Abstract
The pivotal role of cysteine-rich receptor-like kinases (CRKs) in modulating growth, development, and responses to stress has been widely acknowledged in Arabidopsis. However, the function and regulation of CRK41 has remained unclear. In this study, we demonstrate that CRK41 is critical for modulating microtubule depolymerization in response to salt stress. The crk41 mutant exhibited increased tolerance, while overexpression of CRK41 led to hypersensitivity to salt. Further analysis revealed that CRK41 interacts directly with the MAP kinase3 (MPK3), but not with MPK6. Inactivation of either MPK3 or MPK6 could abrogate the salt tolerance of the crk41 mutant. Upon NaCl treatment, microtubule depolymerization was heightened in the crk41 mutant, yet alleviated in the crk41mpk3 and crk41mpk6 double mutants, indicating that CRK41 suppresses MAPK-mediated microtubule depolymerizations. Collectively, these results reveal that CRK41 plays a crucial role in regulating microtubule depolymerization triggered by salt stress through coordination with MPK3/MPK6 signalling pathways, which are key factors in maintaining microtubule stability and conferring salt stress resistance in plants.
1. Introduction
Plants have developed a wide array of receptor-like protein kinases (RLKs) in response to different environmental and internal stimuli. RLKs are a type of serine–threonine protein kinase that consists of three distinct parts: an extracellular domain, a transmembrane helix, and a conserved intracellular protein kinase catalytic domain [1]. RLKs can convert extracellular signals into intracellular signals and activate downstream signal pathways to regulate a number of defensive and developmental processes. Cysteine-rich receptor-like kinases (CRKs) are a large family of RLKs, containing 46 members, which are thought to be linked to the control of plant growth and development, as well as the reactions to different environmental stimuli [1,2].
In Arabidopsis thaliana, CRK family members are crucial in regulating the responses to biotic and abiotic stressors. When confronted with biotic stress and pathogen associated molecular pattern (PAMP) treatments, CRKs exhibit a rapid and drastic increase in the expression level. CRK4, CRK6, CRK13, CRK20, CRK28, CRK29, CRK36, and CRK45 are resistant to bacteria such as Pseudomonas syringae Pv. Tomato DC3000 [3,4,5], and CRK36 also contributes to responsive to necrotrophic fungal pathogen Alternaria brassicicola in Arabidopsis [6]. ALS1, a typical CRK in rice, is involved in Salicylic acid (SA), jasmonate, and NH1-mediated defense responses [7], while NH1-mediated immunological responses are mediated by CRK6 and CRK10 [8]. TaCRK1 in wheat mediates the defense response against Rhizoctonia cerealis through ABA signaling pathway [9], and HvCRK1 in barley can regulate the defense response against saprophytic fungal barley powdery mildew [10]. GbCRK18 affected the resistance of Gossypium barbadense to Verticillium dahlia [11].
Research has also demonstrated that CRKs play a role in controlling abiotic stress tolerance. Overexpression of CRK4, CRK5, CRK19, and CRK45 enhances ABA sensitivity of stomatal movement and drought tolerance in early seedlings [12]. CRK5 affects the release of reactive oxygen species (ROS) in response to ultraviolet light [13], and CRK45 acts as a positive regulator in seed germination and seedling growth in response to salt stress [14]. Additionally, CRK6, CRK7, CRK8, CRK10, and CRK15 show increased transcript levels after exposure to O3 and work together to enable a suitable response to oxidative stress induced by O3 (which induces extracellular ROS production) in Arabidopsis thaliana [1,15]. Salt stress also leads to an increase in ROS levels, resulting in oxidative stress. Excessive production of ROS causes serious oxidative damage, while appropriate production of ROS acts as a defense signal to improve stress tolerance [16]. Moreover, TaCRK41 was involved in response to salt and drought stress in wheat [17]. Study has revealed that CRK41 has a suppressive impact on salt tolerance in Arabidopsis [18]. Consequently, it is evident that CRKs are essential for the reaction to biotic and abiotic stresses, but their functional studies focus mainly on resistance to biotic stress, which is only rarely reported on abiotic stress resistance in Arabidopsis thaliana. However, there is limited understanding about the molecular regulatory processes of CRK41 in plants when exposed to salt stress.
Microtubules, which are highly dynamic polymers that can depolymerize and polymerize, are involved in various cellular processes [19]. The polymerization–depolymerization dynamics of microtubules are influenced by a range of developmental processes, as well as environmental stimuli and stress conditions [19,20,21]. Microtubules have been identified as playing a role in the amplification of signals, as well as being integral components in the processing of stress signals, which are closely related to salt stress [20]. Studies also have demonstrated that salt stress can alter the organization of cortical microtubules [22]. Moreover, microtubule stability and dynamic affect salt tolerance in Arabidopsis. Research has found that the application of paclitaxel, a stabilizer of microtubules, decreases the seedlings survival rate of Arabidopsis during salt stress, whereas propyzamide and oryzalin, which are known to disrupt microtubules, increase the survival rate [23]. Thus, rapid microtubule depolymerization is advantageous in helping plants to tolerate salt stress [20,21,22,23].
Furthermore, MAPK signalling pathways, initially thought to be responsible for the regulation of microtubule organization and dynamic instability, have been found to be involved in the growth, development and stress response of plants [24,25,26,27]. MAPKs can be activated when Arabidopsis is exposed to biotic and abiotic stresses [28]. MAPKs phosphorylate microtubule-associated proteins (MAPs), which can assist the coordinated action of the assembly and disassembly dynamics of microtubule and modulate the affinity of proteins to microtubule surface [29,30,31]. Studies have revealed a connection among three Arabidopsis MAPKs, MPK4, MPK6, and MPK13, and microtubule organization and dynamics [27]. These MAPKs have been observed to interact with MAPs, with MPK4 and MPK13 interacting with MAP65-1, MAP65-2, and MAP65-3, and MPK6 interacting with MAP65-1 [27]. Additionally, MAP65-1 has been determined to be a protein that can be effectively phosphorylated by MPK4 and MPK6 [25,32]. MPK3 can be colocalized with MAP65-1, as well as microtubules [23,33]. Investigation has shown that phosphorylation of MAP65-1 reduces its capability to form microtubule bundles in vitro, which could consequently initiate the depolymerization of microtubules [23,34].
In general, salt stress signals can be transduced into cells by plasma membrane proteins, which result in dynamic modifications of microtubules. To date, little is known about the function of plasma membrane protein CRK41 during salt stress; it was further explored in this study by using wild type, crk41 mutant, and overexpressed lines of CRK41. Moreover, the mechanism of CRK41’s regulated responses to salt stress was also elucidated.
2. Results
2.1. The Basic Characteristics of CRK41
We firstly analyzed the functional domain composition of CRK41 to determine the fundamental characteristics of CRK41. The result revealed that CRK41 contained the DUF26 domain, protein kinase domain, and transmembrane domain, which was made up of 665 amino acids (aa) (Figure 1A). To assess its cellular localization, we transformed wild type with a 35S:CRK41-GFP vector to create 35S:CRK41-GFP lines. Results indicated that CRK41 was found to be situated on the plasma membrane in the root cells of Arabidopsis (Figure 1B). Therefore, we speculate that the function of CRK41 is primarily to sense the changes in environmental signals.
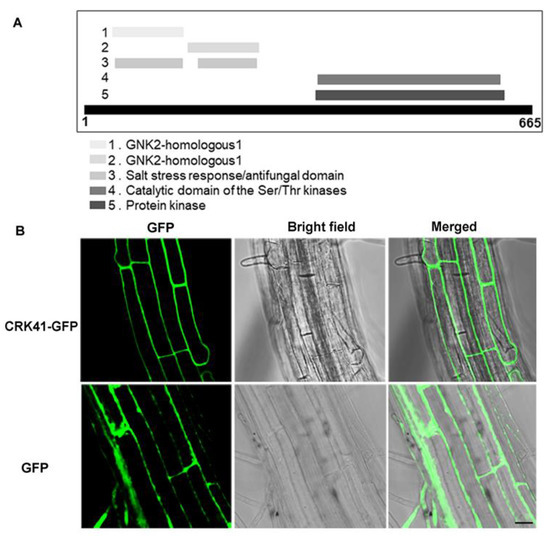
Figure 1.
