Genetic and Metabolite Variability among Commercial Varieties and Advanced Lines of Vicia faba L.
Abstract
1. Introduction
2. Results
2.1. Genetic Diversity Estimated by SCoT Markers
2.2. Correlation Analysis
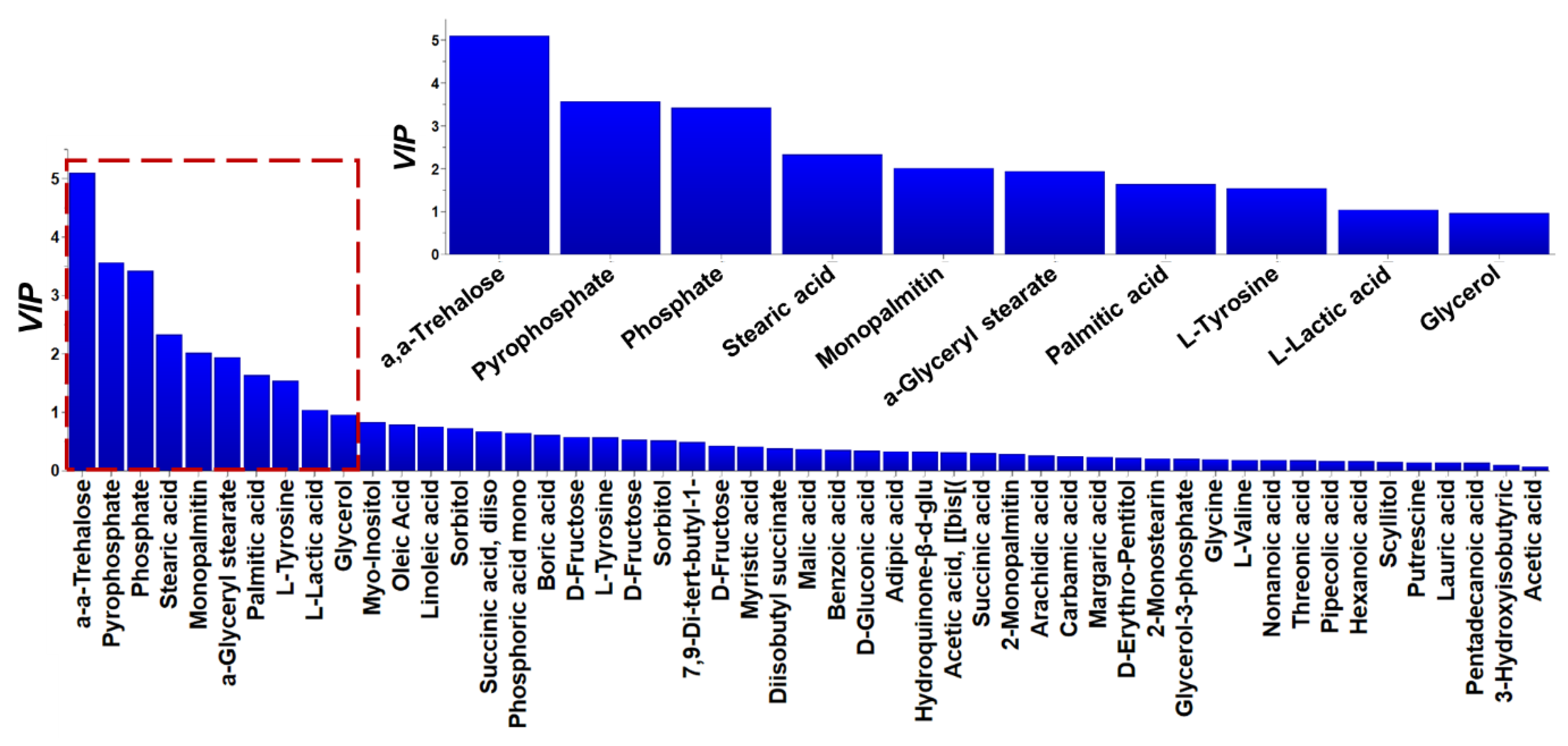
2.3. Metabolomics Analysis
3. Discussion
4. Materials and Methods
4.1. DNA Isolation and Markers Analysis
4.2. Data Analysis
4.3. Plant Material and Experimental Conditions for Metabolomic Analysis
4.4. Extraction of Seeds’ Metabolites and Sample Preparation for GC/EI/MS Metabolomics Analysis
4.5. GC/EI/MS Metabolomics Analysis of Seed Extracts
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leelambika, M.; Mahesh, S.; Jaheer, M.; Tripathi, P.K.; Ranjith Kumar, P.; Sathyanarayana, N. Targeted Metabolic and Genomic Profiling Reveals Parents for L-Dopa Breeding in Mucuna pruriens (L.) DC. Trop. Plant Biol. 2016, 9, 239–251. [Google Scholar] [CrossRef]
- Laurentin, H.; Ratzinger, A.; Karlovsky, P. Relationship between metabolic and genomic diversity in sesame (Sesamum indicum L.). BMC Genom. 2008, 9, 250. [Google Scholar] [CrossRef]
- Tanno, K.; Willcox, G. The origins of cultivation of Cicer arietinum L. and Vicia faba L.: Early finds from Tell el-Kerkh, north-west Syria, late 10th millennium B.P. Veg. Hist. Archaeobotany 2006, 15, 197–204. [Google Scholar] [CrossRef]
- Zahran, M.A.; Willis, A.J. The History of the Vegetation: Its Salient Features and Future Study. Veg. Egypt 2008, 2, 305–318. [Google Scholar]
- Mekky, R.H.; Thabet, M.M.; Rodriguez-Pérez, C.; Elnaggar, D.M.Y.; Marhous, E.A.; Segura-Carretero, A.; Abdel-Sattar, E. Comparative metabolite profiling and antioxidant potentials of seeds and sprouts of three Egyptian cultivars of Vicia faba L. Food Res. Int. 2020, 136, 109537. [Google Scholar] [CrossRef] [PubMed]
- Warsame, A.O.; O’Sullivan, D.M.; Tosi, P. Seed Storage Proteins of Faba Bean (Vicia faba L.): Current Status and Prospects for Genetic Improvement. J. Agric. Food Chem. 2018, 66, 12617–12626. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Perdomo, E.; Vidal, A.; Kreplak, J.; Duborjal, H.; Leveugle, M.; Duarte, J.; Desmetz, C.; Deulvot, C.; Raffiot, B.; Marget, P.; et al. Development of new genetic resources for faba bean (Vicia faba L.) breeding through the discovery of gene-based SNP markers and the construction of a high-density consensus map. Sci. Rep. 2020, 10, 6790. [Google Scholar] [CrossRef]
- Pociecha, E.; Kościelniak, J.; Filek, W. Acta Physiologiae Plantarum; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Katerji, N.; Mastrorilli, M.; Lahmer, F.Z.; Maalouf, F.; Oweis, T. Faba bean productivity in saline-drought conditions. Eur. J. Agron. 2011, 35, 2–12. [Google Scholar] [CrossRef]
- Zhou, R.; Hyldgaard, B.; Yu, X.; Rosenqvist, E.; Ugarte, R.M.; Yu, S.; Wu, Z.; Ottosen, C.O.; Zhao, T. Phenotyping of faba beans (Vicia faba L.) under cold and heat stresses using chlorophyll fluorescence. Euphytica 2018, 214, 68. [Google Scholar] [CrossRef]
