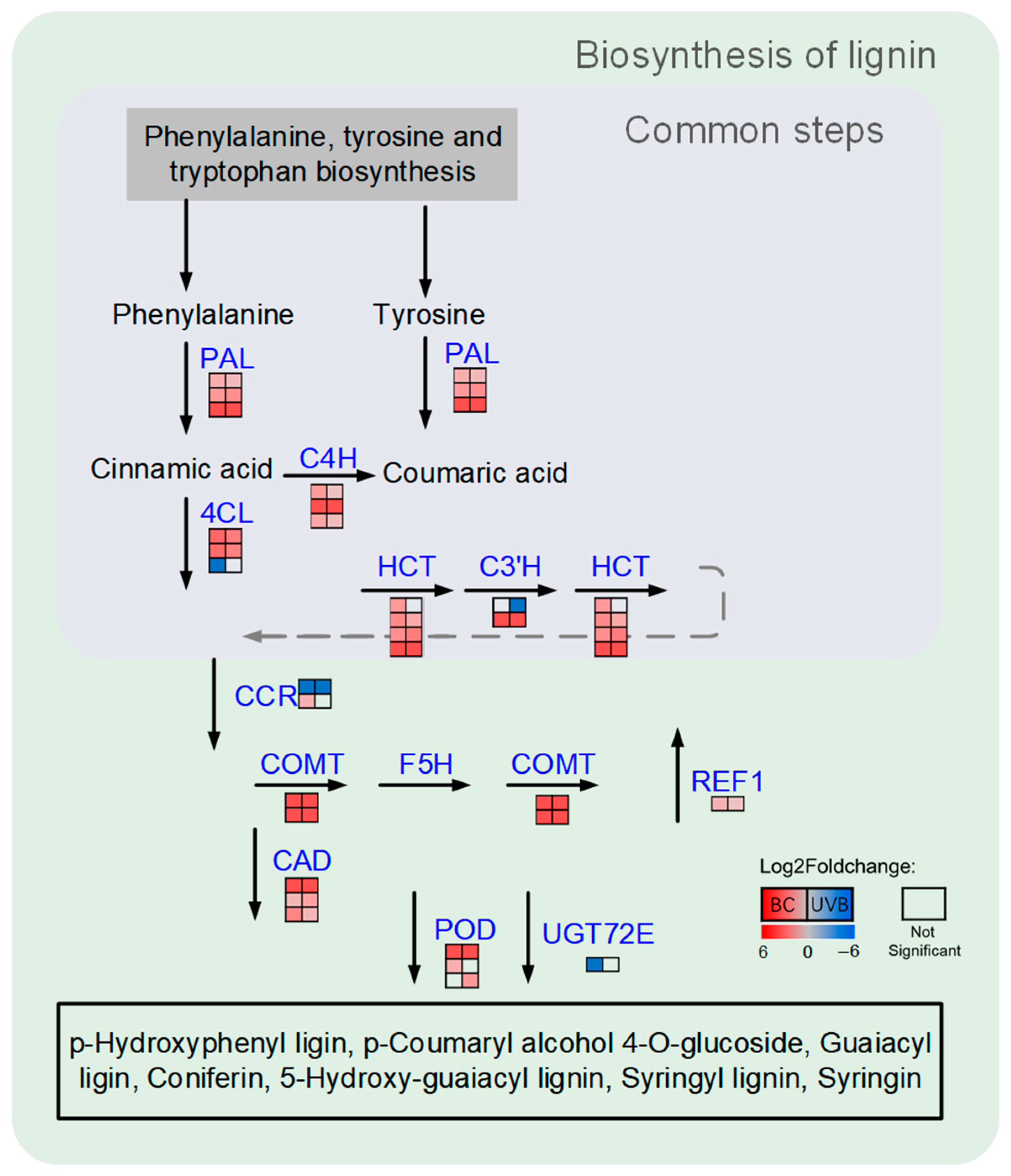
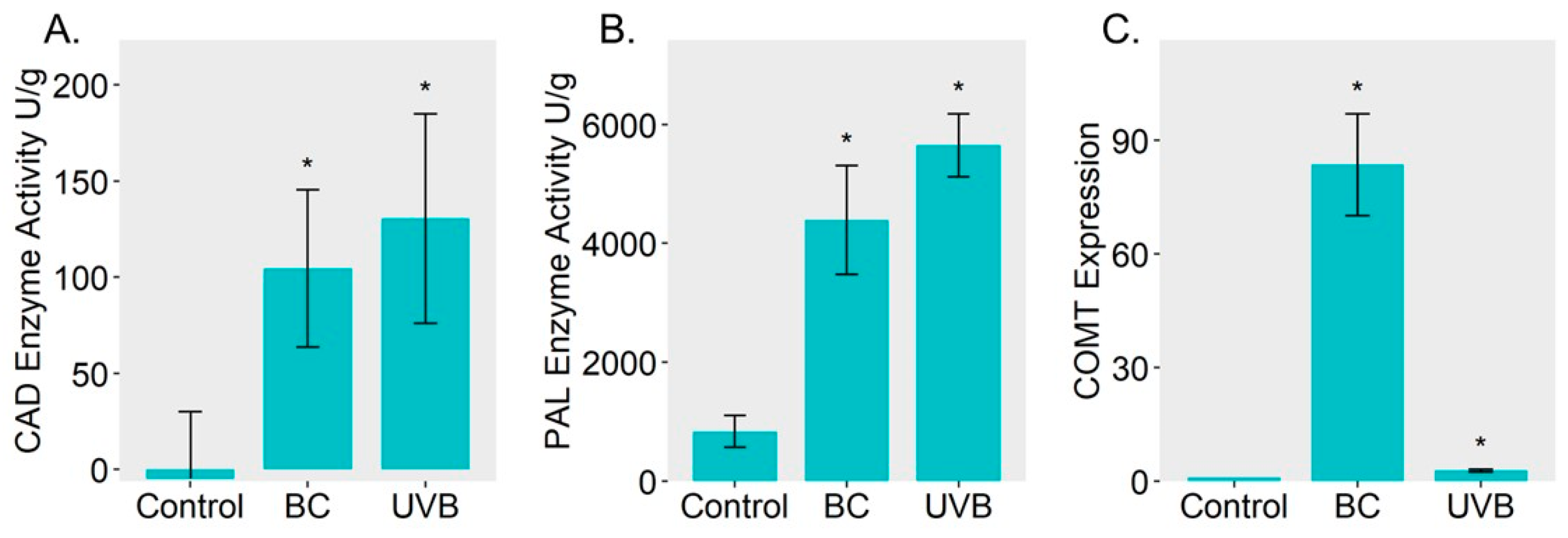
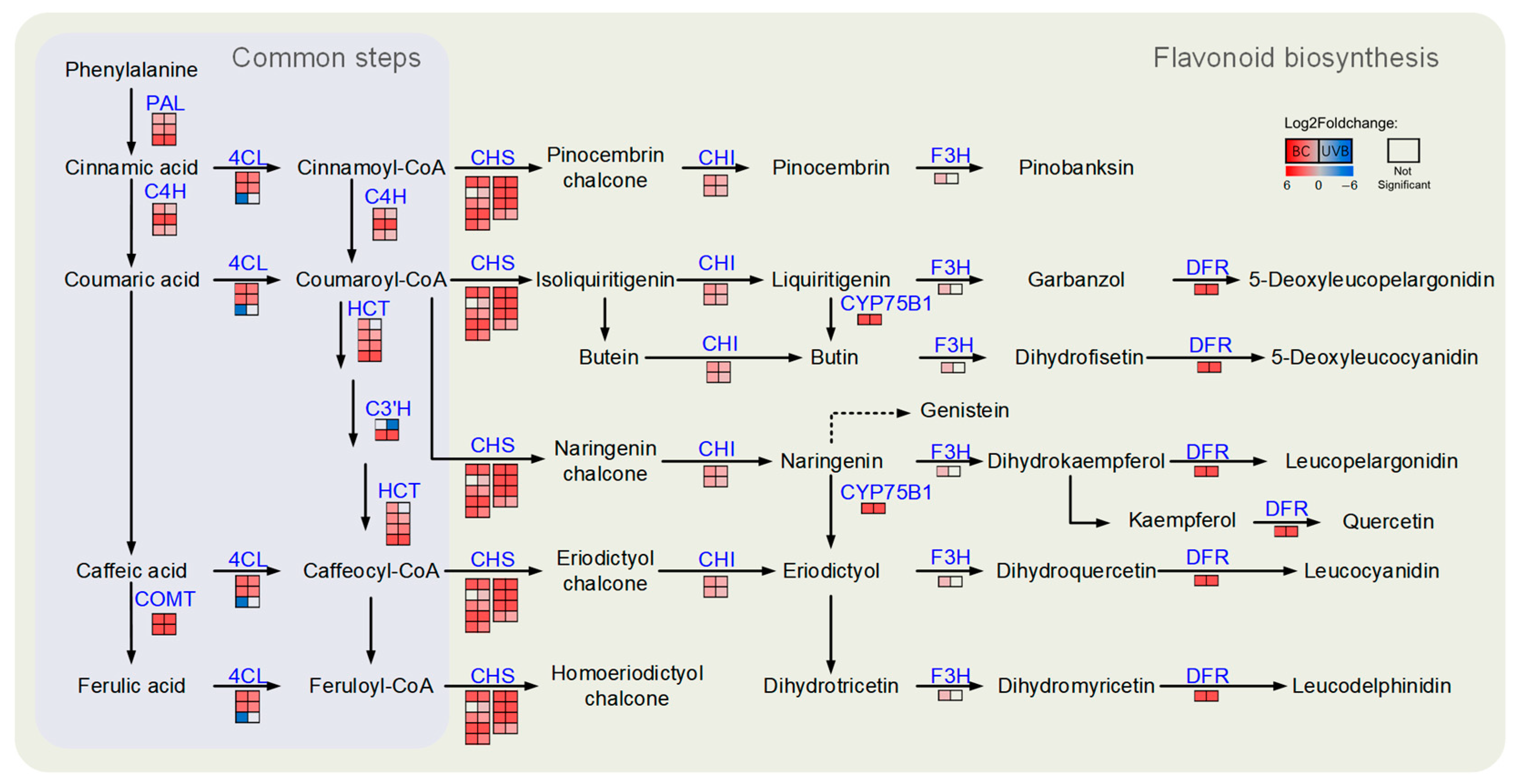
2.4. Lignin Biosynthesis Was Enhanced under Either BC or UVB Stress
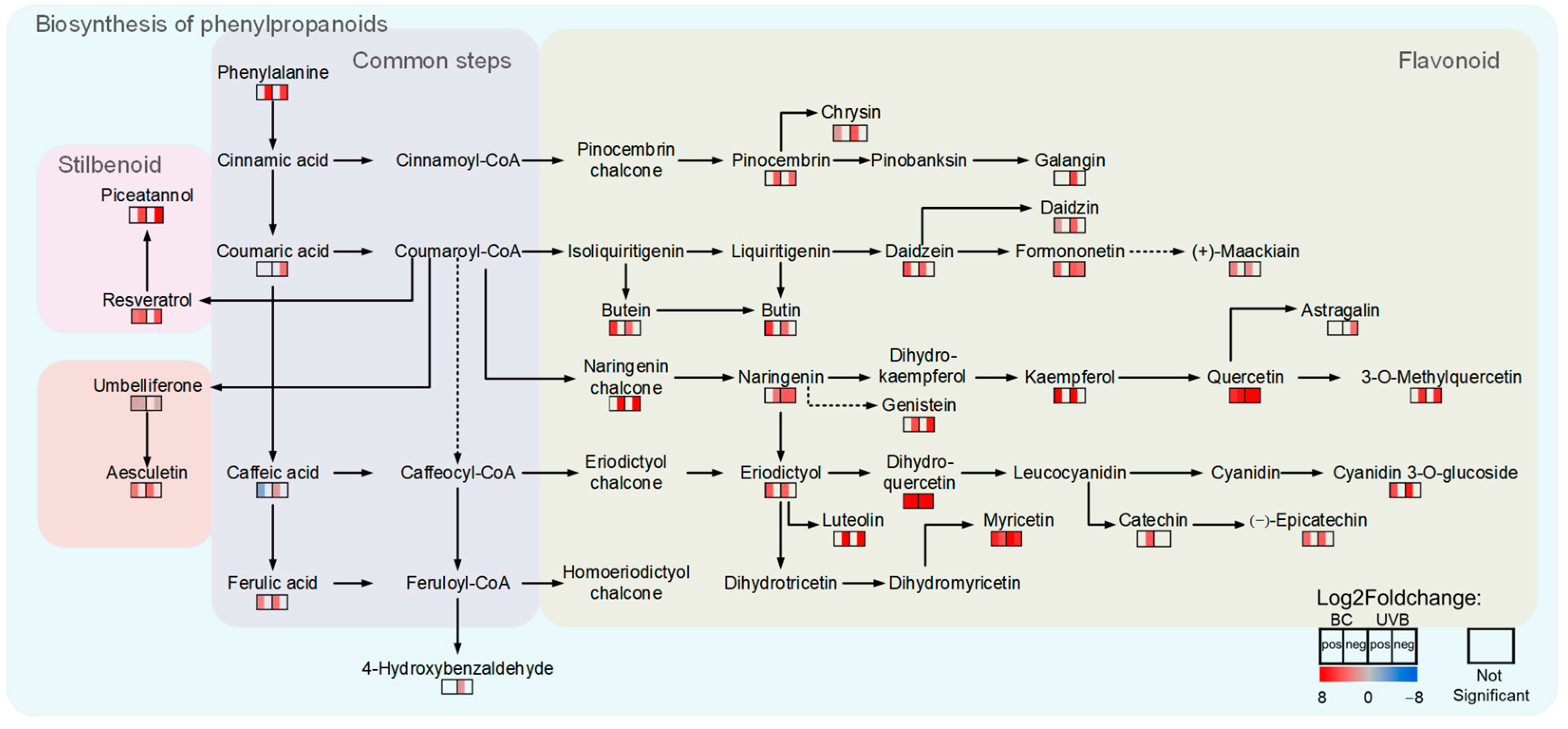
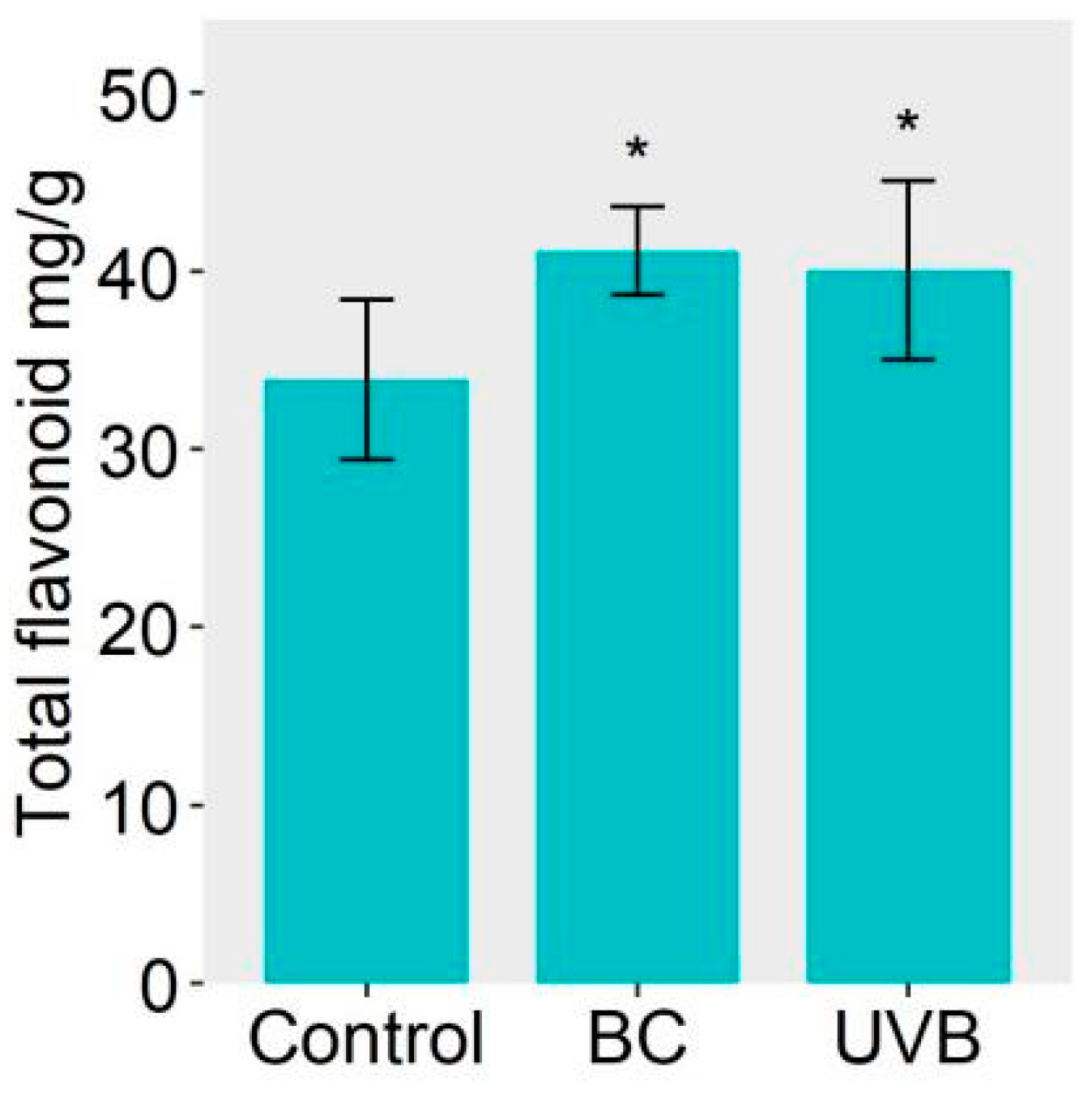
2.5. Flavonoid Biosynthesis Was Enhanced under Either BC or UVB Stress
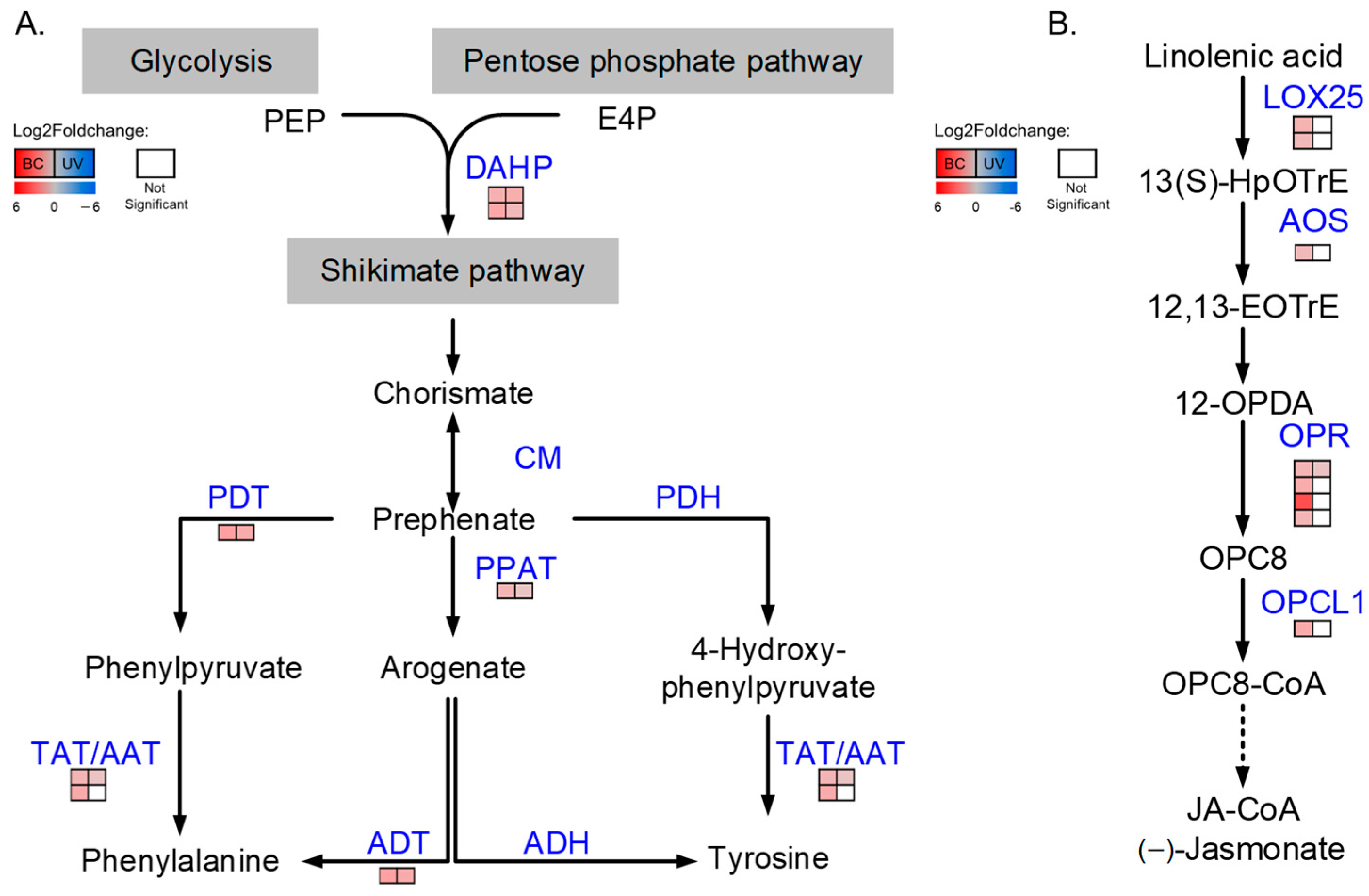
2.6. Proteins Involved in the Biosynthesis of Phenylalanine and Tyrosine Were Increased under BC and UVB Stress
2.7. Proteins Related to Jasmonic Acid Biosynthesis Were Altered in the BC Group
3. Discussion
3.1. Lignin and Flavonoid Biosynthesis Were Both Enhanced under B. cinerea Infection and UV-B Stress
3.2. Accumulation of Flavonoids Contributes to B. cinerea Infection Resistance and UV-B Resistance in M. alba Leaves
3.3. Lignin Biosynthesis Contributes to B. cinerea Resistance and UV-B Resistance in M. alba Leaves
4. Materials and Methods
4.1. Materials and Treatments
4.2. Protein Extraction, Purification, and Digestion
4.3. Nanoflow Liquid Chromatography-Tandem Mass Spectrometry Analysis of Proteomics
4.4. Protein Identification and Data Processing
4.5. GO and KEGG Enrichment of DEPs
4.6. Metabolite Extraction from M. alba Leaves and Untargeted Metabolomic Analyses
4.7. Metabolomic Data Processing
4.8. Quantitative Real-Time PCR
