Genome-Wide Identification of the U-Box E3 Ubiquitin Ligase Gene Family in Cabbage (Brassica oleracea var. capitata) and Its Expression Analysis in Response to Cold Stress and Pathogen Infection
Abstract
1. Introduction
2. Results
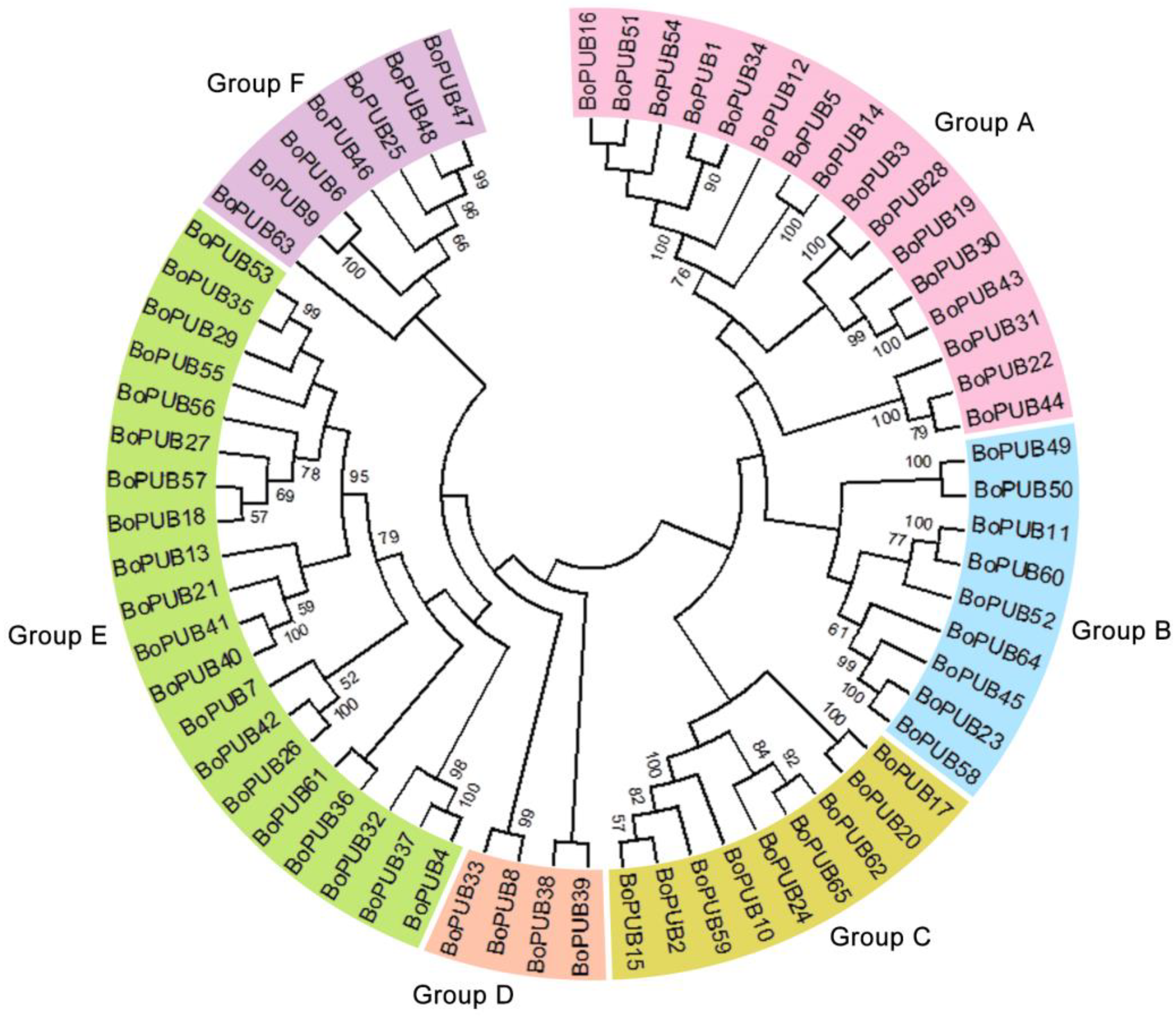
2.1. Genome-Wide Identification and Phylogenetic Relationships of BoPUB Genes
2.2. Conserved Domains and Exon-Intron Structures of BoPUB Genes
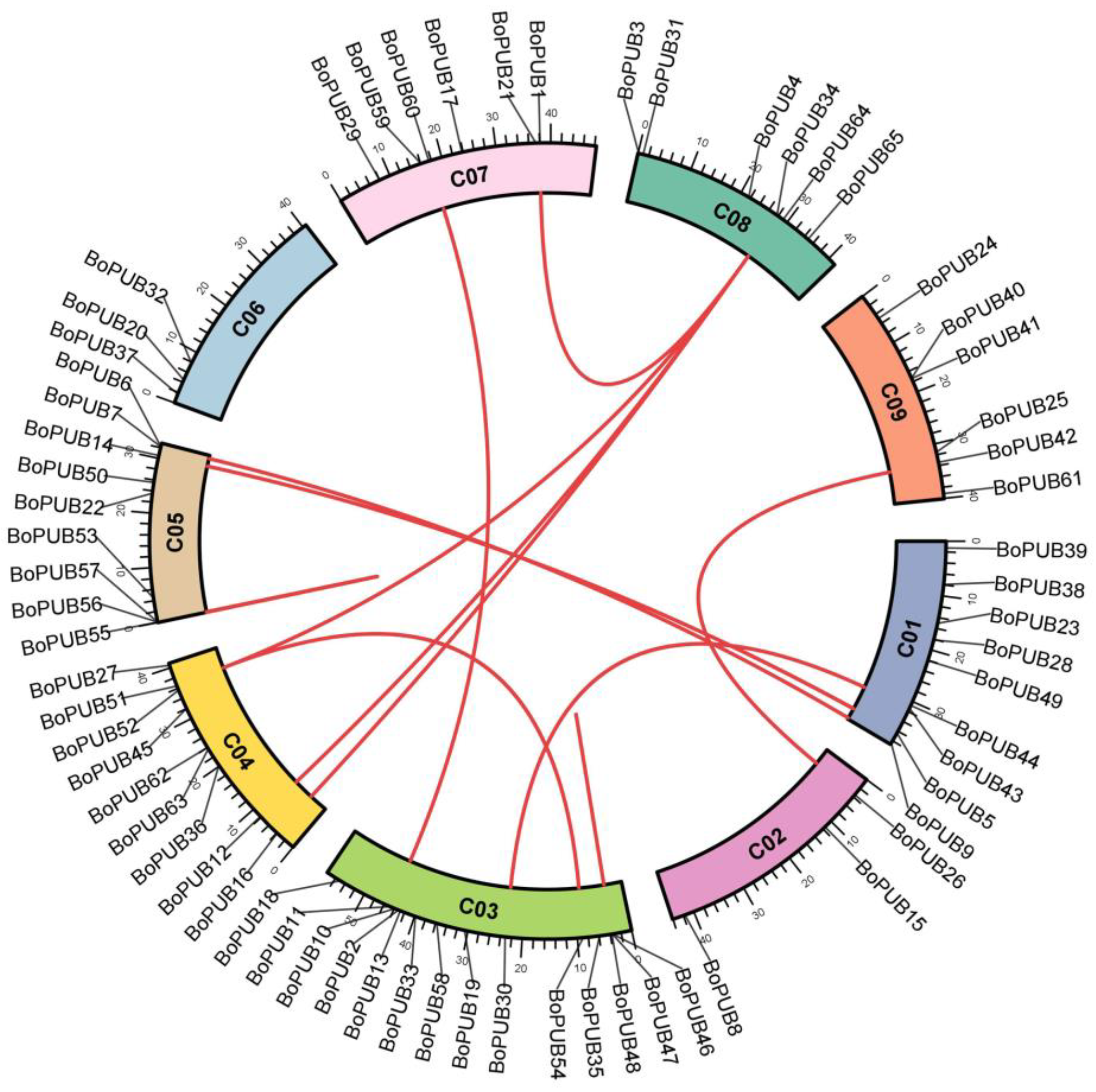
2.3. Chromosomal Localization and Duplication of BoPUB Genes
2.4. Interaction Relationships between BoPUB Proteins
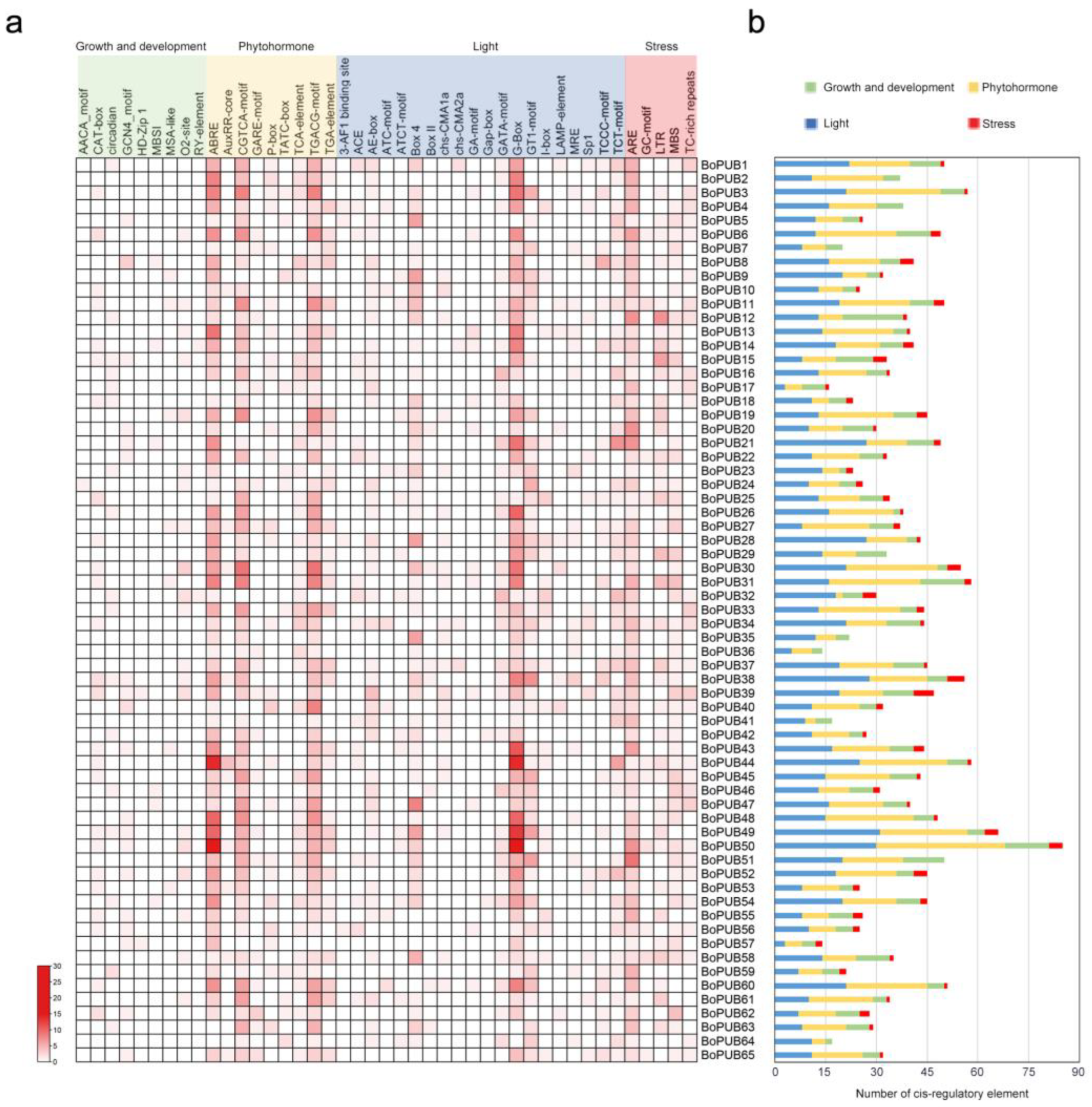
2.5. Cis-Regulatory Elements in the Promoters of BoPUB Genes
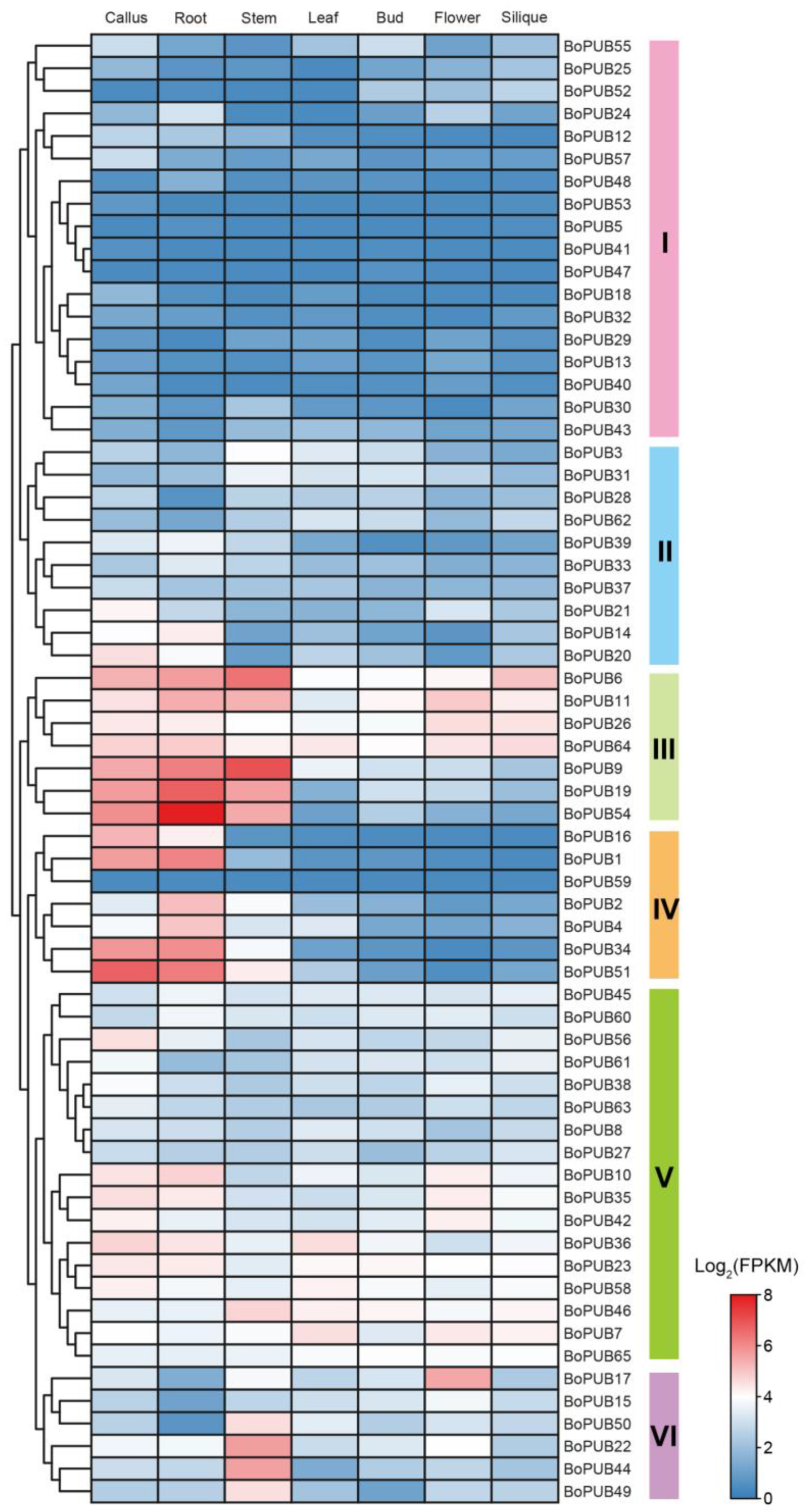
2.6. Spatio-Temporal Expression Patterns of BoPUB Genes in Different Tissues
2.7. Expression Patterns of BoPUB Genes Involved in Cuticular Wax Biosynthesis
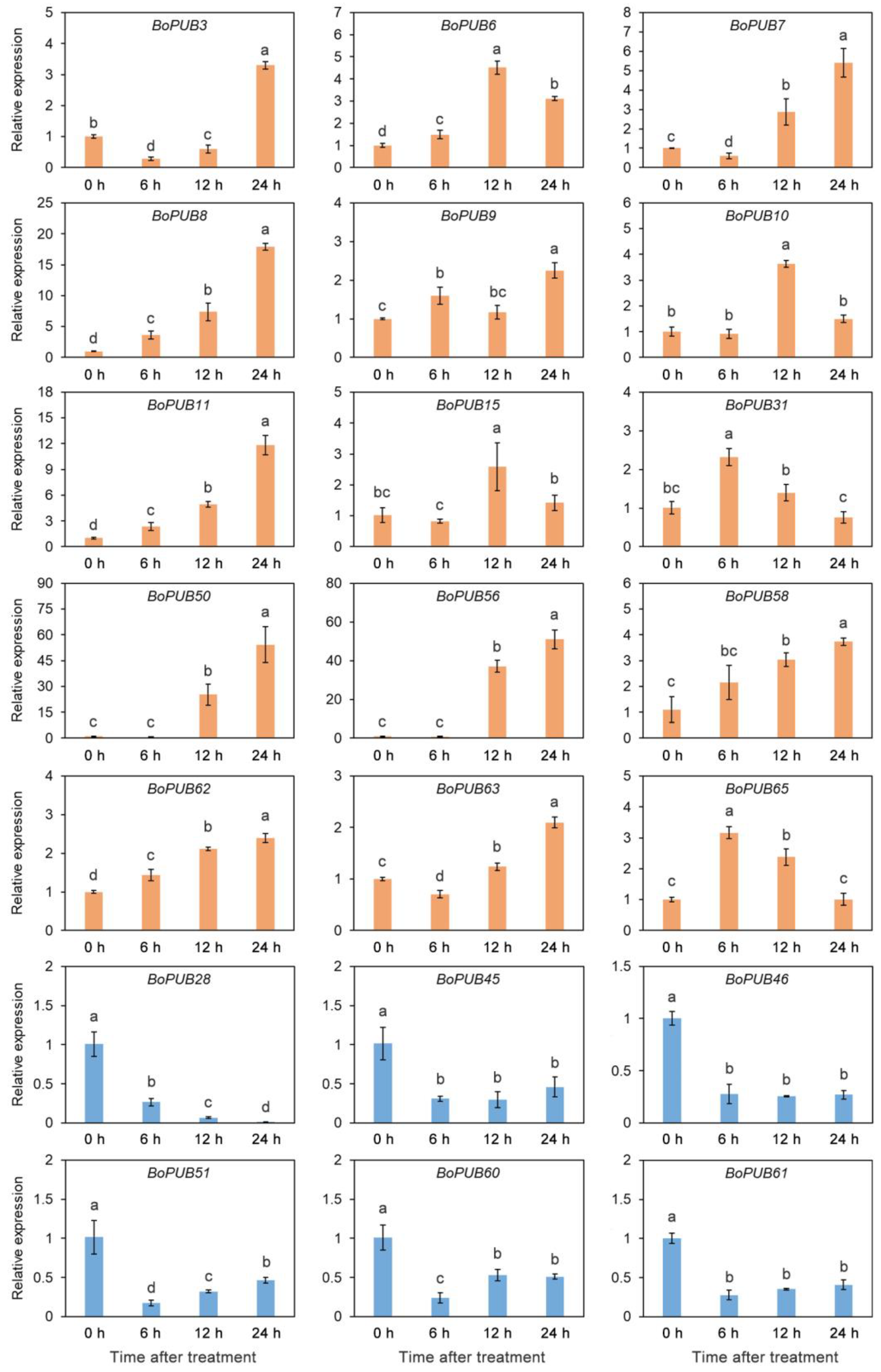
2.8. Expression Patterns of BoPUB Genes in Response to Cold Stress
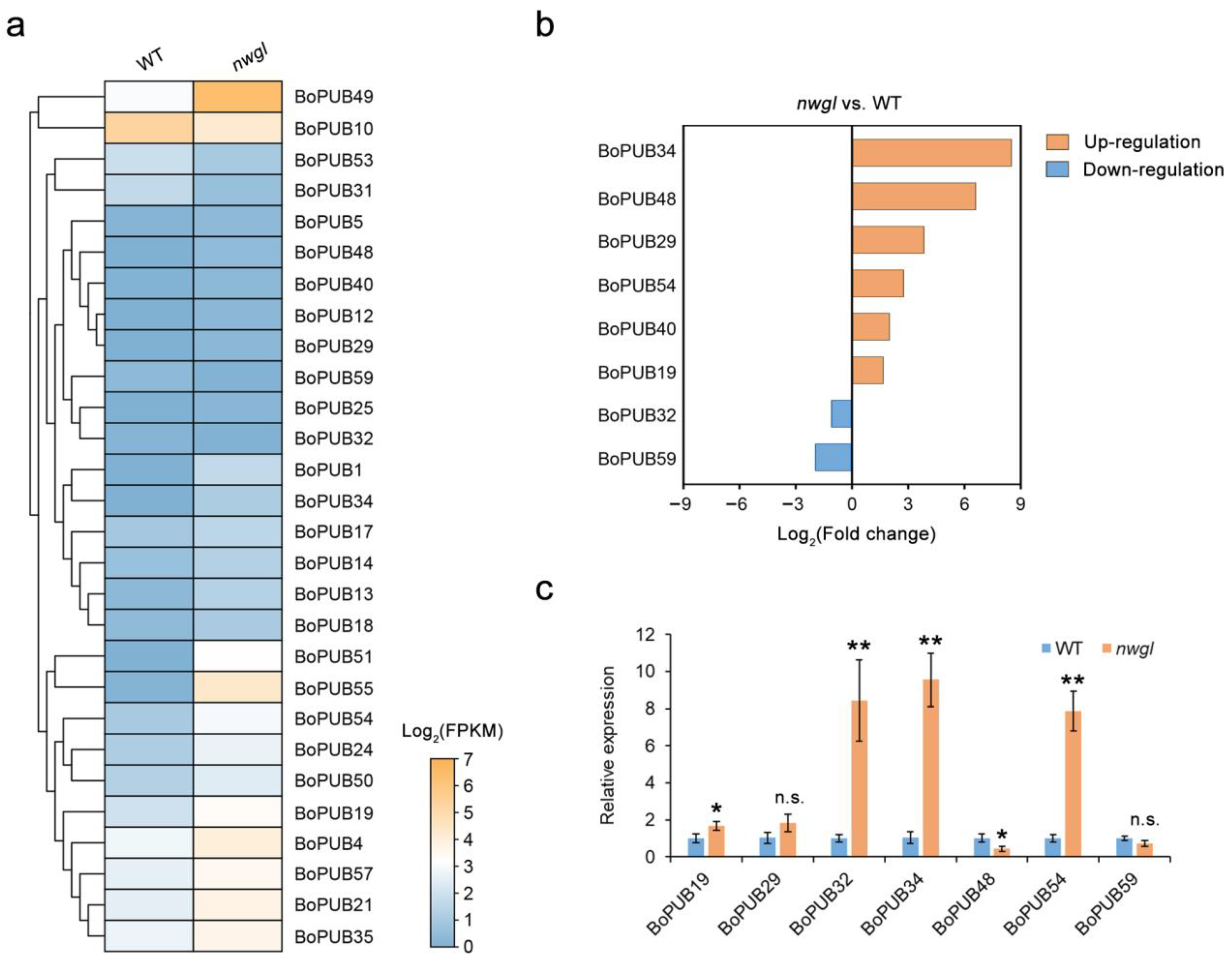
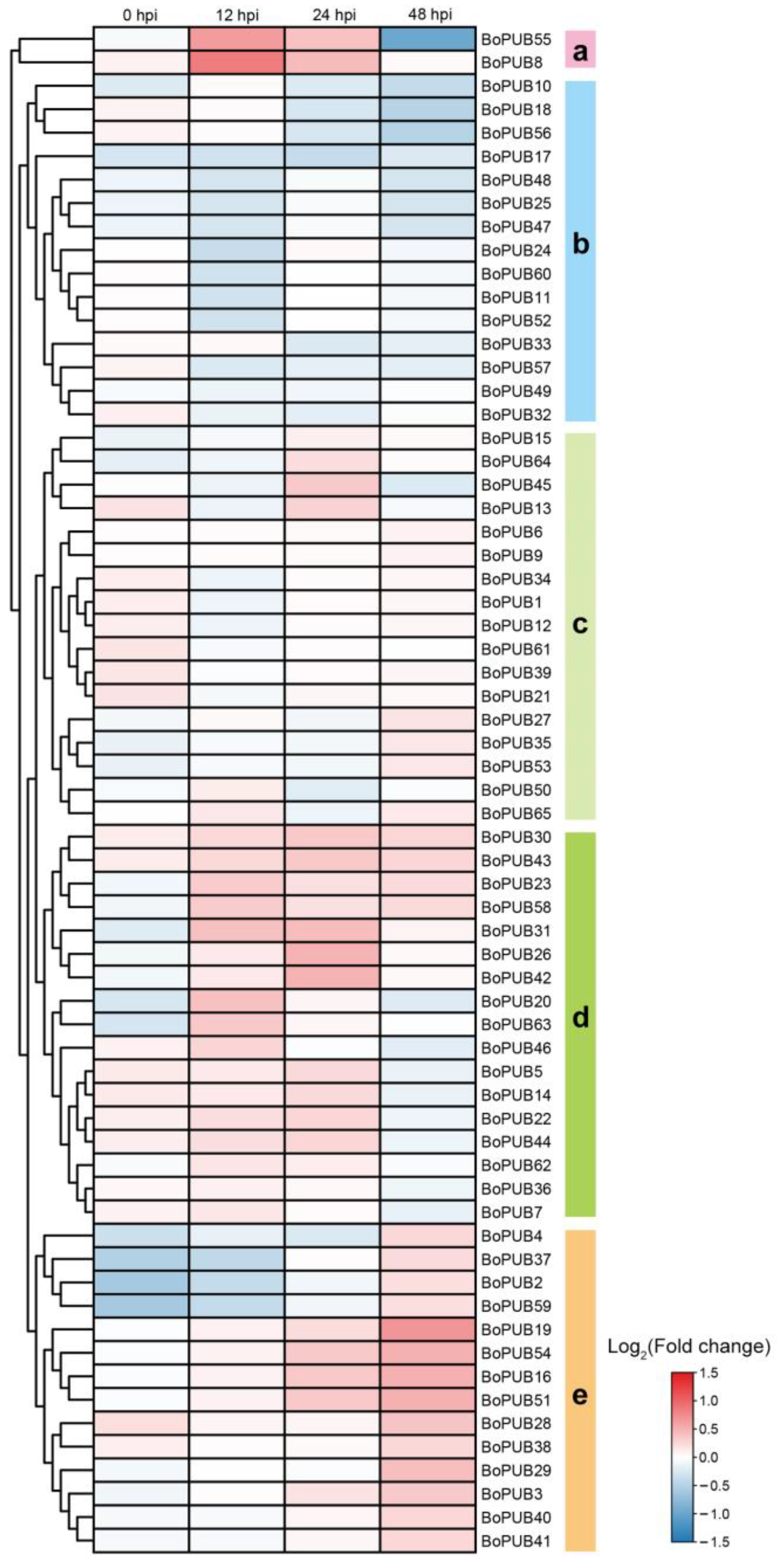
2.9. Expression Patterns of BoPUB Genes in Response to Alternaria brassicicola Infection
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Cold Treatment
4.2. Identification of BoPUB Genes
4.3. Phylogenetic Analysis
4.4. Conserved Domain and Gene Structure Analysis
4.5. Chromosomal Localization and Synteny Analysis
4.6. Protein Interaction Prediction
4.7. Cis-Regulatory Element Prediction
4.8. Expression Analysis of BoPUB Genes in Different Tissues
4.9. Expression Analysis of BoPUB Genes Involved in Cuticular Wax Biosynthesis
4.10. RNA Isolation and Quantitative Real-Time PCR
4.11. Expression Analysis of BoPUB Genes after Alternaria brassicicola Infection
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.W.; Elledge, S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Ben-Neriah, Y. Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu. Rev. Immunol. 2000, 18, 621–663. [Google Scholar] [CrossRef] [PubMed]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef]
- Liu, T.B.; Xue, C. The ubiquitin-proteasome system and F-box proteins in pathogenic fungi. Mycobiology 2011, 39, 243–248. [Google Scholar] [CrossRef]