The basic characteristics of CRK41. (A) The functional domain composition analysis of CRK41 was from the website: https://www.ebi.ac.uk/interpro/protein/UniProt/O23081/ (accessed on 8 November 2022). (B) The localization of CRK41 in roots cells (bar = 20 μm). Three replicates of similar results are obtained in the experiments.
2.2. crk41 Mutant Increased Salt Tolerance
An examination of the role of CRK41 in salt stress was carried out through the measurement of its gene expression level in Col-0 following NaCl treatment. Real-time PCR analysis showed an increase expression level of CRK41 upon exposure to different times and concentrations of NaCl (Figure 2A,B). The transcript level of CRK41 was also detected after different concentrations of mannitol. Osmotic stress caused by mannitol had no effect on the expression level of CRK41 (Figure S1). This indicates that CRK41 can be induced by salt stress, and the response of CRK41 is specific to NaCl.
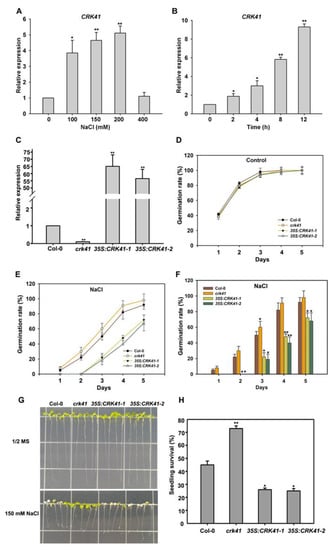
Figure 2.
CRK41 has an inhibitory effect on salt stress tolerance. (A,B) The relative expression level of CRK41 was determined after NaCl treatment for different concentrations (0, 100, 150, and 200 mM) (A) and times (0, 2, 4, 8, and 12 h) (B). The standard deviation (SD) is represented by the error bars; n = 3. * p < 0.05; ** p < 0.01 vs. Control. (C) The relative expression level of CRK41 in Col-0, crk41 mutant, 35S:CRK41-1, and 35S:CRK41-2 under normal condition. SD is represented by the error bars; n = 3. ** p < 0.01 vs. wild type. (D–F) The seed germination rate of four lines, including crk41 mutant, 35S:CRK41-1, and 35S:CRK41-2, was assessed on 1/2 MS medium without (D) or with 120 mM NaCl (E,F). Observations were made daily post-transfer to assess germination rate. SD is represented by the error bars; n = 3 (80 seeds were measured in per genotype of each replicate). * p < 0.05; ** p < 0.01 vs. wild type. (G,H) An analysis of salt sensitivity (G) and seedling survival rate (H) were conducted on wild type, crk41 mutant, 35S: CRK41-1, and 35S: CRK41-2 after 150 mM NaCl treatment. SD is represented by the error bars; n = 3 (at least 60 seeds were measured in per genotype of each replicate). * p < 0.05; ** p < 0.01 vs. wild type.
Furthermore, to investigate the influence of CRK41 in salt stress tolerance, two CRK41 overexpression lines, 35S: CRK41-1 and 35S: CRK41-2, were produced by introducing the 35S: CRK41 vector into Col-0. Then, the gene expression level of CRK41 in the crk41 mutants, 35S: CRK41-1 and 35S: CRK41-2, was determined. Quantitative PCR results showed a notable increase in the expression level of CRK41 in 35S: CRK41-1 and 35S: CRK41-2, whereas CRK41 was barely expressed in the crk41 mutant. The results indicated that the crk41 mutant and CRK41 overexpression lines 35S: CRK41-1 and 35S: CRK41-2 could be used for phenotypic analysis. Then, the seed germination of Col-0, crk41, 35S: CRK41-1, and 35S: CRK41-2 was observed after treatment with NaCl. The seeds of the indicated genotypes were sown on 1/2 MS plates with 120 mM NaCl, the seeds germination was observed in 5 days, and the seed germination rate was counted. Without NaCl, the genotype of the seed had no impact on the rate of germination (Figure 2D). The germination rate of crk41 was greater than that of Col-0 when the medium contained 120 mM NaCl, whereas the germination rate of 35S: CRK41-1 and 35S: CRK41-2 was decreased compared with Col-0 (Figure 2E,F). This indicates that the crk41 mutant had greater salt resistance.
In order to provide further evidence that CRK41 was implicated in salt stress, a phenotypic assessment of Col-0, crk41, 35S: CRK41-1, and 35S: CRK41-2 after NaCl treatment was conducted. When grown without NaCl, the lengths of roots and leaves of Col-0, crk41 mutant, 35S: CRK41-1, and 35S: CRK41-2 showed no significant difference. However, after transferring 6-day-old seedlings to a plate with 150 mM NaCl, the leaves of 35S: CRK41-1 and 35S: CRK41-2 were drastically discolored and bleached compared with Col-0 after 3 d NaCl treatment. Interestingly, the crk41 mutant showed lesser bleaching than the wild type (Figure 2G). We further evaluated the survival of seedlings by examining leaf bleaching during salt stress, a sign of seedling death. If two cotyledons both had turned white, the seedling was determined to be dead, while all other seedlings were classified as living. The survival rate was calculated on the basis of the ratio of living seedlings to the total number of seedlings. The survival rate of wild type seedlings was substantially lower than that of crk41 mutant and higher than that of 35S: CRK41-1 and 35S: CRK41-2 after NaCl treatment (Figure 2H).
Trypan blue staining was used to assess the effect of CRK41 on leaf cell-death-related phenotypes. In comparison with Col-0, the crk41 mutant exhibited fewer cells stained by trypan blue after salt treatment, indicating reduced cell death (Figure S2). Additionally, electrolyte leakage was measured in Col-0, crk41 mutant, and CRK41 overexpression lines after treatment with medium containing 150 mM NaCl. The results revealed that the plasma membrane of the CRK41 overexpression lines had more severe damage than that of Col-0, while the crk41 mutant had only mild damage (Figure S3).
To confirm that the salt-tolerant phenotype in crk41 mutant was a result of the disruption of the CRK41 gene, the crk41 complemented lines CRK41/crk41-5 and CRK41/crk41-6 were utilized, which restored the salt-sensitive phenotype and were similar to that of the wild type (Figure S4).
Our findings suggested that CRK41 was a vital factor for the resistance to salt stress. Results from seed germination, phenotypic observation, leaf cell death, plasma membrane damage, and gene expression study indicated that CRK41 has an inhibitory effect on salt stress tolerance.
2.3. CRK41 Has Been Identified as a Factor in the Alteration of Microtubule Depolymerization When Exposed to Salt Stress
Research has revealed that microtubule depolymerization is a significant factor in the signaling pathways of both biotic and abiotic stress [21,22,27,34,35]. Moreover, microtubule dynamic instability is required for salt stress tolerance [35,36,37]. A rapid depolymerization of microtubules is beneficial to plant survival under salt stress [21,36]. Therefore, we determined whether CRK41 was a crucial factor for controlling the depolymerization of microtubules during salt stress. The relationship between CRK41 and microtubule disassembly during salt stress was investigated using Col-0, crk41 mutant, 35S: CRK41-1, and 35S: CRK41-2. The microtubules were visualized directly in Col-0, crk41 mutant, 35S: CRK41-1, and 35S: CRK41-2 seedlings after 150 mM NaCl treatment at various times (0, 15, 30, 45, and 60 min). No substantial difference was observed in terms of microtubule organization and density prior to salt application. After treatment with 150 mM NaCl, the cortical microtubules were depolymerized, and the level of depolymerization varied among Col-0, crk41 mutant, and CRK41 overexpression lines. Wild type cotyledon pavement cells displayed a dramatic depolymerization in cortical microtubule with extended treatment duration; in contrast, the 35S:CRK41 overexpression line showed a weaker depolymerization in cortical microtubule (Figure 3). Moreover, the crk41 mutant demonstrated a more extreme and rapid depolymerization of cortical microtubules compared with wild type (Figure 3). The results suggest that CRK41 appears to be a key factor for controlling microtubule depolymerization in response to salt stress.