- Lyu, J.; Ramekar, R.; Kim, J.M.; Hung, N.N.; Seo, J.S.; Kim, J.B.; Choi, I.K.; Park, K.C.; Kwon, S.J. Unraveling the complexity of faba bean (Vicia faba L.) transcriptome to reveal cold-stress-responsive genes using long-read isoform sequencing technology. Sci. Rep. 2021, 11, 21094. [Google Scholar] [CrossRef]
- Baddeley, J.A.; Jones, S.; Topp, C.F.E.; Watson, C.A.; Helming, J.; Stoddard, F.L. Biological nitrogen fixation (BNF) by legume crops in Europe. Legume Futur. 2014, 1, 27. [Google Scholar]
- Duc, G. Faba bean (Vicia faba L.). Field Crops Res. 1997, 53, 99–109. [Google Scholar] [CrossRef]
- Mohseni, M.S.M.; Golshani, B. Simultaneous determination of levodopa and carbidopa from fava bean, green peas and green beans by high performance liquid gas chromatography. J. Clin. Diagn. Res. 2013, 7, 1004–1007. [Google Scholar]
- Etemadi, F.; Hashemi, M.; Randhir, R.; ZandVakili, O.; Ebadi, A. Accumulation of L-DOPA in various organs of faba bean and influence of drought, nitrogen stress, and processing methods on L-DOPA yield. Crop J. 2018, 6, 426–434. [Google Scholar] [CrossRef]
- Etemadi, F.; Hashemi, M.; Barker, A.V.; Zandvakili, O.R.; Liu, X. Agronomy, Nutritional Value, and Medicinal Application of Faba Bean (Vicia faba L.). Hortic. Plant J. 2019, 5, 170–182. [Google Scholar] [CrossRef]
- Horstmann, C.; Schlesier, B.; Otto, A.; Kostka, S.; Müntz, K. Polymorphism of legumin subunits from field bean (Vicia faba L. var. minor) and its relation to the corresponding multigene family. Theor. Appl. Genet. 1993, 86, 867–874. [Google Scholar] [CrossRef]
- Fuchs, J.; Schubert, I. Localization of seed protein genes on metaphase chromosomes of Vicia faba via fluorescence In Situ hybridization. Chromosome Res. 1995, 3, 94–100. [Google Scholar] [CrossRef]
- Müntz, K.; Horstmann, C.; Schlesier, B. Vicia Globulins; Springer: Dordrecht, The Netherlands, 1999; pp. 259–284. [Google Scholar]
- Khazaei, H.; O’Sullivan, D.M.; Jones, H.; Pitts, N.; Sillanpää, M.J.; Pärssinen, P.; Manninen, O.; Stoddard, F.L. Flanking SNP markers for vicine–convicine concentration in faba bean (Vicia faba L.). Mol. Breed. 2015, 35, 38. [Google Scholar] [CrossRef]
- Khazaei, H.; Purves, R.W.; Song, M.; Stonehouse, R.; Bett, K.E.; Stoddard, F.L.; Vandenberg, A. Development and validation of a robust, breeder-friendly molecular marker for the vc—Locus in faba bean. Mol. Breed. 2017, 37, 140. [Google Scholar] [CrossRef]
- Khazaei, H.; Purves, R.W.; Hughes, J.; Link, W.; O’Sullivan, D.M.; Schulman, A.H.; Björnsdotter, E.; Geu-Flores, F.; Nadjieza, M.; Andersen, S.U.; et al. Eliminating vicine and convicine, the main anti-nutritional factors restricting faba bean usage. Trends Food Sci. Technol. 2019, 91, 549–556. [Google Scholar] [CrossRef]
- Gutierrez, N.; Avila, C.M.; Moreno, M.T.; Torres, A.M. Development of SCAR markers linked to zt-2, one of the genes controlling absence of tannins in faba bean. Aust. J. Agric. Res. 2008, 59, 62–68. [Google Scholar] [CrossRef]