4.9. Analysis of Physiological Changes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, N.-Q.; Lin, H.-X. Contribution of phenylpropanoid metabolis to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Savarese, C.; Cozzolino, V.; Verrillo, M.; Vinci, G.; De Martino, A.; Piccolo, A. Combination of humic biostimulants with a microbial inoculum improves lettuce productivity, nutrient uptake, and primary and secondary metabolism. Plant Soil 2022, 481, 285–314. [Google Scholar] [CrossRef]
- Clayton, W.A.; Albert, N.W.; Thrimawithana, A.H.; McGhie, T.K.; Deroles, S.C.; Schwinn, K.E.; Warren, B.A.; McLachlan, A.R.G.; Bowman, J.L.; Jordan, B.R.; et al. UVR8-mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants. Plant J. 2018, 96, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Gutiérrez-Albanchez, E.; Gradillas, A.; García, A.; García-Villaraco, A.; Gutierrez-Mañero, F.J.; Ramos-Solano, B. Elicitation with Bacillus QV15 reveals a pivotal role of F3H on flavonoid metabolism improving adaptation to biotic stress in blackberry. PLoS ONE 2020, 15, e0232626. [Google Scholar] [CrossRef]
- Schenke, D.; Utami, H.P.; Zhou, Z.; Gallegos, M.-T.; Cai, D. Suppression of UV-B stress induced flavonoids by biotic stress: Is there reciprocal crosstalk? Plant Physiol. Biochem. 2019, 134, 53–63. [Google Scholar] [CrossRef]
- Ding, Y.; Yu, S.; Wang, J.; Li, M.; Qu, C.; Li, J.; Liu, L. Comparative transcriptomic analysis of seed coats with high and low lignin contents reveals lignin and flavonoid biosynthesis in Brassica napus. BMC Plant Biol. 2021, 21, 246. [Google Scholar] [CrossRef]