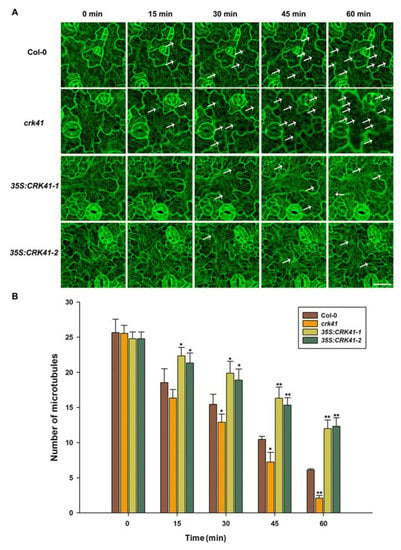
Figure 3.
CRK41 has been identified as a regulator of cortical microtubule depolymerization when exposed to 150 mM NaCl. (A) Analysis of the microtubule modifications caused by 150 mM NaCl of Col-0, crk41 mutant, 35S:CRK41-1, and 35S:CRK41-2 was conducted through a series of sequential images (scale bar = 20 μm). The arrows indicated representative microtubule depolymerization. (B) Image tool software was utilized to quantify cortical microtubules of Col-0, crk41 mutant, 35S:CRK41-1, and 35S:CRK41-2, with a minimum of 18 cells from 3 samples being analyzed. SD is represented by the error bars. * p < 0.05; ** p < 0.01 vs. wild type in the same condition.
2.4. CRK41 Interacts with MPK3
MAPKs were initially perceived as a mechanism to control the arrangement and activity of microtubules; however, over time, it was determined that MAPKs were responsible for mediating defence reactions to biotic and abiotic stresses in plants [28]. Studies in Arabidopsis have revealed that MAPKs MPK3/MPK6 and MPK4 phosphorylate HSFA4A and SOS1, two heat shock factors, are found to enhance salt tolerance [38]. Furthermore, evidence suggested that MAPK cascades played a critical role in the downstream pathways of RLKs [39]. Previous research has also demonstrated that the YODA-MKK4/5-MPK3/MPK6 signaling pathway is regulated by ERECTA (ER), a RLK that controls localized cell proliferation in plants [32,40]. Consequently, we aimed to determine whether CRK41 physically interacts with MAPKs signaling in plants.
In order to determine the interaction between CRK41 and MPK3/MPK6, a yeast two-hybrid assay was carried out. It was observed that CRK41 interacted with MPK3 in yeast (Figure 4A). A Luciferase complementation imaging (LCI) assay was then conducted on Nicotiana tabacum leaves to investigate the interaction between CRK41 and MPK3/MPK6 in plants. This revealed that CRK41 interacted with MPK3 in plants (Figure 4B). Nevertheless, neither the yeast two-hybrid assay nor the LCI assay detected any interaction between CRK41 and MPK6 (Figure 4).
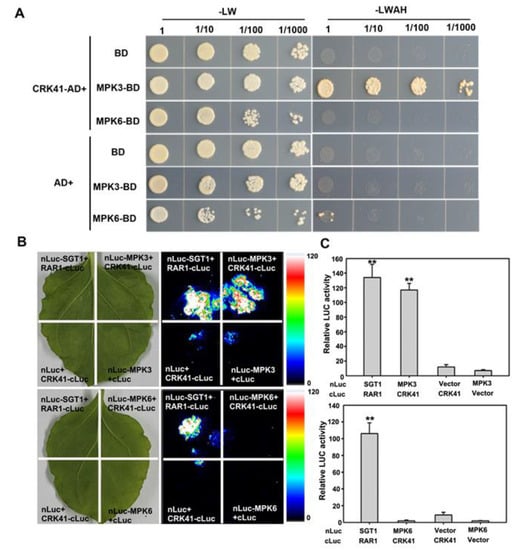
Figure 4.
CRK41 interacts with MPK3. (A,B) Evidence of the interaction among CRK41 and MPK3 and MPK6 was obtained via a yeast two-hybrid assay (A) and a LCI assay (B). SGT1-nLUC and RAR1-cLUC served as a positive control, while MPK3-nLUC + GUS-cLUC, MPK6-nLUC + GUS-cLUC, CRK41-cLUC + GUS-nLUC, and CRK41-cLUC + GUS-nLUC served as negative controls in (B). (C) The relative luminescence of the sample in (B) was measured. SD is represented by the error bars, n = 3. ** p < 0.01 vs. MPK3-nLUC + GUS-cLUC/MPK6-nLUC + GUS-cLUC group.
2.5. CRK41 Is Responsible for the Regulation of MPK3 and MKP6, Both of Which Are Integral in Salt Stress Tolerance
To further explore whether CRK41 impacted MPK3 and MPK6 when exposed to salt stress, real-time PCR was used to measure MPK3 and MPK6 expression level in Col-0, crk41 mutant, 35S: CRK41-1, and 35S: CRK41-2 seedlings. Following NaCl treatment, the transcript levels of MPK3 and MPK6 were significantly elevated in Col-0 and crk41 mutant, with the expression level of MPK3 and MPK6 in crk41 mutant exhibiting a greater increase than that of Col-0. Conversely, no increase in the transcript levels of MPK3 and MPK6 was observed in 35S: CRK41-1 or 35S: CRK41-2 (Figure 5A,B).
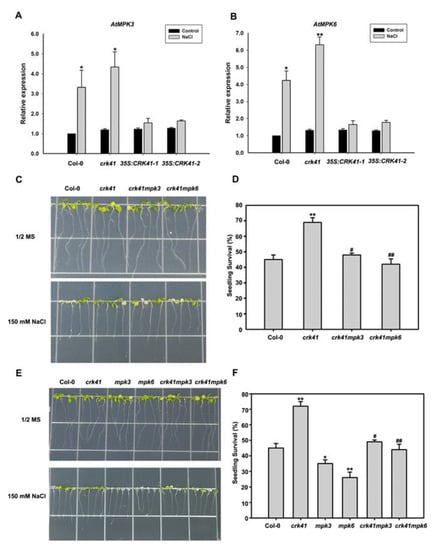
Figure 5.
MPK3 and MKP6 are required in crk41 mutant during salt stress. (A,B) The relative expression levels of MPK3 and MPK6 were determined using real-time PCR analysis after ddH2O or 150 mM NaCl treatment for 12 h in wild type. The seedlings treated with ddH2O were used as control. SD is represented by the error bars; n = 3. * p < 0.05; ** p < 0.01 vs. Control in the same genotype. (C,E) The growth phenotypes of the seedlings in Col-0, crk41, crk41mpk3, crk41mpk6, mpk3, and mpk6 mutants after 150 mM NaCl treatments. (D,F) Seedling survival rates for Col-0, crk41, crk41mpk3, crk41mpk6, mpk3, and mpk6 mutants. The survival rate of seedlings was then measured after NaCl treatment for 5 d. SD is represented by the error bars; n = 3 (at least 60 seeds were measured in per genotype of each replicate). * p < 0.05; ** p < 0.01 vs. wild type; # p < 0.05; ## p < 0.01 vs. crk41 mutant.