- Chen, L.; Hellmann, H. Plant E3 ligases: Flexible enzymes in a sessile world. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies Mulan, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, X.; Liu, S.; Su, T.; Huang, H.; Ren, H.; Gao, Z.; Wang, X.; Lin, D.; Wohlschlegel, J.A.; et al. Regulation of Arabidopsis photoreceptor CRY2 by two distinct E3 ubiquitin ligases. Nat. Commun. 2021, 12, 2155. [Google Scholar] [CrossRef]
- Chen, H.; Ma, B.; Zhou, Y.; He, S.J.; Tang, S.Y.; Lu, X.; Xie, Q.; Chen, S.Y.; Zhang, J.S. E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of AUX/IAA protein. Proc. Natl. Acad. Sci. USA 2018, 115, 4513–4518. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, H.; Su, T.; Wu, W.H.; Chen, Y.F. The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low-Pi stress in Arabidopsis. Plant Cell 2018, 30, 1062–1076. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.C.; Kim, S.H.; Jeon, Y.A.; Lee, H.S.; Adeva, C.; Kang, J.W.; Kim, H.J.; Tai, T.H.; Ahn, S.N. A RING-type E3 ubiquitin ligase, OsGW2, controls chlorophyll content and dark-induced senescence in rice. Int. J. Mol. Sci. 2020, 21, 1704. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M. News from the PUB: Plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 2018, 69, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.; Kragelund, B.B.; Olsen, A.N.; Larsen, F.H.; Chua, N.H.; Poulsen, F.M.; Skriver, K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 2004, 279, 40053–40061. [Google Scholar] [CrossRef]
- Mudgil, Y.; Shiu, S.H.; Stone, S.L.; Salt, J.N.; Goring, D.R. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004, 134, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, A.; ten Hove, C.A.; Tabata, R.; Yamada, M.; Shimizu, N.; Ishida, T.; Yamaguchi, K.; Shigenobu, S.; Takebayashi, Y.; Iuchi, S.; et al. A plant U-box protein; PUB4; regulates asymmetric cell division and cell proliferation in the root meristem. Development 2015, 142, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.J.; Yi, Y.B.; Lee, J.H.; Chung, Y.S.; Jung, H.W.; Yun, D.J.; Kim, K.M.; Park, D.S.; Kim, D.H. Molecular cloning and characterization of OsUPS, a U-box containing E3 ligase gene that respond to phosphate starvation in rice (Oryza sativa). Mol. Biol. Rep. 2012, 39, 5883–5888. [Google Scholar] [CrossRef]
