Studies on the Composition and Diversity of Seagrass Ruppia sinensis Rhizosphere Mmicroorganisms in the Yellow River Delta
Abstract
1. Introduction
2. Results
2.1. Characteristics of Environmental Factors in Seagrass Beds
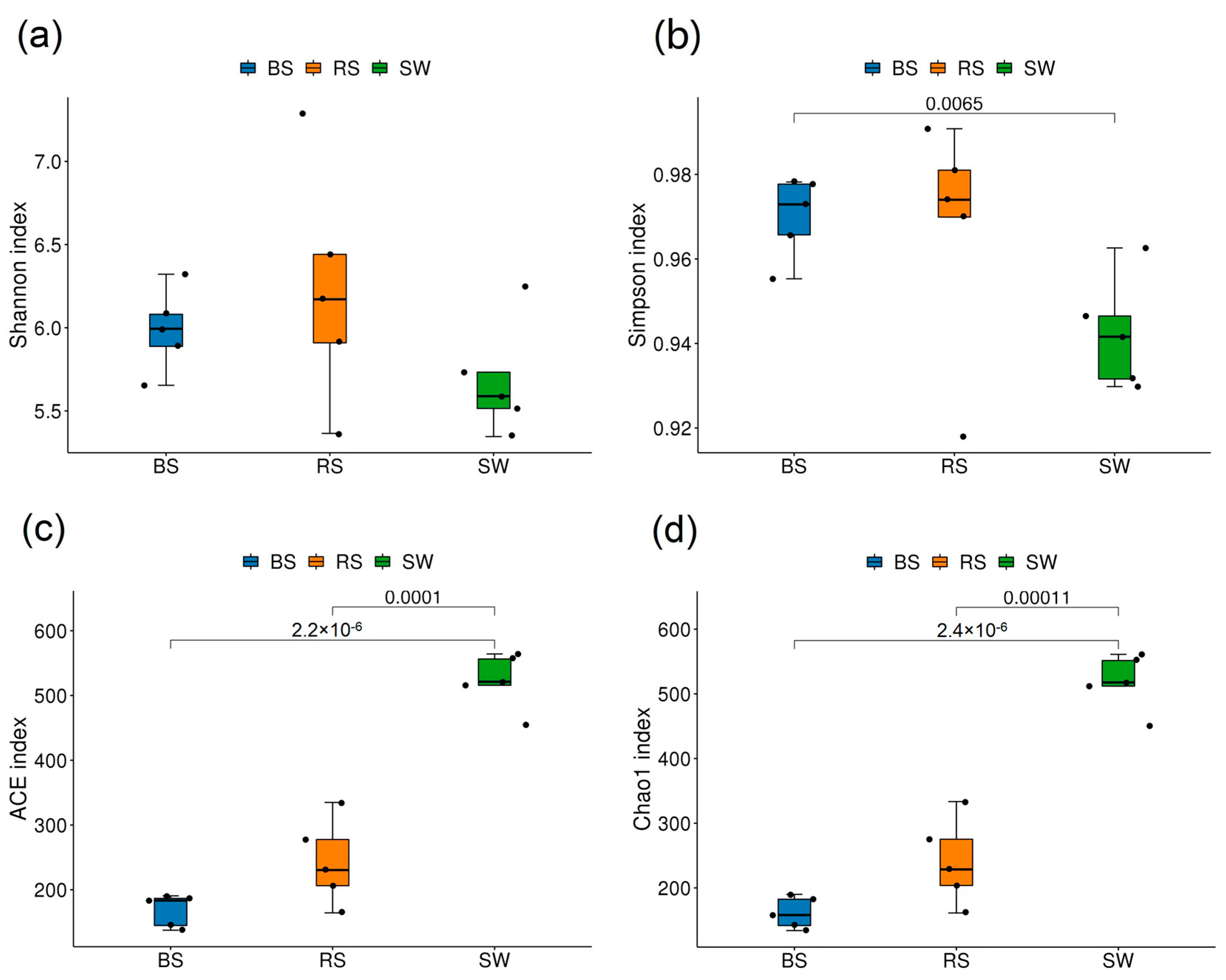
2.2. Alpha Diversity
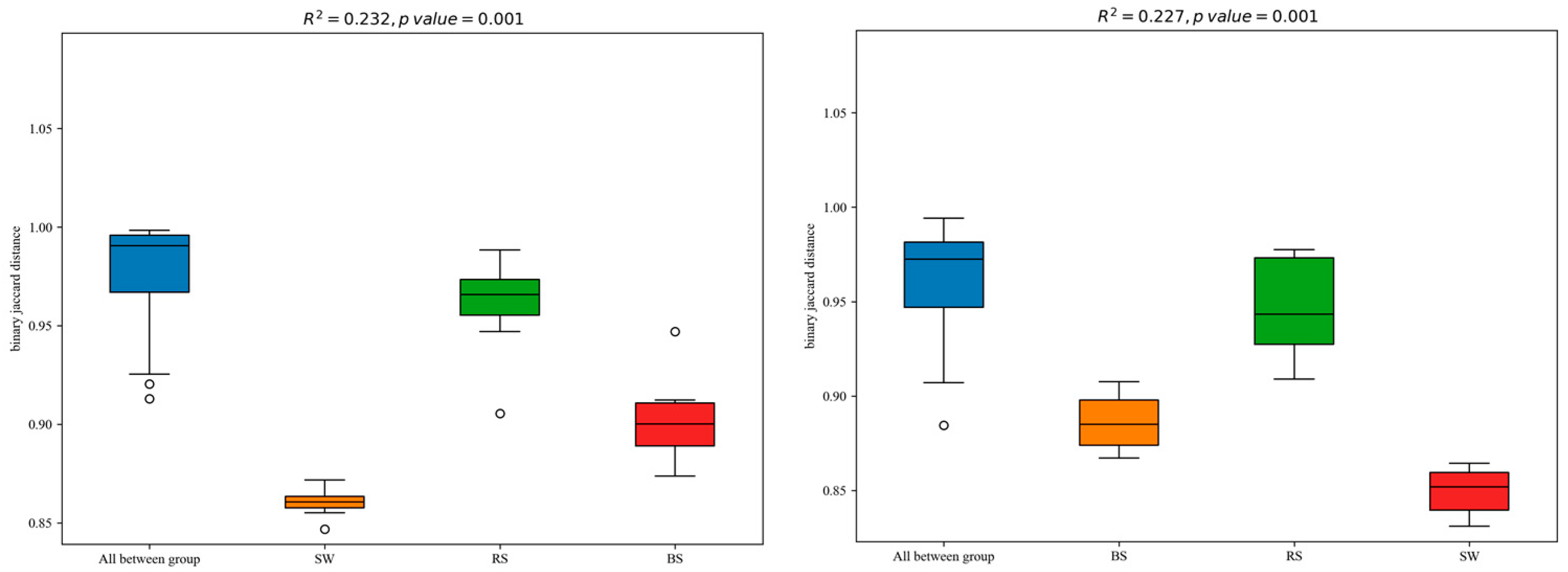
2.3. Beta Diversity
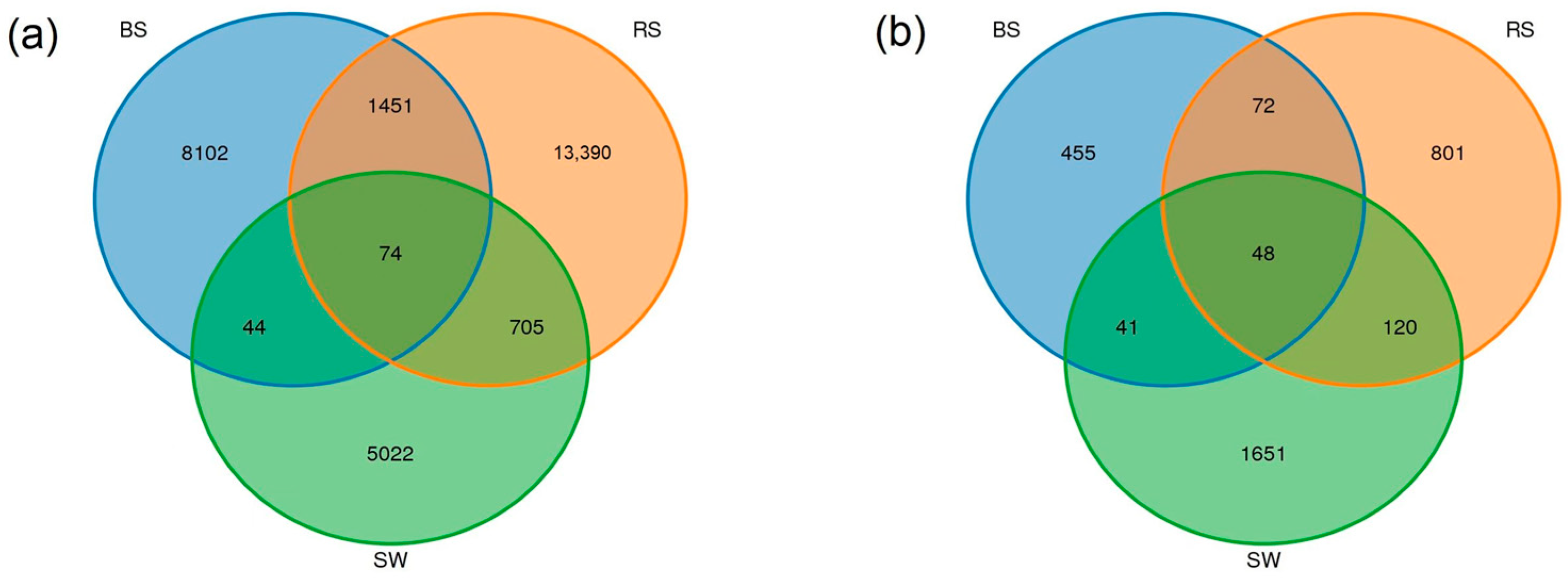
2.4. Sequence Variants and Abundant Genera
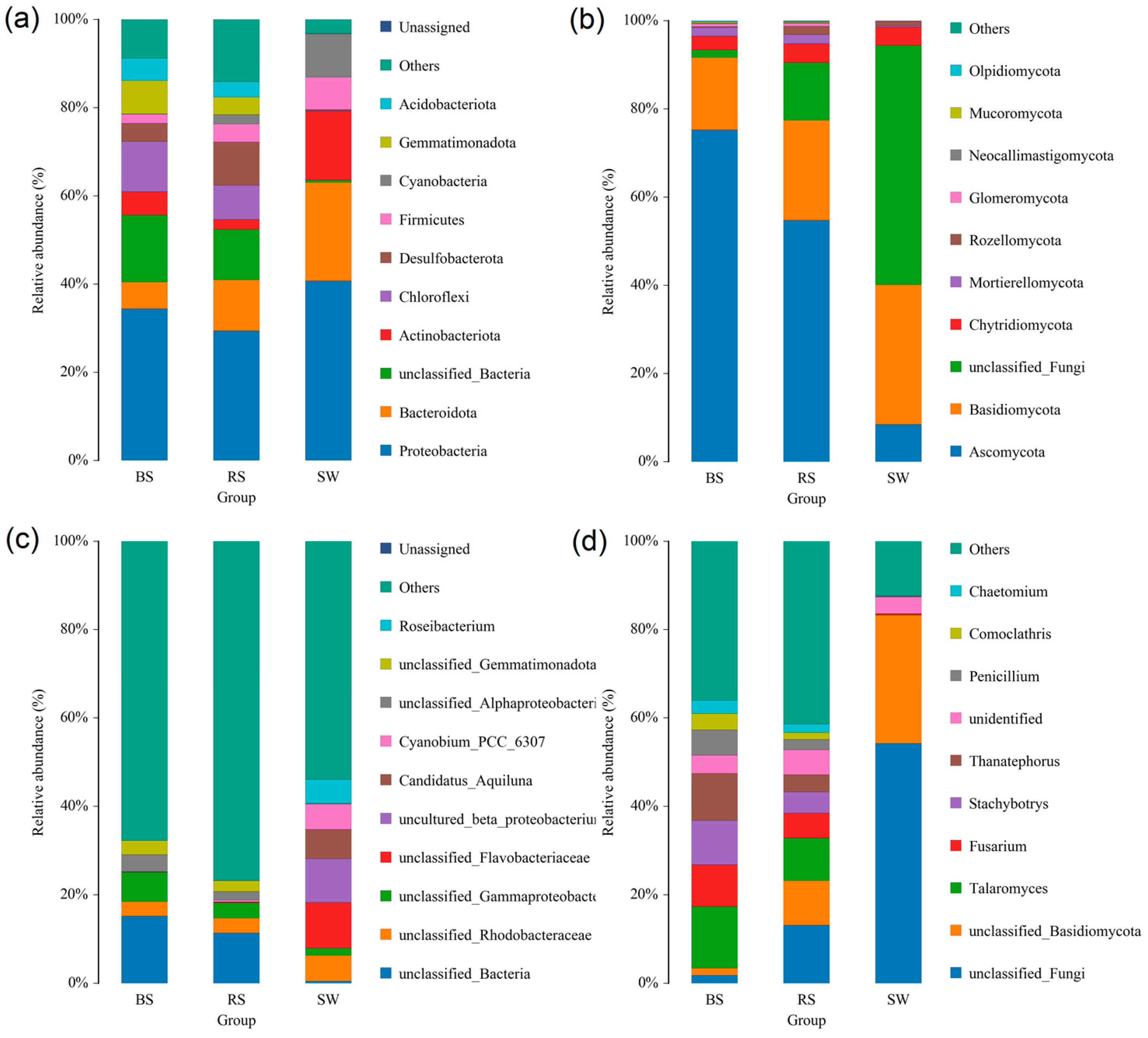
2.5. Composition of the Rhizosphere Microbial Community
3. Discussion
4. Materials and Methods
4.1. Study Area
4.2. Sediment and Seawater Physicochemical Properties Analysis
4.3. Molecular Methods
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Les, D.H.; Cleland, M.A.; Waycott, M. Phylogenetic studies in alismatidae, II: Evolution of marine angiosperms (seagrasses) and hydrophily. Syst. Bot. 1997, 22, 443–463. [Google Scholar] [CrossRef]
- Orth, R.J.; Carruthers, T.J.B.; Dennison, W.C.; Duarte, C.M.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A global crisis for seagrass ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef]
- Fourqurean, J.W.; Duarte, C.M.; Kennedy, H.; Marba, N.; Holmer, M.; Mateo, M.A.; Apostolaki, E.T.; Kendrick, G.A.; Krause-Jensen, D.; McGlathery, K.J.; et al. Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 2012, 5, 505–509. [Google Scholar] [CrossRef]
- Yue, S.D.; Zhou, Y.; Zhang, Y.; Xu, S.C.; Gu, R.T.; Xu, S.; Zhang, X.M.; Zhao, P. Effects of salinity and temperature on seed germination and seedling establishment in the endangered seagrass Zostera japonica Asch. & Graebn. in northern China. Mar. Pollut. Bull. 2019, 146, 848–856. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant-microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef]