In order to investigate whether the altered gene expression of MPK3 and MPK6 was the cause for the salt tolerance of crk41, crk41mpk3 and crk41mpk6 were generated by crossing crk41 with mpk3 and mpk6. Six-day-old seedlings of wild type, mpk3, mpk6, crk41mpk3, and crk41mpk6 mutants were moved to plates containing either no NaCl or 150 mM NaCl. After 3 days of the NaCl treatment, phenotypic analyses revealed that the mpk3 and mpk6 mutants had much more severe chlorosis and bleaching of their leaves than the wild type, while the leaves in crk41 mutant showed less bleaching than the wild type (Figure 5E). However, the crk41mpk3 and crk41mpk6 mutants did not demonstrate any remarkable variations in comparison with the wild type (Figure 5C,E). Moreover, the survival rate mpk3 and mpk6 was notably reduced when compared with wild type, whereas the survival rate of crk41mpk3 and crk41mpk6 mutants was comparable to that of wild type (Figure 5D,F).
Results showed that crk41 mutant had a greater resistance to salt stress, while deletion of MPK3 and MPK6 genes in the crk41 mutant restored its salt tolerance phenotype (Figure 5C–F). These results demonstrate that MPK3 and MPK6 are required to regulate salt stress tolerance caused by the mutation of CRK41.
2.6. MPK3 and MKP6 Are Essential Components in CRK41-Modulating Salt-Stress-Induced Microtubule Depolymerization
To confirm the molecular mechanism of MPK3 and MPK6 in reaction to NaCl, we determined whether MPK3 and MPK6 affected microtubule depolymerization. The effects of NaCl treatment on microtubule depolymerization were observed in Col-0, mpk3, and mpk6 mutant seedlings. Prior to the application of salt, no observable disparities were seen in the microtubule arrangement and concentrations of the mpk3 and mpk6 mutants in comparison with those of the wild type. The depolymerization of cortical microtubules was less pronounced in the mpk6 mutant than in Col-0, while the mpk3 mutant had a slightly reduced depolymerization compared with the wild type (Figure S5). Thus, it appears that MPK3 and MPK6 are both implicated in the control of microtubules depolymerization when exposed to salt stress, with MPK6 playing a particularly significant role.
In addition, to further determine whether MPK3 and MPK6 modulated microtubule depolymerization in crk41 mutant in response to salt stress, the microtubule dynamics of seedlings in wild type, mpk3, mpk6, crk41mpk3, and crk41mpk6 mutants were investigated after NaCl treatment. With NaCl treatment, microtubule depolymerization occurred, and microtubule density decreased in wild type, crk41mpk3, and crk41mpk6 mutants. The crk41mpk3 showed a similar level of microtubule depolymerization to wild type; however, the crk41mpk6 mutant exhibited a weakened depolymerization (Figure 6).
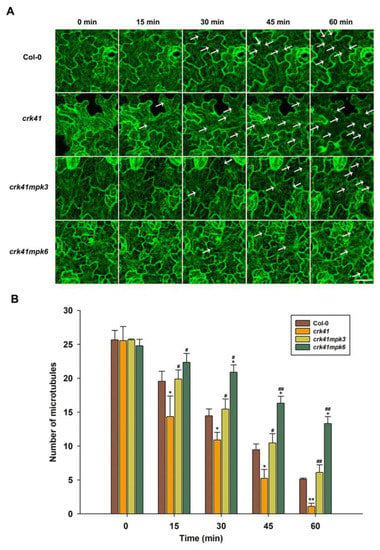
Figure 6.
MPK3 and MPK6 are essential components in CRK41-modulating microtubule depolymerization induced by NaCl. (A) Analysis of the microtubule modifications in Col-0, crk41, crk41mpk3, and crk41mpk6 was conducted induced by 150 mM NaCl through a series of sequential images (scale bar = 20 μm). The arrows indicated representative microtubule depolymerization. (B) Image tool software was utilized to quantify cortical microtubules of Col-0, crk41, crk41mpk3, and crk41mpk6, with a minimum of 18 cells from 3 samples being analyzed. SD is represented by the error bars. * p < 0.05; ** p < 0.01 vs. wild type in the same condition, # p < 0.05; ## p < 0.01 vs. crk41 mutant in the same condition.
The findings indicated that both MPK3 and MPK6, especially MPK6, promote microtubule depolymerization of the crk41 mutant, and deletion of MPK3 and MPK6 inhibits depolymerization of crk41 mutant microtubules. These data revealed that MPK3 and MPK6 are connected to regulating the reaction to salt stress by depolymerizing microtubules in the crk41 mutant.
Our data indicate that CRK41 modulates the expression levels of MPK3 and MPK6 when exposed to salt stress and appears to be an inhibitory regulator of these genes. It is clear that MPK3 and MPK6 are vital in the microtubule depolymerization of the crk41 mutant in the presence of salt stress.
3. Discussion
CRKs have been found to be essential in controlling growth and development, in addition to the modulation of defence or immune responses and abiotic stress [1,14,41,42,43]. Recent studies have demonstrated that the elimination of CRK41 enhances the salt tolerance of Arabidopsis, while its overexpression increases the plant’s sensitivity to salt stress [18]. However, there have been no genetic data to support the role that CRK41 plays in salt stress tolerance. Our research demonstrated that that a loss-of-function mutation in the Arabidopsis gene CRK41 results in increased expression of MPK3 and MPK6, as well as alterations in depolymerization rate of microtubules.
Previous study indicated that a novel wheat CRK gene, CRK41, has an effect on the germination of Arabidopsis thaliana seeds when exposed to osmotic stress, and TaCRK41 product is deposited in the cytoplasm [17]. A recent study revealed that CRK41 was co-localized with endocytic membrane markers of FM4−64 in Arabidopsis root cells as analyzed by genetic transformation systems, indicating that CRK41 is localized in the plasma membrane [18]. Consistent with previous studies, our research also demonstrated that CRK41 product is deposited in plasma membrane and contains transmembrane domain (Figure 1). We speculate that the role of CRK41 in salt stress tolerance is primarily to sense salt-sensitive signals and transfer extracellular signals into intracellular signals.
In eukaryotic cells, the cytoskeleton is constantly undergoing disassembly and rearrangement due to the triggering of a variety of signaling pathways. The cortical microtubules, which are integral components of the cytoskeleton, perform a range of roles in cell development and plants’ response to various abiotic stress, particularly salt stress [35,36,37]. Several studies have indicated that depolymerization and rearrangement of cortical microtubules are strategies employed by plants to boost their salt tolerance, and the rapid and global microtubule depolymerization promoted plant resistance to salt stress [35,36,37,44]. Recent studies have shown that the changes in microtubule dynamics can also be beneficial for suspension cells to adapt to salt stress [45]. Moreover, we previously demonstrated that H2Bub1 plays a role as a facilitator in plant salt tolerance by stimulating the depolymerization of cortical microtubules [35]. We also determined that MAP65-1 modulates and accelerates the degradation of microtubules in reaction to salt stress [23]. Our research revealed that absence of CRK41 caused a significant depolymerization in the microtubules, while CRK41 overexpression caused a milder depolymerization (Figure 3). These results imply that CRK41 appears to be able to influence the microtubule depolymerization when exposed salt stress. Results from the molecular genetic analysis revealed that the crk41 mutant had an increased capacity to withstand salt stress, as indicated by its loss-of-function. In contrast, CRK41 overexpression lines, 35S:CRK41-1 and 35S:CRK41-2, were more hypersensitive to salt than wild type (Figure 2). Moreover, studies have shown that CRK41 also took part in salt-induced oxidative stress. The crk41 mutant displayed reduced ROS accumulation when compared with Col-0 after NaCl treatment, whereas overexpression lines of CRK41 resulted in increased levels [18]. Our results determined that the membrane damage after oxidative stress was more severe in CRK41 overexpression lines, while the plasma membrane of the crk41 mutant was only slightly affected (Figure S3). These results suggested that CRK41 was involved in multiple aspects of salt stress response. Among them, microtubule dynamics are a fundamental element of plant cells and are necessary for the successful implementation of cellular processes to help plants better adapt and tolerate salt stress. The microtubule depolymerization phenotype of crk41 mutant and CRK41 overexpression lines in this study further demonstrated that plants enhance salt tolerance by managing the depolymerization and rearrangement of cortical microtubules [44,45].