- Stone, S.L.; Arnoldo, M.; Goring, D.R. A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 1999, 286, 1729–1731. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, L. The tomato U-box type E3 ligase PUB13 acts with group III ubiquitin E2 enzymes to modulate FLS2-mediated immune signaling. Front. Plant Sci. 2018, 9, 615. [Google Scholar] [CrossRef]
- Han, P.L.; Dong, Y.H.; Gu, K.D.; Yu, J.Q.; Hu, D.G.; Hao, Y.J. The apple U-box E3 ubiquitin ligase MdPUB29 contributes to activate plant immune response to the fungal pathogen Botryosphaeria dothidea. Planta 2019, 249, 1177–1188. [Google Scholar] [CrossRef]
- Wang, C.; Duan, W.; Riquicho, A.R.; Jing, Z.; Liu, T.; Hou, X.; Li, Y. Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol. Genet. Genom. 2015, 290, 2241–2260. [Google Scholar] [CrossRef]
- Yu, Y.H.; Li, X.Z.; Guo, D.L.; Zhang, H.L.; Li, G.R.; Li, X.Q.; Zhang, G.H. Genome-wide identification and analysis of the U-box family of E3 ligases in grapevine. Russ. J. Plant Physiol. 2016, 63, 835–848. [Google Scholar] [CrossRef]
- Sharma, B.; Taganna, J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 2020, 10, 9581. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Song, B.; Dai, Y.; Zhang, S.; Huang, X. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 235. [Google Scholar] [CrossRef]
- Hu, D.; Xie, Q.; Liu, Q.; Zuo, T.; Zhang, H.; Zhang, Y.; Lian, X.; Zhu, L. Genome-wide distribution, expression and function analysis of the U-box gene family in Brassica oleracea L. Genes 2019, 10, 1000. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Van Eck, J.; Zhou, X.; Lopez, A.B.; O’Halloran, D.M.; Cosman, K.M.; Conlin, B.J.; Paolillo, D.J.; Garvin, D.F.; Vrebalov, J.; et al. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 2006, 18, 3594–3605. [Google Scholar] [CrossRef]
- Zeng, J.; Gao, Q.; Shi, S.; Lian, X.; Converse, R.; Zhang, H.; Yang, X.; Ren, X.; Chen, S.; Zhu, L. Dissecting pistil responses to incompatible and compatible pollen in self-incompatibility Brassica oleracea using comparative proteomics. Protein J. 2017, 36, 123–137. [Google Scholar] [CrossRef]