- Zhang, P.; Nie, M.; Li, B.; Wu, J.H. The transfer and allocation of newly fixed C by invasive Spartina alterniflora and native Phragmites australis to soil microbiota. Soil Biol. Biochem. 2017, 113, 231–239. [Google Scholar] [CrossRef]
- Qin, Y.; Druzhinina, I.S.; Pan, X.Y.; Yuan, Z.L. Microbially Mediated Plant Salt Tolerance and Microbiome-Based Solutions for Saline Agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Christiaen, B.; McDonald, A.; Cebrian, J.; Ortmann, A.C. Response of the microbial community to environmental change during seagrass transplantation. Aquat. Bot. 2013, 109, 31–38. [Google Scholar] [CrossRef]
- Markovski, M.; Najdek, M.; Herndl, G.J.; Korlevic, M. Compositional stability of sediment microbial communities during a seagrass meadow decline. Front. Mar. Sci. 2022, 9, 966070. [Google Scholar] [CrossRef]
- Cole, L.W.; McGlathery, K.J. Nitrogen fixation in restored eelgrass meadows. Mar. Ecol. Prog. Ser. 2012, 448, 235–246. [Google Scholar] [CrossRef]
- Martin, B.C.; Bougoure, J.; Ryan, M.H.; Bennett, W.W.; Colmer, T.D.; Joyce, N.K.; Olsen, Y.S.; Kendrick, G.A. Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME J. 2019, 13, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Ugarelli, K.; Chakrabarti, S.; Laas, P.; Stingl, U. The Seagrass Holobiont and Its Microbiome. Microorganisms 