Plant resilience to stress tolerance conditions is contingent upon a multitude of factors, among which MAPK signalling is a key regulatory mechanism [46]. MAPK signals, particularly those reliant on MPK3/MPK6, are essential for the response to a variety of biotic and abiotic stressors in plants [47,48,49]. MKK5-MPK3/MPK6 can be activated after NaCl treatment and mediates to regulate the expression of iron superoxide dismutase genes to adapt to salt stress in Arabidopsis. Genetic evidence showed that MKK5-RNAi plants presented a salt-sensitive phenotype [50]. MKK5DD triggered MPK3 and MPK6 activity, which enhanced salt tolerance [51]. Exposing plants to high salinity activates MPK3/MPK6, which phosphorylate and destabilize downstream factors, eventually promoting salt tolerance in Arabidopsis [51,52,53]. Our preceding study found that activation of MPK3 and MPK6 is triggered in response to salt stress and is a key factor in salt stress tolerance [23,35]. However, there were few reports about the association between CRK41 and MPK3/MPK6 in salt stress. Our findings indicated that CRK41 was able to interact with MPK3 in vitro and in vivo (Figure 4), and CRK41 participated in the regulation of the expression of MPK3 and MPK6 (Figure 5A,B). Molecular genetic analysis showed that loss functions of mpk3 and mpk6 mutants were hypersensitive to NaCl treatment, compared with wild type (Figure 5E). These results indicated that CRK41 may mediate salt stress responses through MPK3 and MPK6 signalling module in Arabidopsis.
Increasing evidence has connected MAPK activation to conditional and developmental microtubule rearrangements. MPK4 and MPK6 are involved in regulating cytoskeletal in Arabidopsis, and MPK6 colocalizes with microtubules [26,54]. MPK4 and MPK18 have been identified as regulating the stability of microtubules, as loss-of-function mpk18-1 mutant has more enduring and drug-resistant cortical microtubules. Moreover, the mpk4 mutant exhibits a more severe phenotype [25,55]. Our research revealed that MPK3 and MPK6, especially MPK6, affected microtubule dynamics when exposed to salt stress (Figure S5). Genetic analysis showed that loss of mpk3 and mpk6 mutants alleviated the rapid microtubule depolymerization of the crk41 mutant (Figure 6). As shown in Figure 4, CRK41 can interact with MPK3. Our previous research revealed that MPK3 and MPK6 interact with MAP65-1, a microtubule binding protein that influences microtubule stability [23]. We hypothesized that CRK41 could have an effect on MAP65-1 through MPK3, thus impacting microtubule stability. Thus, MPK3 and MPK6 play essential roles in regulating microtubule depolymerization during salt stress response of CRK41. Salt-sensitive phenotypes were also analyzed in crk41mpk3 and crk41mpk6 double mutants. crk41mpk3 and crk41mpk6 mutants restored the salt tolerance phenotype of crk41, and the salt tolerance of crk41mpk3 was comparable to the wild type, while crk41mpk6 was slightly more responsive to NaCl treatment compared with the wild type (Figure 5). The phenotypic results were consistent with those of microtubule depolymerization, and further proved that salt tolerance requires microtubule dynamics.
4. Materials and Methods
4.1. Plant Materials
Wild type (Col-0), crk41 (SALK_056649.45.60.x) [18], 35S:CRK41-1, 35S:CRK41-2, CRK41/crk41-5, CRK41/crk41-6, mpk3 (SALK_100651), mpk6(SALK_062471) [56], crk41mpk3, and crk41mpk6 mutants (all with a Col-0 background) were used in this study. The T-DNA insertion mutants crk41, mpk3, and mpk6 were acquired from State Key Laboratory of Plant Physiology and Biochemistry Arabidopsis seeds platform. The crk41, mpk3, and mpk6 mutants were identified as homozygous mutants by PCR. The mpk3 and mpk6 mutants were confirmed by activity analysis of MPK3 and MPK6 [35]. The 35S:CRK41-1 and 35S:CRK41-2 were generated by transforming the wild type with 35S:CRK41 vector. The crk41 complemented lines CRK41/crk41-5 and CRK41/crk41-6 were generated by transforming the crk41 mutant with 35S:CRK41 vector. The crk41mpk3 and crk41mpk6 mutants were generated by crossing crk41 with mpk3 and mpk6.
Seeds were subjected to a sterilization process using 0.2% sodium hypochlorite prior to sowing, and 1/2 MS medium with 1% (w/v) sucrose and 0.8% (w/v) agar was used as the growing medium in Petri dishes. The plants were grown under a 16 h light/8 h dark photoperiod and 70% relative humidity in a growth cabinet.
4.2. Subcellular Localization
For subcellular localization analyses of CRK41, full-length cDNA of CRK41 was incorporated into 35S:CRK41-GFP vector. The homozygous transgenic lines of 35S:CRK41-GFP were produced by introducing Col-0 with 35S:CRK41-GFP vector. The deposition of CRK41 was examined using a laser confocal microscope Zeiss LSM710 (Carl Zeiss Micro Imaging GmbH, Oberkochen, Germany).
4.3. Salt Sensitivity Assay
For the seed germination assay, 80 seeds of wild type, crk41, 35S:CRK41-1, and 35S:CRK41-2 were planted on 1/2 MS medium with or without 120 mM NaCl for 5 d. Germination (defined as the apparent emergence of radicles) was monitored at the various times.
Additionally, seeds of the wild type, crk41, 35S:CRK41-1, 35S:CRK41-2, mpk3, mpk6, crk41mpk3, and crk41mpk6 mutants were sown on plates for 6 days. Then, at least 60 seedlings of each genotype were transplanted to plates containing either 150 mM NaCl or no NaCl for 3 d to analyze the phenotypic. The percentage of seedlings surviving was determined after treatment for 5 days.
4.4. Detection of Cell Death in Leaves
The cell death in leaves was quantified by staining with trypan blue, following the protocol as described by Bowling et al. [57]. Three independent biological replicates were performed.
4.5. Ion Leakage Assays
Seven-day-old seedlings were transferred from 1/2 MS medium to 1/2 MS medium containing 150 mM NaCl. After 0, 2, 4, 8, and 12 h, seedlings were removed from plates, washed with deionized water, and placed in tubes containing 5 mL of deionized water. Then, ion leakage was measured and calculated as described previously [35].
4.6. Quantitative Real-Time PCR
Arabidopsis total RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and subjected to DNaseI (Invitrogen) treatment. The reverse-transcribed cDNA was synthesized with PrimeScript Reverse Transcription kit (TaKaRa, Dalian, China). Real-time PCR analysis was conducted using cDNA as the template, with SYBR Premix Ex Taq Kit (Takara) and an ABI 7500 Real-Time PCR machine (Applied Biosystem, Foster City, CA, USA). Primers for real-time PCR to detect CRK41 (AT4G00970), MPK3 (AT3G45640), and MPK6 (AT2G43790) were provided in Supplementary Table S1, with ACTIN2 as the internal control.
4.7. Yeast Two-Hybrid Assays
Yeast culture, transformation, and analysis were conducted using methods previously documented in [58]. The full-length cDNAs of MPK3 and MPK6 were inserted into pGBKT7 (Clontech, Mountain View, CA, USA) as the bait vector, while CRK41 full-length cDNA was inserted into pGADT7 (Clontech) as the prey vector. Primers in Supplementary Table S1 were used to amplify CRK41, MPK3, and MPK6 cDNAs. Co-transformation of pGBKT7-MPK3/pGBKT7-MPK6 and pGADT7-CRK41 into AH109 competent cells was conducted, and the cells were grown in SD-Trp-Leu medium. Negative controls were picked using the empty plasmids of pGBKT7 and pGADT7. Positive clones that appeared were picked and inoculated onto SD-Trp-Leu-His-Ade plates. Three independent biological replicates were performed.