- Abuyusuf, M.; Nath, U.K.; Kim, H.T.; Islam, M.R.; Park, J.I.; Nou, I.S. Molecular markers based on sequence variation in BoFLC1.C9 for characterizing early- and late-flowering cabbage genotypes. BMC Genet. 2019, 20, 42. [Google Scholar] [CrossRef]
- Xiao, Z.; Xing, M.; Liu, X.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Y.; Zhuang, M.; Lv, H. An efficient virus-induced gene silencing (VIGS) system for functional genomics in Brassicas using a cabbage leaf curl virus (CaLCuV)-based vector. Planta 2020, 252, 42. [Google Scholar] [CrossRef]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef] [PubMed]
- Lyu, M.; Iftikhar, J.; Guo, R.; Wu, B. Patterns of expansion and expression divergence of the polygalacturonase gene family in Brassica oleracea. Int. J. Mol. Sci. 2020, 21, 5706. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.Y.; Xia, E.H.; Liu, F.H.; Gao, L.Z. Genome-wide identification and comparative expression analysis reveal a rapid expansion and functional divergence of duplicated genes in the WRKY gene family of cabbage; Brassica oleracea var. capitata. Gene 2015, 557, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Furlan, G.; Nakagami, H.; Eschen-Lippold, L.; Jiang, X.; Majovsky, P.; Kowarschik, K.; Hoehenwarter, W.; Lee, J.; Trujillo, M. Changes in PUB22 ubiquitination modes triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 dampen the immune response. Plant Cell 2017, 29, 726–745. [Google Scholar] [CrossRef]
- Maston, G.A.; Evans, S.K.; Green, M.R. Transcriptional regulatory elements in the human genome. Annu. Rev. Genom. Hum. Genet. 2006, 7, 29–59. [Google Scholar] [CrossRef]
- Yang, Z.; Dai, Z.; Chen, X.; Xie, D.; Tang, Q.; Cheng, C.; Xu, Y.; Deng, C.; Liu, C.; Chen, J.; et al. Gene coexpression network analysis and tissue-specific profiling of gene expression in jute (Corchorus capsularis L.). BMC Genom. 2020, 21, 406. [Google Scholar] [CrossRef]
- Lee, H.G.; Kim, J.; Suh, M.C.; Seo, P.J. The MIEL1 E3 ubiquitin ligase negatively regulates cuticular wax biosynthesis in Arabidopsis stems. Plant Cell Physiol. 2017, 58, 1249–1259. [Google Scholar] [CrossRef]