2017, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.T.; Zhou, Y.; Song, X.Y.; Xu, S.C.; Zhang, X.M.; Lin, H.Y.; Xu, S.; Zhu, S.Y. Effects of temperature and salinity on Ruppia sinensis seed germination, seedling establishment, and seedling growth. Mar. Pollut. Bull. 2018, 134, 177–185. [Google Scholar] [CrossRef]
- Bai, J.H.; Xiao, R.; Zhang, K.J.; Gao, H.F. Arsenic and heavy metal pollution in wetland soils from tidal freshwater and salt marshes before and after the flow-sediment regulation regime in the Yellow River Delta, China. J. Hydrol. 2012, 450, 244–253. [Google Scholar] [CrossRef]
- Song, Z.L.; Sun, Y.Y.; Liu, P.Y.; Wang, Y.B.; Huang, Y.Y.; Gao, Y.; Hu, X.K. Invasion of Spartina alterniflora on Zostera japonica enhances the abundances of bacteria by absolute quantification sequencing analysis. Ecol. Evol. 2022, 12, e8939. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.P.; Li, P.P.; Wang, G.B.; Wu, C.F.; Ge, A.H.; et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef]
- Wang, S.P.; Yan, Z.G.; Wang, P.Y.; Zheng, X.; Fan, J.T. Comparative metagenomics reveals the microbial diversity and metabolic potentials in the sediments and surrounding seawaters of Qinhuangdao mariculture area. PLoS ONE 2020, 15, e0234128. [Google Scholar] [CrossRef]
- Ugarelli, K.; Laas, P.; Stingl, U. The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites. Microorganisms 2019, 7, 4. [Google Scholar] [CrossRef]