- Li, D.; Yang, J.; Pak, S.; Zeng, M.; Sun, J.; Yu, S.; He, Y.; Li, C. PuC3H35 confers drought tolerance by enhancing lignin and proanthocyanidin biosynthesis in the roots of Populus ussuriensis. New Phytol. 2022, 233, 390–408. [Google Scholar] [CrossRef]
- Monties, B.; Fukushima, K. Occurrence, function and biosynthesis of lignins. In Biopolymers Online; Steinbuchel, A., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Schenke, D.; Böttcher, C.; Scheel, D. Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ. 2011, 34, 1849–1864. [Google Scholar] [CrossRef]
- McLusky, S.R.; Bennett, M.H.; Beale, M.H.; Lewis, M.J.; Gaskin, P.; Mansfield, J.W. Cell wall alterations and localized accumulation of feruloyl-3′-methoxytyramine in onion epidermis at sites of attempted penetration by Botrytis allii are associated with actin polarisation, peroxidase activity and suppression of flavonoid biosynthesis. Plant J. 1999, 17, 523–534. [Google Scholar] [CrossRef]
- Yuan, Q.; Xie, Y.; Wang, W.; Yan, Y.; Ye, H.; Jabbar, S.; Zeng, X. Extraction optimization, characterization and antioxidant activity in vitro of polysaccharides from mulberry (Morus alba L.) leaves. Carbohydr. Polym. 2015, 128, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Culenova, M.; Sychrova, A.; Hassan, S.; Berchova-Bimova, K.; Svobodova, P.; Helclova, A.; Michnova, H.; Hosek, J.; Vasilev, H.; Suchy, P. Multiple in vitro biological effects of phenolic compounds from Morus alba root bark. J. Ethnopharmacol. 2020, 248, 112296. [Google Scholar] [CrossRef]