- Liu, S.; Tong, M.; Zhao, L.; Li, X.; Kunst, L. The ARRE RING-type E3 ubiquitin ligase negatively regulates cuticular wax biosynthesis in Arabidopsis thaliana by controlling ECERIFERUM1 and ECERIFERUM3 protein levels. Front. Plant Sci. 2021, 12, 752309. [Google Scholar] [CrossRef]
- Wang, Z.; Tian, X.; Zhao, Q.; Liu, Z.; Li, X.; Ren, Y.; Tang, J.; Fang, J.; Xu, Q.; Bu, Q. The E3 ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice. Plant Cell 2018, 30, 228–244. [Google Scholar] [CrossRef]
- Zhu, X.; Tai, X.; Ren, Y.; Chen, J.; Bo, T. Genome-wide analysis of coding and long non-coding RNAs involved in cuticular wax biosynthesis in cabbage (Brassica oleracea L. Var. capitata). Int. J. Mol. Sci. 2019, 20, 2820. [Google Scholar] [CrossRef]
- Wang, X.; Ding, Y.; Li, Z.; Shi, Y.; Wang, J.; Hua, J.; Gong, Z.; Zhou, J.M.; Yang, S. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev. Cell 2019, 51, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Byun, M.Y.; Cui, L.H.; Oh, T.K.; Jung, Y.J.; Lee, A.; Park, K.Y.; Kang, B.G.; Kim, W.T. Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Mo, X.; Yang, H.; Yue, L.; Song, J.; Mo, B. The U-box family genes in Medicago truncatula: Key elements in response to salt, cold, and drought stresses. PLoS ONE 2017, 12, e0182402. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Zhang, Y.; Lu, J. Overexpression of a stress-responsive U-box protein gene VaPUB affects the accumulation of resistance related proteins in Vitis vinifera ‘thompson seedless’. Plant Physiol. Biochem. 2017, 112, 53–63. [Google Scholar] [CrossRef]
- Jacobs, S.; Zechmann, B.; Molitor, A.; Trujillo, M.; Petutschnig, E.; Lipka, V.; Kogel, K.H.; Schäfer, P. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 2011, 156, 726–740. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef]
- Chen, Y.C.; Wong, C.L.; Muzzi, F.; Vlaardingerbroek, I.; Kidd, B.N.; Schenk, P.M. Root defense analysis against Fusarium oxysporum reveals new regulators to confer resistance. Sci. Rep. 2014, 4, 5584. [Google Scholar] [CrossRef]
- Macioszek, V.K.; Gapińska, M.; Zmienko, A.; Sobczak, M.; Skoczowski, A.; Oliwa, J.; Kononowicz, A.K. Complexity of Brassica oleracea-Alternaria brassicicola susceptible interaction reveals downregulation of photosynthesis at ultrastructural, transcriptional, and physiological levels. Cells 2020, 9, 2329. [Google Scholar] [CrossRef]