- Sun, Y.Y.; Song, Z.L.; Zhang, H.K.; Liu, P.Y.; Hu, X.K. Seagrass vegetation affect the vertical organization of microbial communities in sediment. Mar. Environ. Res. 2020, 162, 105174. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, C.Y.; Yu, S.; Jiang, Z.J.; Liu, S.L.; Wu, Y.C.; Huang, X.P. Rhizosphere Microbial Community Structure Is Selected by Habitat but Not Plant Species in Two Tropical Seagrass Beds. Front. Microbiol. 2020, 11, 161. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Voerman, S.E.; Lang, J.M.; Stachowicz, J.J.; Eisen, J.A. Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 2017, 5, e3246. [Google Scholar] [CrossRef] [PubMed]
- Behera, P.; Mahapatra, S.; Mohapatra, M.; Kim, J.Y.; Adhya, T.K.; Raina, V.; Suar, M.; Pattnaik, A.K.; Rastogi, G. Salinity and macrophyte drive the biogeography of the sedimentary bacterial communities in a brackish water tropical coastal lagoon. Sci. Total Environ. 2017, 595, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, K.C.; Karaoz, U.; Hanson, C.A.; Santee, C.A.; Bradford, M.A.; Treseder, K.K.; Wallenstein, M.D.; Brodie, E.L. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2011, 2, 94. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.F.; Zhang, X.L.; Zhang, Q.Q.; Liu, F.H.; Zhang, J.P.; Gong, J. Seagrass (Zostera marina) Colonization Promotes the Accumulation of Diazotrophic Bacteria and Alters the Relative Abundances of Specific Bacterial Lineages Involved in Benthic Carbon and Sulfur Cycling. Appl. Environ. Microbiol. 2015, 81, 6901–6914. [Google Scholar] [CrossRef]
- Holmer, M.; Nielsen, S.L. Sediment sulfur dynamics related to biomass-density patterns in Zostera marina (eelgrass) beds. Mar. Ecol. Prog. Ser. 1997, 146, 163–171. [Google Scholar] [CrossRef]