4.8. LCI Assays
Amplification and recombination of the MPK3 and MPK6 was performed to generate the nLUC plasmid, while CRK41 were amplified and recombined into cLUC plasmid. Agrobacterium tumefaciens strain GV3101 was utilized to transiently transfect the ligated nLuc-MPK3/nLuc-MPK6 and CRK41-cLuc into 5-week-old N.benthamiana, and then leaves were collected after 2–3 days. Primers in Supplementary Table S1 were used to amplify of CRK41-, MPK3-, and MPK6-coding sequences. A positive control was picked using the plasmid of nLuc-SGT1 and RAR1-cLuc. Negative controls were picked using the empty plasmids of nLUC and cLUC. The LCI assay and relative LUC activity was performed [59].
4.9. Observation of Microtubule Organization under Salt Stress
Wild type, crk41, 35S:CRK41-1, 35S:CRK41-2, mpk3, mpk6, crk41mpk3, and crk41mpk6 mutants stably expressing GFP–tubulin were used for this assay. The leaves of 7-day-old seedlings were incubated with 150 mM NaCl, then the disassembly and reorganization of the cortical microtubules the organization and disassembly of the cortical microtubules was observed over time using a Zeiss LSM 710 microscope (Carl Zeiss Micro Imaging GmbH). The procedure and protocol were described by Shi et al. [60]. Microtubules were analyzed in z stacks of images at 1.5 μm intervals. Zeiss LSM 710 software was used to acquire fluorescent images of microtubules, which were then converted to TIFF format and further edited in Adobe Photoshop 5.0. The number of microtubules was calculated, as described previously [61]. Microtubule number was counted along a fixed line (~50 μm) perpendicular to the direction of most cortical microtubules in the cell using ImageJ software (NIH, Bethesda, MD, USA).
4.10. Statistical Analysis
Statistical analysis was conducted using Student’s t-test and one-way ANOVA in SPSS 26.0. A statistical significance was noted if the p-value was less than 0.05, which was denoted by * and # (*,# = p < 0.05, **,## = p < 0.01).
5. Conclusions
The current research indicates that CRK41 has a negative effect on Arabidopsis salt tolerance due to its influence on microtubule dynamic instability. MPK3/MPK6 are essential for the reaction of crk41 mutant in salt stress. CRK41 is associated with MPK3, not MPK6. MPK3 and MPK6, especially MPK6, are essential for the microtubule depolymerization, which is necessary for salt tolerance in the crk41 mutant (Figure 7). Microtubule dynamics can also be used as a signal, transmitting salt signals into cell nucleus, and influencing the expression of salt-related genes. Eventually, Arabidopsis can adapt to salt stress.
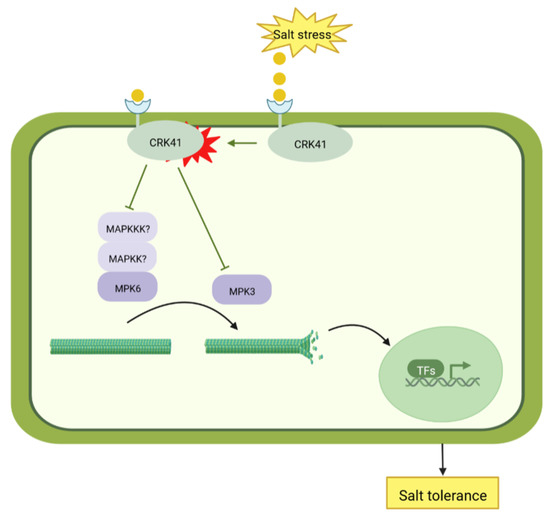
Figure 7.
Model of CRK41-mediated salt tolerance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12061285/s1, Table S1: Primers used in this work; Figure S1: The relative expression level of CRK41 was determined after different concentrations of mannitol treatment; Figure S2: Cell death induced by different concentrations of NaCl in leaves of Col-0 and crk41 mutant; Figure S3: Ion leakage in seedlings of Col-0, crk41 mutant, 35S:CRK41-1, and 35S:CRK41-2 treated with NaCl; Figure S4: The salt-sensitive phenotype could be restored in the crk41 complemented lines; Figure S5: MPK3 and MKP6 have been identified as regulators of cortical microtubule depolymerization when exposed to salt stress.
Author Contributions
Conceptualization, project administration, funding acquisition, writing—review and editing, W.M. and B.P.; writing—original draft preparation and methodology, S.Z. and Q.L.; software, S.Z., Q.L. and Z.N.; validation, C.W., W.Z. and Y.H.; formal analysis, S.Z. and Q.L.; investigation, J.Z.; resources, W.M. and S.Z.; data curation, Z.N., C.W., W.Z. and Y.H.; supervision, B.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31900233), Shandong Key R&D Program (2019GSF107088), the Outstanding Youth Innovation Team of Shandong High Universities (2019kJe117), and the open funds of the State Key Laboratory of Plant Physiology and Biochemistry (SKLPPBKF2201).
Data Availability Statement
The data presented in this study are available within the article.
Acknowledgments
The authors acknowledge the assistance of Jiaping Zhou from Research Centre of Modern Analytical Technology, Tianjin University of Science and Technology, China, who offered great help in the microtubule observation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wrzaczek, M.; Brosché, M.; Salojärvi, J.; Kangasjärvi, S.; Idänheimo, N.; Mersmann, S.; Robatzek, S.; Karpiński, S.; Karpińska, B.; Kangasjärvi, J. Transcriptional Regulation of the CRK/DUF26 Group of Receptor-like Protein Kinases by Ozone and Plant Hormones in Arabidopsis. BMC Plant Biol. 2010, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. A Superfamily of Proteins with Novel Cysteine-Rich Repeats. Plant Physiol. 2001, 126, 473–476. [Google Scholar] [CrossRef]
- Yeh, Y.-H.; Chang, Y.-H.; Huang, P.-Y.; Huang, J.-B.; Zimmerli, L. Enhanced Arabidopsis Pattern-Triggered Immunity by Overexpression of Cysteine-Rich Receptor-like Kinases. Front. Plant Sci. 2015, 6, 322. [Google Scholar] [CrossRef] [PubMed]
- Ederli, L.; Madeo, L.; Calderini, O.; Gehring, C.; Moretti, C.; Buonaurio, R.; Paolocci, F.; Pasqualini, S. The Arabidopsis Thaliana Cysteine-Rich Receptor-like Kinase CRK20 Modulates Host Responses to Pseudomonas Syringae Pv. Tomato DC3000 Infection. J. Plant Physiol. 2011, 168, 1784–1794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like Cytoplasmic Kinases Integrate Signaling from Multiple Plant Immune Receptors and Are Targeted by a Pseudomonas Syringae Effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Kim, Y.C.; Kwon, S.J.; Ryu, C.-M.; Park, O.K. The Arabidopsis Cysteine-Rich Receptor-Like Kinase CRK36 Regulates Immunity through Interaction with the Cytoplasmic Kinase BIK1. Front. Plant Sci. 2017, 8, 1856. [Google Scholar] [CrossRef]
- Du, D.; Liu, M.; Xing, Y.; Chen, X.; Zhang, Y.; Zhu, M.; Lu, X.; Zhang, Q.; Ling, Y.; Sang, X.; et al. Semi-Dominant Mutation in the Cysteine-Rich Receptor-like Kinase Gene, ALS1, Conducts Constitutive Defence Response in Rice. Plant Biol. 2019, 21, 25–34. [Google Scholar] [CrossRef]
- Chern, M.; Xu, Q.; Bart, R.S.; Bai, W.; Ruan, D.; Sze-To, W.H.; Canlas, P.E.; Jain, R.; Chen, X.; Ronald, P.C. A Genetic Screen Identifies a Requirement for Cysteine-Rich-Receptor-Like Kinases in Rice NH1 (OsNPR1)-Mediated Immunity. PLoS Genet. 2016, 12, e1006049. [Google Scholar] [CrossRef]