- Kuk, E.B.; Jo, A.R.; Oh, S.I.; Sohn, H.S.; Seong, S.H.; Roy, A.; Choi, J.S.; Jung, H.A. Anti-Alzheimer’s disease activity of compounds from the root bark of Morus alba L. Arch. Pharmacal. Res. 2017, 40, 338–349. [Google Scholar] [CrossRef]
- Gan, W.-J.; Gao, C.-L.; Zhang, W.-Q.; Gu, J.-L.; Zhao, T.-T.; Guo, H.-L.; Zhou, H.; Xu, Y.; Yu, L.-L.; Li, L.-F.; et al. Kuwanon G protects HT22 cells from advanced glycation end product-induced damage. Exp. Ther. Med. 2021, 21, 425. [Google Scholar] [CrossRef]
- Bagnasco, D.; Povero, M.; Pradelli, L.; Brussino, L.; Rolla, G.; Caminati, M.; Menzella, F.; Heffler, E.; Canonica, G.W.; Paggiaro, P.; et al. Economic impact of mepolizumab in uncontrolled severe eosinophilic asthma, in real life. World Allergy Organ. J. 2021, 14, 100509. [Google Scholar] [CrossRef]
- Fukai, T.; Kaitou, K.; Terada, S. Antimicrobial activity of 2-arylbenzofurans from Morus species against methicillin-resistant Staphylococcus aureus. Fitoterapia 2005, 76, 708–711. [Google Scholar] [CrossRef]
- Ishikawa, A.; Kitamura, Y.; Ozeki, Y.; Itoh, Y.; Yamada, A.; Watanabe, M. Post-stress metabolism involves umbelliferone production in anthocyanin-producing and non-producing cells of Glehnia littoralis suspension cultures. J. Plant Physiol. 2005, 162, 703–710. [Google Scholar] [CrossRef]
- Saijo, Y.; Tintor, N.; Lu, X.; Rauf, P.; Pajerowska-Mukhtar, K.; Häweker, H.; Dong, X.; Robatzek, S.; Schulze-Lefert, P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009, 28, 3439–3449. [Google Scholar] [CrossRef]