- Holmer, M.; Andersen, F.O.; Nielsen, S.L.; Boschker, H.T.S. The importance of mineralization based on sulfate reduction for nutrient regeneration in tropical seagrass sediments. Aquat. Bot. 2001, 71, 1–17. [Google Scholar] [CrossRef]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xu, J.; Riera, N.; Jin, T.; Li, J.Y.; Wang, N. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 2017, 5, 97. [Google Scholar] [CrossRef]
- Fahimipour, A.K.; Kardish, M.R.; Lang, J.M.; Green, J.L.; Eisen, J.A.; Stachowicz, J.J. Global-Scale Structure of the Eelgrass Microbiome. Appl. Environ. Microbiol. 2017, 83, e03391-16. [Google Scholar] [CrossRef]
- Liu, F.D.; Mo, X.; Kong, W.J.; Song, Y. Soil bacterial diversity, structure, and function of Suaeda salsa in rhizosphere and non-rhizosphere soils in various habitats in the Yellow River Delta, China. Sci. Total Environ. 2020, 740, 140144. [Google Scholar] [CrossRef]
- Shang, S.; Li, L.Y.; Zhang, Z.W.; Zang, Y.; Chen, J.; Wang, J.; Wu, T.; Xia, J.B.; Tang, X.X. The Effects of Secondary Growth of Spartina alterniflora after Treatment on Sediment Microorganisms in the Yellow River Delta. Microorganisms 2022, 10, 1722. [Google Scholar] [CrossRef] [PubMed]
- Logue, J.B.; Stedmon, C.A.; Kellerman, A.M.; Nielsen, N.J.; Andersson, A.F.; Laudon, H.; Lindstrom, E.S.; Kritzberg, E.S. Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 2016, 10, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Hu, S.X.; Liu, X.X.; Zang, Y.; Chen, J.; Gao, N.; Li, L.Y.; Wang, J.; Liu, L.X.; Xu, J.K.; et al. Effects of Spartina alterniflora invasion on the community structure and diversity of wetland soil bacteria in the Yellow River Delta. Ecol. Evol. 2022, 12, e8905. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Erratum in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]