- Yang, K.; Rong, W.; Qi, L.; Li, J.; Wei, X.; Zhang, Z. Isolation and Characterization of a Novel Wheat Cysteine-Rich Receptor-like Kinase Gene Induced by Rhizoctonia Cerealis. Sci. Rep. 2013, 3, 3021. [Google Scholar] [CrossRef]
- Rayapuram, C.; Jensen, M.K.; Maiser, F.; Shanir, J.V.; Hornshøj, H.; Rung, J.H.; Gregersen, P.L.; Schweizer, P.; Collinge, D.B.; Lyngkjaer, M.F. Regulation of Basal Resistance by a Powdery Mildew-Induced Cysteine-Rich Receptor-like Protein Kinase in Barley: CRK-Regulated Basal Resistance in Barley. Mol. Plant Pathol. 2012, 13, 135–147. [Google Scholar] [CrossRef]
- Li, T.-G.; Zhang, D.-D.; Zhou, L.; Kong, Z.-Q.; Hussaini, A.S.; Wang, D.; Li, J.-J.; Short, D.P.G.; Dhar, N.; Klosterman, S.J.; et al. Genome-Wide Identification and Functional Analyses of the CRK Gene Family in Cotton Reveals GbCRK18 Confers Verticillium Wilt Resistance in Gossypium Barbadense. Front. Plant Sci. 2018, 9, 1266. [Google Scholar] [CrossRef]
- Lu, K.; Liang, S.; Wu, Z.; Bi, C.; Yu, Y.-T.; Wang, X.-F.; Zhang, D.-P. Overexpression of an Arabidopsis Cysteine-Rich Receptor-like Protein Kinase, CRK5, Enhances Abscisic Acid Sensitivity and Confers Drought Tolerance. J. Exp. Bot. 2016, 67, 5009–5027. [Google Scholar] [CrossRef] [PubMed]
- Burdiak, P.; Rusaczonek, A.; Witoń, D.; Głów, D.; Karpiński, S. Cysteine-Rich Receptor-like Kinase CRK5 as a Regulator of Growth, Development, and Ultraviolet Radiation Responses in Arabidopsis Thaliana. J. Exp. Bot. 2015, 66, 3325–3337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, G.; Shi, R.; Han, X.; Qi, L.; Wang, R.; Xiong, L.; Li, G. Arabidopsis Cysteine-Rich Receptor-like Kinase 45 Functions in the Responses to Abscisic Acid and Abiotic Stresses. Plant Physiol. Biochem. 2013, 67, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Idänheimo, N.; Gauthier, A.; Salojärvi, J.; Siligato, R.; Brosché, M.; Kollist, H.; Mähönen, A.P.; Kangasjärvi, J.; Wrzaczek, M. The Arabidopsis Thaliana Cysteine-Rich Receptor-like Kinases CRK6 and CRK7 Protect against Apoplastic Oxidative Stress. Biochem. Biophys. Res. Commun. 2014, 445, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Unraveling Salt Stress Signaling in Plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Chen, D.; Wu, J.; Zhao, M.; Ma, X.; Zhang, W.; Xia, G.; Wang, M. A Novel Wheat Cysteine-Rich Receptor-like Kinase Gene CRK41 Is Involved in the Regulation of Seed Germination under Osmotic Stress in Arabidopsis Thaliana. J. Plant Biol. 2017, 60, 571–581. [Google Scholar] [CrossRef]
- Li, X.; Zhao, J.; Sun, Y.; Li, Y. Arabidopsis Thaliana CRK41 Negatively Regulates Salt Tolerance via H2O2 and ABA Cross-Linked Networks. Environ. Exp. Bot. 2020, 179, 104210. [Google Scholar] [CrossRef]
- Wasteneys, G.O. Progress in Understanding the Role of Microtubules in Plant Cells. Curr. Opin. Plant Biol. 2004, 7, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Elliott, A.; Shaw, S.L. Update: Plant Cortical Microtubule Arrays. Plant Physiol. 2018, 176, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kurepa, J.; Hashimoto, T.; Smalle, J.A. Salt Stress-Induced Disassembly of Arabidopsis Cortical Microtubule Arrays Involves 26S Proteasome-Dependent Degradation of SPIRAL1. Plant Cell 2011, 23, 3412–3427. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Suzuki, K.; Abe, T.; Kaneko, Y.; Shi, H.; Zhu, J.-K.; Rus, A.; Hasegawa, P.M.; Hashimoto, T. Salt Stress Affects Cortical Microtubule Organization and Helical Growth in Arabidopsis. Plant Cell Physiol. 2006, 47, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Chen, Q.; Li, X.; Li, Y. MAP65-1 Is Required for the Depolymerization and Reorganization of Cortical Microtubules in the Response to Salt Stress in Arabidopsis. Plant Sci. 2017, 264, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Komis, G.; Müller, J.; Menzel, D.; Samaj, J. Arabidopsis Homologs of Nucleus- and Phragmoplast-Localized Kinase 2 and 3 and Mitogen-Activated Protein Kinase 4 Are Essential for Microtubule Organization. Plant Cell 2010, 22, 755–771. [Google Scholar] [CrossRef]
- Kosetsu, K.; Matsunaga, S.; Nakagami, H.; Colcombet, J.; Sasabe, M.; Soyano, T.; Takahashi, Y.; Hirt, H.; Machida, Y. The MAP Kinase MPK4 Is Required for Cytokinesis in Arabidopsis Thaliana. Plant Cell 2010, 22, 3778–3790. [Google Scholar] [CrossRef]
- Müller, J.; Beck, M.; Mettbach, U.; Komis, G.; Hause, G.; Menzel, D.; Samaj, J. Arabidopsis MPK6 Is Involved in Cell Division Plane Control during Early Root Development, and Localizes to the Pre-Prophase Band, Phragmoplast, Trans-Golgi Network and Plasma Membrane. Plant J. 2010, 61, 234–248. [Google Scholar] [CrossRef]
- Komis, G.; Illés, P.; Beck, M.; Šamaj, J. Microtubules and Mitogen-Activated Protein Kinase Signalling. Curr. Opin. Plant Biol. 2011, 14, 650–657. [Google Scholar] [CrossRef]
- Hirt, H. MAP Kinases in Plant Signal Transduction. Results Probl. Cell Differ. 2000, 27, 1–9. [Google Scholar] [CrossRef]
- Smertenko, A.P.; Chang, H.-Y.; Sonobe, S.; Fenyk, S.I.; Weingartner, M.; Bögre, L.; Hussey, P.J. Control of the AtMAP65-1 Interaction with Microtubules through the Cell Cycle. J. Cell Sci. 2006, 119, 3227–3237. [Google Scholar] [CrossRef]
- Sasabe, M.; Machida, Y. MAP65: A Bridge Linking a MAP Kinase to Microtubule Turnover. Curr. Opin. Plant Biol. 2006, 9, 563–570. [Google Scholar] [CrossRef]
- Cassimeris, L.; Spittle, C. Regulation of Microtubule-Associated Proteins. Int. Rev. Cytol. 2001, 210, 163–226. [Google Scholar] [CrossRef] [PubMed]
- Smékalová, V.; Luptovčiak, I.; Komis, G.; Šamajová, O.; Ovečka, M.; Doskočilová, A.; Takáč, T.; Vadovič, P.; Novák, O.; Pechan, T.; et al. Involvement of YODA and Mitogen Activated Protein Kinase 6 in Arabidopsis Post-Embryogenic Root Development through Auxin up-Regulation and Cell Division Plane Orientation. New Phytol. 2014, 203, 1175–1193. [Google Scholar] [CrossRef] [PubMed]
- Samajová, O.; Komis, G.; Samaj, J. Immunofluorescent Localization of MAPKs and Colocalization with Microtubules in Arabidopsis Seedling Whole-Mount Probes. Methods Mol. Biol. 2014, 1171, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, F.; Mao, T.; Nie, J.; Yan, M.; Yuan, M.; Zhang, W. Phosphatidic Acid Regulates Microtubule Organization by Interacting with MAP65-1 in Response to Salt Stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576. [Google Scholar] [CrossRef]