- Liu, S.; Zhong, Z.; Sun, Z.; Tian, J.; Sulaiman, K.; Shawky, E.; Fu, H.; Zhu, W. De novo transcriptome analysis revealed the putative pathway genes involved in biosynthesis of moracins in Morus alba L. ACS Omega 2022, 7, 11343–11352. [Google Scholar] [CrossRef]
- Baozhu, L.; Ruonan, F.; Yanting, F.; Runan, L.; Hui, Z.; Tingting, C.; Jiong, L.; Han, L.; Xiang, Z.; Chun-peng, S. The flavonoid biosynthesis regulator PFG3 confers drought stress tolerance in plants by promoting flavonoid accumulation. Environ. Exp. Bot. 2022, 196, 104792. [Google Scholar] [CrossRef]
- Ortuño, A.; Arcas, M.C.; Botía, J.M.; Fuster, M.D.; Del Río, J.A. Increasing resistance against Phytophthora citrophthora in tangelo Nova fruits by modulating polymethoxyflavones levels. J. Agric. Food Chem. 2002, 50, 2836–2839. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Su, C.; Du, X.; Wang, R.; Chen, S.; Zhou, Y.; Liu, C.; Liu, X.; Tian, R.; Zhang, L.; et al. FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis. Nat. Chem. 2020, 12, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Mascarello, A.; Menegatti, A.C.O.; Calcaterra, A.; Martins, P.G.A.; Chiaradia-Delatorre, L.D.; D’Acquarica, I.; Ferrari, F.; Pau, V.; Sanna, A.; De Logu, A.; et al. Naturally occurring Diels-Alder-type adducts from Morus nigra as potent inhibitors of Mycobacterium tuberculosis protein tyrosine phosphatase B. Eur. J. Med. Chem. 2018, 144, 277–288. [Google Scholar] [CrossRef]
- Wu, S.C.; Han, F.; Song, M.R.; Chen, S.; Li, Q.; Zhang, Q.; Zhu, K.; Shen, J.Z. Natural flavones from Morus alba against methicillin-resistant Staphylococcus aureus via targeting the proton motive force and membrane permeability. J. Agric. Food Chem. 2019, 67, 10222–10234. [Google Scholar] [CrossRef]