- Wiborg, J.; O’Shea, C.; Skriver, K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef]
- Zeng, L.R.; Park, C.H.; Venu, R.C.; Gough, J.; Wang, G.L. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 2008, 1, 800–815. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, Y.; Cong, Y.; Wang, T.; Zhong, X.; Yang, S.; Li, Y.; Gai, J. Genome-wide identification of soybean U-box E3 ubiquitin ligases and roles of GmPUB8 in negative regulation of drought stress response in Arabidopsis. Plant Cell Physiol. 2016, 57, 1189–1209. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shu, N.; Wang, D.; Wang, J.; Chen, X.; Zhang, B.; Wang, S.; Guo, L.; Chen, C.; Ye, W. Genome-wide identification and expression analysis of PUB genes in cotton. BMC Genom. 2020, 21, 213. [Google Scholar] [CrossRef]
- Kim, D.Y.; Lee, Y.J.; Hong, M.J.; Kim, J.H.; Seo, Y.W. Genome wide analysis of U-box E3 ubiquitin ligases in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 2699. [Google Scholar] [CrossRef]
- Raab, S.; Drechsel, G.; Zarepour, M.; Hartung, W.; Koshiba, T.; Bittner, F.; Hoth, S. Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 2009, 59, 39–51. [Google Scholar] [CrossRef]
- Luo, J.; Shen, G.; Yan, J.; He, C.; Zhang, H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 2006, 46, 649–657. [Google Scholar] [CrossRef]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef]
- Samuel, M.A.; Mudgil, Y.; Salt, J.N.; Delmas, F.; Ramachandran, S.; Chilelli, A.; Goring, D.R. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 2008, 147, 2084–2095. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.H.; Ryu, M.Y.; Jammes, F.; Hwang, J.H.; Turek, M.; Kang, B.G.; Kwak, J.M.; Kim, W.T. Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 2012, 160, 556–568. [Google Scholar] [CrossRef]