| Groups | TOC (g/kg) | TS (g/kg) | EC | pH |
|---|---|---|---|---|
| BS | 3.208 ± 0.297 a | 0.44 ± 0.19 a | 66.22 ± 6.40 a | 7.81 ± 0.48 b |
| RS | 3.096 ± 0.735 a | 1.25 ± 0.40 a | 37.66 ± 7.03 b | 7.74 ± 0.16 b |
| SW | - | - | 13.46 ± 0.40 c | 8.60 ± 0.09 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, S.; Li, L.; Xiao, H.; Chen, J.; Zang, Y.; Wang, J.; Tang, X. Studies on the Composition and Diversity of Seagrass Ruppia sinensis Rhizosphere Mmicroorganisms in the Yellow River Delta. Plants 2023, 12, 1435. https://doi.org/10.3390/plants12071435
Shang S, Li L, Xiao H, Chen J, Zang Y, Wang J, Tang X. Studies on the Composition and Diversity of Seagrass Ruppia sinensis Rhizosphere Mmicroorganisms in the Yellow River Delta. Plants. 2023; 12(7):1435. https://doi.org/10.3390/plants12071435
Chicago/Turabian StyleShang, Shuai, Liangyu Li, Hui Xiao, Jun Chen, Yu Zang, Jun Wang, and Xuexi Tang. 2023. "Studies on the Composition and Diversity of Seagrass Ruppia sinensis Rhizosphere Mmicroorganisms in the Yellow River Delta" Plants 12, no. 7: 1435. https://doi.org/10.3390/plants12071435
APA StyleShang, S., Li, L., Xiao, H., Chen, J., Zang, Y., Wang, J., & Tang, X. (2023). Studies on the Composition and Diversity of Seagrass Ruppia sinensis Rhizosphere Mmicroorganisms in the Yellow River Delta. Plants, 12(7), 1435. https://doi.org/10.3390/plants12071435