- Hu, S.; Chen, F.; Wang, M. Photoprotective effects of oxyresveratrol and Kuwanon O on DNA damage induced by UVA in human epidermal keratinocytes. Chem. Res. Toxicol. 2015, 28, 541–548. [Google Scholar] [CrossRef]
- Sharma, R.; Sharma, A.; Shono, T.; Takasugi, M.; Shirata, A.; Fujimura, T.; Machii, H. Mulberry moracins: Scavengers of UV stress-generated free radicals. Biosci. Biotechnol. Biochem. 2001, 65, 1402–1405. [Google Scholar] [CrossRef]
- Wong, T.M.; Sullivan, J.H.; Eisenstein, E. Acclimation and compensating metabolite responses to UV-B radiation in natural and transgenic Populus spp. defective in lignin biosynthesis. Metabolites 2022, 12, 767. [Google Scholar] [CrossRef]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]
- Yang, C.; Liang, Y.; Qiu, D.; Zeng, H.; Yuan, J.; Yang, X. Lignin metabolism involves Botrytis cinerea BcGs1- induced defense response in tomato. BMC Plant Biol. 2018, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.H.; Phan, D.-P.; Lee, E.Y. Review on lignin modifications toward natural UV protection ingredient for lignin-based sunscreens. Green Chem. 2021, 23, 4633–4646. [Google Scholar] [CrossRef]
- Landry, L.G.; Chapple, C.; Last, R.L. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995, 109, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Fornalé, S.; Shi, X.; Chai, C.; Encina, A.; Irar, S.; Capellades, M.; Fuguet, E.; Torres, J.-L.; Rovira, P.; Puigdomènech, P.; et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 2010, 64, 633–644. [Google Scholar] [CrossRef]