- Jung, C.; Zhao, P.; Seo, J.S.; Mitsuda, N.; Deng, S.; Chua, N.H. PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 2015, 27, 2016–2031. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, M.; Guo, Y. RING/U-box protein AtUSR1 functions in promoting leaf senescence through JA signaling pathway in Arabidopsis. Front. Plant Sci. 2020, 11, 608589. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Lü, S.; Zhao, H.; Des Marais, D.L.; Parsons, E.P.; Wen, X.; Xu, X.; Bangarusamy, D.K.; Wang, G.; Rowland, O.; Juenger, T.; et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yu, S.I.; Jung, S.H.; Lee, B.H.; Suh, M.C. The F-box protein SAGL1 and ECERIFERUM3 regulate cuticular wax biosynthesis in response to changes in humidity in Arabidopsis. Plant Cell 2019, 31, 2223–2240. [Google Scholar] [CrossRef]
- Khan, T.A.; Fariduddin, Q.; Yusuf, M. Low-temperature stress: Is phytohormones application a remedy? Environ. Sci. Pollut. Res. 2017, 24, 21574–21590. [Google Scholar] [CrossRef]
- Min, H.J.; Jung, Y.J.; Kang, B.G.; Kim, W.T. CaPUB1; a hot pepper U-box E3 ubiquitin ligase, confers enhanced cold stress tolerance and decreased drought stress tolerance in transgenic rice (Oryza sativa L.). Mol. Cells 2016, 39, 250–257. [Google Scholar]
- Yan, J.; Wang, J.; Li, Q.; Hwang, J.R.; Patterson, C.; Zhang, H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 2003, 132, 861–869. [Google Scholar] [CrossRef]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef]
- Huang, Y.; Minaker, S.; Roth, C.; Huang, S.; Hieter, P.; Lipka, V.; Wiermer, M.; Li, X. An E4 ligase facilitates polyubiquitination of plant immune receptor resistance proteins in Arabidopsis. Plant Cell 2014, 26, 485–496. [Google Scholar] [CrossRef]
- Tong, M.; Kotur, T.; Liang, W.; Vogelmann, K.; Kleine, T.; Leister, D.; Brieske, C.; Yang, S.; Lüdke, D.; Wiermer, M.; et al. E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 2017, 215, 1516–1532. [Google Scholar] [CrossRef]
- Monaghan, J.; Xu, F.; Gao, M.; Zhao, Q.; Palma, K.; Long, C.; Chen, S.; Zhang, Y.; Li, X. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 2009, 5, e1000526. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Gene Expression Omnibus. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68670 (accessed on 20 May 2022).