- Zhou, S.; Chen, Q.; Sun, Y.; Li, Y. Histone H2B Monoubiquitination Regulates Salt Stress-Induced Microtubule Depolymerization in Arabidopsis. Plant Cell Environ. 2017, 40, 1512–1530. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, J.; Yuan, M. Salt Tolerance Requires Cortical Microtubule Reorganization in Arabidopsis. Plant Cell Physiol. 2007, 48, 1534–1547. [Google Scholar] [CrossRef] [PubMed]
- Fujita, S.; Pytela, J.; Hotta, T.; Kato, T.; Hamada, T.; Akamatsu, R.; Ishida, Y.; Kutsuna, N.; Hasezawa, S.; Nomura, Y.; et al. An Atypical Tubulin Kinase Mediates Stress-Induced Microtubule Depolymerization in Arabidopsis. Curr. Biol. 2013, 23, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Salamó, I.; Papdi, C.; Rigó, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horváth, B.; Domoki, M.; Darula, Z.; et al. The Heat Shock Factor A4A Confers Salt Tolerance and Is Regulated by Oxidative Stress and the Mitogen-Activated Protein Kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, S. Mitogen-Activated Protein Kinase Cascades in Signaling Plant Growth and Development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, H.; He, Y.; Liu, Y.; Walker, J.C.; Torii, K.U.; Zhang, S. A MAPK Cascade Downstream of ERECTA Receptor-like Protein Kinase Regulates Arabidopsis Inflorescence Architecture by Promoting Localized Cell Proliferation. Plant Cell 2012, 24, 4948–4960. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Osakabe, Y.; Katsura, S.; Mizuno, S.; Maruyama, K.; Kusakabe, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abiotic Stress-Inducible Receptor-like Kinases Negatively Control ABA Signaling in Arabidopsis: Receptor-like Kinases in ABA Signaling. Plant J. 2012, 70, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Waszczak, C.; Hunter, K.; Wrzaczek, M. Bound by Fate: The Role of Reactive Oxygen Species in Receptor-Like Kinase Signaling. Plant Cell 2017, 29, 638–654. [Google Scholar] [CrossRef]
- Arellano-Villagómez, F.C.; Guevara-Olvera, L.; Zuñiga-Mayo, V.M.; Cerbantez-Bueno, V.E.; Verdugo-Perales, M.; Medina, H.R.; De Folter, S.; Acosta-García, G. Arabidopsis Cysteine-Rich Receptor-like Protein Kinase CRK33 Affects Stomatal Density and Drought Tolerance. Plant Signal. Behav. 2021, 16, 1905335. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liu, M. The Microtubule Cytoskeleton Acts as a Sensor for Stress Response Signaling in Plants. Mol. Biol. Rep. 2019, 46, 5603–5608. [Google Scholar] [CrossRef] [PubMed]
- Chun, H.J.; Baek, D.; Jin, B.J.; Cho, H.M.; Park, M.S.; Lee, S.H.; Lim, L.H.; Cha, Y.J.; Bae, D.-W.; Kim, S.T.; et al. Microtubule Dynamics Plays a Vital Role in Plant Adaptation and Tolerance to Salt Stress. Int. J. Mol. Sci. 2021, 22, 5957. [Google Scholar] [CrossRef] [PubMed]
- Šamajová, O.; Plíhal, O.; Al-Yousif, M.; Hirt, H.; Šamaj, J. Improvement of Stress Tolerance in Plants by Genetic Manipulation of Mitogen-Activated Protein Kinases. Biotechnol. Adv. 2013, 31, 118–128. [Google Scholar] [CrossRef]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A Complex Signalling Network Involved in Multiple Biological Processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef] [PubMed]
- González Besteiro, M.A.; Bartels, S.; Albert, A.; Ulm, R. Arabidopsis MAP Kinase Phosphatase 1 and Its Target MAP Kinases 3 and 6 Antagonistically Determine UV-B Stress Tolerance, Independent of the UVR8 Photoreceptor Pathway. Plant J. 2011, 68, 727–737. [Google Scholar] [CrossRef]
- Ichimura, K.; Mizoguchi, T.; Yoshida, R.; Yuasa, T.; Shinozaki, K. Various Abiotic Stresses Rapidly Activate Arabidopsis MAP Kinases ATMPK4 and ATMPK6. Plant J. 2000, 24, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Chen, W.; Jia, W.; Zhang, J. Mitogen-Activated Protein Kinase Kinase 5 (MKK5)-Mediated Signalling Cascade Regulates Expression of Iron Superoxide Dismutase Gene in Arabidopsis under Salinity Stress. J. Exp. Bot. 2015, 66, 5971–5981. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, J.; Wang, F.; Xie, C.; Lv, B.; Yu, Z.; Dai, S.; Liu, X.; Xia, G.; Tian, H.; et al. MPK3/6-Induced Degradation of ARR1/10/12 Promotes Salt Tolerance in Arabidopsis. EMBO Rep. 2021, 22, e52457. [Google Scholar] [CrossRef] [PubMed]
- Droillard, M.; Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Different Protein Kinase Families Are Activated by Osmotic Stresses in Arabidopsis Thaliana Cell Suspensions. Involvement of the MAP Kinases AtMPK3 and AtMPK6. FEBS Lett. 2002, 527, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic Acid Mediates Salt Stress Response by Regulation of MPK6 in Arabidopsis Thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Komis, G.; Ziemann, A.; Menzel, D.; Šamaj, J. Mitogen-Activated Protein Kinase 4 Is Involved in the Regulation of Mitotic and Cytokinetic Microtubule Transitions in Arabidopsis Thaliana. New Phytol. 2011, 189, 1069–1083. [Google Scholar] [CrossRef]
- Walia, A.; Lee, J.S.; Wasteneys, G.; Ellis, B. Arabidopsis Mitogen-Activated Protein Kinase MPK18 Mediates Cortical Microtubule Functions in Plant Cells. Plant J. 2009, 59, 565–575. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.; Lei, L.; Yang, H.; Liu, G.; Ren, D. Activation of MAPK Kinase 9 Induces Ethylene and Camalexin Biosynthesis and Enhances Sensitivity to Salt Stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef] [PubMed]
- Bowling, S.A.; Clarke, J.D.; Liu, Y.; Klessig, D.F.; Dong, X. The Cpr5 Mutant of Arabidopsis Expresses Both NPR1-Dependent and NPR1-Independent Resistance. Plant Cell 1997, 9, 1573–1584. [Google Scholar] [CrossRef]
- Xie, Q.; Sanz-Burgos, A.P.; Guo, H.; García, J.A.; Gutiérrez, C. GRAB Proteins, Novel Members of the NAC Domain Family, Isolated by Their Interaction with a Geminivirus Protein. Plant Mol. Biol. 1999, 39, 647–656. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.-M. Firefly Luciferase Complementation Imaging Assay for Protein-Protein Interactions in Plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.-M.; Yao, L.-L.; Pei, B.-L.; Zhou, Q.; Li, X.-L.; Li, Y.; Li, Y.-Z. Cortical Microtubule as a Sensor and Target of Nitric Oxide Signal during the Defence Responses to Verticillium Dahliae Toxins in Arabidopsis. Plant Cell Environ. 2009, 32, 428–438. [Google Scholar] [CrossRef]
- Hu, M.; Pei, B.-L.; Zhang, L.-F.; Li, Y.-Z. Histone H2B Monoubiquitination Is Involved in Regulating the Dynamics of Microtubules during the Defense Response to Verticillium Dahliae Toxins in Arabidopsis. Plant Physiol. 2014, 164, 1857–1865. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).