- Li, Y.H.; Liu, S.Z.; Eman, S.; Tao, M.L.; Liu, A.M.; Kaisa, S.; Tian, J.K.; Zhu, W. SWATH-based quantitative proteomic analysis of Morus alba L. leaves after exposure to ultraviolet-B radiation and incubation in the dark. J. Photochem. Photobiol. B Biol. 2022, 230, 112443. [Google Scholar] [CrossRef]
- Zhu, W.; Han, H.; Liu, A.; Guan, Q.; Kang, J.; David, L.; Dufresne, C.; Chen, S.; Tian, J. Combined ultraviolet and darkness regulation of medicinal metabolites in Mahonia bealei revealed by proteomics and metabolomics. J. Proteom. 2021, 233, 104081. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Yuan, K.; Li, Q.; Liu, S.; Li, Y.; Tao, M.; Xu, H.; Tian, J.; Guan, S.; Zhu, W. Metabolomics and proteomics revealed the synthesis difference of aroma precursors in tobacco leaves at various growth stages. Plant Physiol. Biochem. 2022, 192, 308–319. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Botstein, D.; Cherry, J.M.; Ashburner, M.; Ball, C.A.; Blake, J.A.; Butler, H.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E.; et al. The Gene Ontology resource: Enriching a Gold mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Liu, S.; Han, S.; Li, Y.; Tao, M.; Liu, A.; He, Q.; Chen, S.; Dufresne, C.; Zhu, W.; et al. Integrative omic analysis reveals the improvement of alkaloid accumulation by ultraviolet-B radiation and its upstream regulation in Catharanthus roseus. Ind. Crops Prod. 2021, 166, 113448. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis working group(CAWG) metabolomics standards initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Liu, S.; Li, Y.; Tao, M.; Han, H.; Zhong, Z.; Zhu, W.; Tian, J. Phosphoproteomics Reveals Regulation of Secondary Metabolites in Mahonia bealei Exposed to Ultraviolet-B Radiation. Front. Plant Sci. 2022, 12, 794906. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Liu, S.; Zhang, D.; Liu, A.; Zhu, W.; Zhang, J.; Yang, B. Integrative Omic Analysis Reveals the Dynamic Change in Phenylpropanoid Metabolism in Morus alba under Different Stress. Plants 2023, 12, 3265. https://doi.org/10.3390/plants12183265
Li Y, Liu S, Zhang D, Liu A, Zhu W, Zhang J, Yang B. Integrative Omic Analysis Reveals the Dynamic Change in Phenylpropanoid Metabolism in Morus alba under Different Stress. Plants. 2023; 12(18):3265. https://doi.org/10.3390/plants12183265
Chicago/Turabian StyleLi, Yaohan, Shengzhi Liu, Di Zhang, Amin Liu, Wei Zhu, Jianbin Zhang, and Bingxian Yang. 2023. "Integrative Omic Analysis Reveals the Dynamic Change in Phenylpropanoid Metabolism in Morus alba under Different Stress" Plants 12, no. 18: 3265. https://doi.org/10.3390/plants12183265
APA StyleLi, Y., Liu, S., Zhang, D., Liu, A., Zhu, W., Zhang, J., & Yang, B. (2023). Integrative Omic Analysis Reveals the Dynamic Change in Phenylpropanoid Metabolism in Morus alba under Different Stress. Plants, 12(18), 3265. https://doi.org/10.3390/plants12183265