- Webb, A.; Cottage, A.; Wood, T.; Khamassi, K.; Hobbs, D.; Gostkiewicz, K.; White, M.; Khazaei, H.; Ali, M.; Street, D.; et al. A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba L.). Plant Biotechnol. J. 2015, 14, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Zanotto, S.; Vandenberg, A.; Khazaei, H. Development and validation of a robust KASP marker for zt2 locus in faba bean (Vicia Faba). Plant Breed. 2020, 139, 375–380. [Google Scholar] [CrossRef]
- Warsame, A.O.; Michael, N.; O’Sullivan, D.M.; Tosi, P. Identification and Quantification of Major Faba Bean Seed Proteins. J. Agric. Food Chem. 2020, 68, 8535–8544. [Google Scholar] [CrossRef]
- Akash, M.W.; Myers, G.O. The development of faba bean expressed sequence tag-simple sequence repeats (EST-SSRs) and their validity in diversity analysis. Plant Breed. 2012, 131, 522–530. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.M.; Sun, Z.H.; Yang, T.; Tan, Z.L.; Wang, B.F.; Han, X.F.; He, Z.X. The growth performance and meat quality of goats fed diets based on maize or wheat grain. J. Anim. Feed Sci. 2012, 21, 587–598. [Google Scholar] [CrossRef]
- Kaur, S.; Kimber, R.B.E.; Cogan, N.O.I.; Materne, M.; Forster, J.W.; Paull, J.G. SNP discovery and high-density genetic mapping in faba bean (Vicia faba L.) permits identification of QTLs for ascochyta blight resistance. Plant Sci. 2014, 217–218, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Duc, G.; Bao, S.; Baum, M.; Redden, B.; Sadiki, M.; Suso, M.J.; Vishniakova, M.; Zong, X. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crops Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- Wang, H.F.; Zong, X.X.; Guan, J.P.; Yang, T.; Sun, X.L.; Ma, Y.; Redden, R. Genetic diversity and relationship of global faba bean (Vicia faba L.) germplasm revealed by ISSR markers. Theoret. Appl. Genet. 2012, 124, 789–797. [Google Scholar] [CrossRef]
- Link, W.; Dixkens, C.; Singh, M.; Schwall, M.; Melchinger, A.E. Genetic diversity in European and Mediterranean faba bean germ plasm revealed by RAPD markers. Theoret. Appl. Genet. 1995, 90, 27–32. [Google Scholar] [CrossRef]
- Avramidou, E.; Ganopoulos, I.; Mylona, P.; Abraham, E.M.; Nianiou-Obeidat, I.; Osathanunkul, M.; Madesis, P. Comparative analysis of the genetic diversity of faba bean (Vicia faba L.). Sustainability 2023, 15, 1016. [Google Scholar] [CrossRef]
- Zong, X.; Liu, X.; Guan, J.; Wang, S.; Liu, Q.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global winter faba bean germplasm. Theoret. Appl. Genet. 2009, 118, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Hu, J.; Coyne, C.J. Genetic diversity and relationship among faba bean (Vicia faba L.) germplasm entries as revealed by TRAP markers. Plant Genet. Resour. 2010, 8, 204–213. [Google Scholar] [CrossRef]