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009–D1014. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Nakai, K.; Horton, P. Computational prediction of subcellular localization. Methods Mol. Biol. 2007, 390, 429–466. [Google Scholar]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Guo, J.; Sun, W.; Wang, Y. Whole genome duplication of intra- and inter-chromosomes in the tomato genome. J. Genet. Genom. 2012, 39, 361–368. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Yu, J.; Tehrim, S.; Zhang, F.; Tong, C.; Huang, J.; Cheng, X.; Dong, C.; Zhou, Y.; Qin, R.; Hua, W.; et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014, 15, 3. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Brulle, F.; Bernard, F.; Vandenbulcke, F.; Cuny, D.; Dumez, S. Identification of suitable qPCR reference genes in leaves of Brassica oleracea under abiotic stresses. Ecotoxicology 2014, 23, 459–471. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Zhu, L.; Li, Z.; Cheng, M.; Chen, X.; Wang, A.; Wang, C.; Zhang, X. Genome-Wide Identification of the U-Box E3 Ubiquitin Ligase Gene Family in Cabbage (Brassica oleracea var. capitata) and Its Expression Analysis in Response to Cold Stress and Pathogen Infection. Plants 2023, 12, 1437. https://doi.org/10.3390/plants12071437
Wang P, Zhu L, Li Z, Cheng M, Chen X, Wang A, Wang C, Zhang X. Genome-Wide Identification of the U-Box E3 Ubiquitin Ligase Gene Family in Cabbage (Brassica oleracea var. capitata) and Its Expression Analysis in Response to Cold Stress and Pathogen Infection. Plants. 2023; 12(7):1437. https://doi.org/10.3390/plants12071437
Chicago/Turabian StyleWang, Peiwen, Lin Zhu, Ziheng Li, Mozhen Cheng, Xiuling Chen, Aoxue Wang, Chao Wang, and Xiaoxuan Zhang. 2023. "Genome-Wide Identification of the U-Box E3 Ubiquitin Ligase Gene Family in Cabbage (Brassica oleracea var. capitata) and Its Expression Analysis in Response to Cold Stress and Pathogen Infection" Plants 12, no. 7: 1437. https://doi.org/10.3390/plants12071437
APA StyleWang, P., Zhu, L., Li, Z., Cheng, M., Chen, X., Wang, A., Wang, C., & Zhang, X. (2023). Genome-Wide Identification of the U-Box E3 Ubiquitin Ligase Gene Family in Cabbage (Brassica oleracea var. capitata) and Its Expression Analysis in Response to Cold Stress and Pathogen Infection. Plants, 12(7), 1437. https://doi.org/10.3390/plants12071437