- Tomas, D.; Dias, A.L.; Silva, M.; Oliveira, H.R.; Suso, M.J.; Viegas, W.; Veloso, M.M. Genetic Diversity Assessment of Portuguese Cultivated Vicia faba L. through IRAP markers. Diversity 2016, 8, 8. [Google Scholar] [CrossRef]
- Akash, M.W.; Al-Awaida, W.; Atiyeh, A.; Al-Debei, H.; Saleh, M.; Zatimeh, A.; Salameh, N.; Alawin, M.; Hasan, S.M. Exploring genetic variations in faba bean (Vicia faba L.) accessions using newly developed EST-SSR markers. Pak. J. Bot. 2017, 49, 667–672. [Google Scholar]
- Terzopoulos, P.J.; Bebeli, P.J. Genetic diversity analysis of Mediterranean faba bean (Vicia faba L.) with ISSR markers. Field Crops Res. 2008, 108, 39–44. [Google Scholar] [CrossRef]
- Salazar-laureles, M.E.; Pérez-lópez, D.D.J.; González-huerta, A. Genetic variability analysis of faba bean accessions using inter-simple sequence repeat (ISSR) markers. Chil. J. Agric. Res. 2015, 75, 122–130. [Google Scholar] [CrossRef]
- Asfaw, B.M.; Dagne, K.; Wakayo, G.K.; Kemal, S.A.; Muleta, K.T. Genetic diversity study of Ethiopian faba bean (Vicia faba L.) varieties based on phenotypic traits and inter simple sequence repeat (ISSR) markers. Afric. J Biotechnol. 2018, 17, 433–446. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A Simple, Novel DNA Marker Technique for Generating Gene-Targeted Markers in Plants. Plant Mol. Biol. Report. 2008, 27, 86–93. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Kumaria, S.; Kumar, S.; Tandon, P. Start Codon Targeted (SCoT) marker reveals genetic diversity of Dendrobium nobile Lindl., an endangered medicinal orchid species. Gene 2013, 529, 21–26. [Google Scholar] [CrossRef]
- Chai, X.; Dong, R.; Liu, W.; Wang, Y.; Liu, Z. Optimizing Sample Size to Assess the Genetic Diversity in Common Vetch (Vicia sativa L.) Populations Using Start Codon Targeted (SCoT) Markers. Molecules 2017, 22, 567. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.-L.; Zhang, J.-Y.; Liu, C.H. Genetic diversity in some grape varieties revealed by SCoT analyses. Mol. Biol. Rep. 2012, 39, 5307–5313. [Google Scholar] [CrossRef] [PubMed]
- Domergue, J.-B.; Abadie, C.; Limami, A.; Way, D.; Tcherkez, G. Seed quality and carbon primary metabolism. Plant Cell Environ. 2019, 42, 2776–2788. [Google Scholar] [CrossRef]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef]
- Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genom. 2001, 2, 155–198. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Bino, R.J.; Hall, R.D.; Fiehn, O.; Kopka, J.; Saito, K.; Draper, J.; Nikolau, B.J.; Mendes, P.; Roessner-Tunali, U.; Beale, M.H.; et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004, 9, 418–425. [Google Scholar] [CrossRef]
- Muscolo, A.; Junker, A.; Klukas, C.; Weigelt-Fischer, K.; Riewe, D.; Altmann, T. Phenotypic and metabolic responses to drought and salinity of four contrasting lentil accessions. J. Exp. Bot. 2015, 66, 5467–5480. [Google Scholar] [CrossRef] [PubMed]
- Razzaq, A.; Sadia, B.; Raza, A.; Hameed, M.K.; Saleem, F. Metabolomics: A way forward for crop improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef]
- Razzaq, A.; Wishart, D.S.; Wani, S.H.; Hameed, M.K.; Mubin, M.; Saleem, F. Advances in Metabolomics-Driven Diagnostic Breeding and Crop Improvement. Metabolites 2022, 12, 511. [Google Scholar] [CrossRef]
- Foti, C.; Kalampokis, I.F.; Aliferis, K.A.; Pavli, O.I. Metabolic responses of two contrasting lentil genotypes to peg-induced drought stress. Agronomy 2021, 11, 1190. [Google Scholar] [CrossRef]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, E.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity stress alters the secondary metabolic profile of M. Sativa, M. Arborea and their hybrid (alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Parida, S.K.; Deshmukh, R.; von Wettberg, E.J.B.; Siddique, K.H.M. Ensuring Global Food Security by Improving Protein Content in Major Grain Legumes Using Breeding and ‘Omics’ Tools. Int. J. Mol. Sci. 2022, 23, 7710. [Google Scholar] [CrossRef]
- Sarrou, E.; Ganopoulos, I.; Xanthopoulou, A.; Masuero, D.; Martens, S.; Madesis, P.; Mavromatis, A.; Chatzopoulou, P. Gernetic diversity and metabolic profile of Salvia officinalis populations: Implications for advanced breeding strategies. Planta 2017, 246, 201–215. [Google Scholar] [CrossRef]
- Yves, G.; Dominique, R.; Catherine, D.; Stphane, B.; Annick, M. New Opportunities in Metabolomics and Biochemical Phenotyping for Plant Systems Biology. In Metabolomics; Roessner, U., Ed.; Intech: London, UK, 2012; pp. 213–240. [Google Scholar] [CrossRef]
- Popoola, J.O.; Aworunse, O.S.; Ojuederie, O.B.; Adewale, B.D.; Ajani, O.C.; Oyatomi, O.A.; Eruemulor, D.I.; Adegboyega, T.T.; Obembe, O.O. The Exploitation of Orphan Legumes for Food, Income, and Nutrition Security in Sub-Saharan Africa. Front Plant Sci. 2022, 13, 782140. (In English) [Google Scholar] [CrossRef] [PubMed]
- Presto, M.H.; Lyberg, K.; Lindberg, J.E. Digestibility of amino acids in organically cultivated white-flowering faba bean and cake from cold-pressed rapeseed, linseed and hemp seed in growing pigs. Arch. Anim. Nutr. 2011, 65, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Olukosi, O.A.; Walker, R.L.; Houdijk, J. Evaluation of the nutritive value of legume alternatives to soybean meal for broiler chickens. Poult. Sci. 2019, 98, 5778–5788. [Google Scholar] [CrossRef]
- Kuoppala, K.; Jaakkola, S.; Garry, B.; Ahvenjärvi, S. Effects of faba bean, blue lupin and rapeseed meal supplementation on nitrogen digestion and utilization of dairy cows fed grass silage-based diets. Animal 2021, 15, 100300. [Google Scholar] [CrossRef]
- Niderkorn, V.; Jayanegara, A. Opportunities Offered by Plant Bioactive Compounds to Improve Silage Quality, Animal Health and Product Quality for Sustainable Ruminant Production: A Review. Agronomy 2021, 11, 86. [Google Scholar] [CrossRef]
- Valente, I.M.; Cabrita, A.; Malushi, N.; Oliveira, H.M.; Papa, L.; Rodrigues, J.A.; Fonseca, A.; Maia, M. Unravelling the phytonutrients and antioxidant properties of European Vicia faba L. seeds. Food Res. Int. (Ott. Ont.) 2019, 116, 888–896. [Google Scholar] [CrossRef]
- Gefrom, A.; Ott, E.M.; Hoedtke, S.; Zeyner, A. Effect of ensiling moist field bean (Vicia faba), pea (Pisum sativum) and lupine (Lupinus spp.) grains on the contents of alkaloids, oligosaccharides and tannins. J. Anim. Physiol. Anim. Nutr. 2013, 97, 1152–1160. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Elshafei, A.A.M.; Amer, M.A.E.; Elenany, M.A.M.; Helal, A.G.A.E. Evaluation of the genetic variability of faba bean (Vicia faba L.) genotypes using agronomic traits and molecular markers. Bull. Natl. Res. Cent. 2019, 43, 106. [Google Scholar] [CrossRef]
- Backouchi, I.Z.; Aouida, M.; Khemiri, N.; Jebara, M. Genetic diversity in Tunisian populations of faba bean (Vicia faba L.) based on morphological traits and molecular markers. Gene Mol. Res. 2015, 14, 7587–7596. [Google Scholar] [CrossRef] [PubMed]
- Ouji, A.; El Bok, S.; Syed, N.H.; Abdellaoui, R.; Rouaissi, M.; Flavell, A.J.; El Gazzah, M. Genetic diversity of faba bean (Vicia faba L.) populations revealed by sequence specific amplified polymorphism (SSAP) markers. Afr. J. Biotechnol. 2012, 11, 2162–2168. [Google Scholar] [CrossRef]
- Stathi, E.; Kougioumoutzis, K.; Abraham, E.M.; Trigas, P.; Ganopoulos, I.; Avramidou, E.V.; Tani, E. Population Genetic Variability and Distribution of the Endangered Greek Endemic Cicer Graecum under Climate Change Scenarios. AoB Plants 2020, 12, plaa007. (In English) [Google Scholar] [CrossRef]
- Gresta, F.; Avola, G.; Albertini, E.; Raggi, L.; Abbate, V. A study of variability in the Sicilian faba bean landrace ‘Larga di Leonforte’. Genet. Resour. Crop Evol. 2010, 57, 523–531. [Google Scholar] [CrossRef]
- Suso, M.J.; Gilsanz, S.; Duc, G.; Marget, P.; Moreno, M.T. Germplasm Management of Faba Bean (Vicia faba L.): Monitoring Intercrossing between Accessions with Inter-plot Barriers. Genet. Resour. Crop Evol. 2006, 53, 1427–1439. [Google Scholar] [CrossRef]
- Smýkal, P.; Horáček, J.; Dostálová, R.; Hýbl, M. Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical, and morphological markers. J. Appl. Genet. 2008, 49, 155–166. [Google Scholar] [CrossRef]
- Emkani, M.; Oliete, B.; Saurel, R. Effect of Lactic Acid Fermentation on Legume Protein Properties, a Review. Fermentation 2022, 8, 244. [Google Scholar] [CrossRef]
- Lo Turco, V.; Potortì, A.G.; Rando, R.; Ravenda, P.; Dugo, G.; Di Bella, G. Functional properties and fatty acids profile of different beans varieties. Nat. Prod. Res. 2016, 30, 2243–2248. [Google Scholar] [CrossRef]
- Renna, M.; Gasmi-Boubaker, A.; Lussiana, C.; Battaglini, L.M.; Belfayez, K.; Fortina, R. Fatty acid composition of the seed oils of selected Vicia L. taxa from Tunisia. Ital. J. Anim. Sci. 2014, 13, 308–316. [Google Scholar] [CrossRef]
- Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Lakhssassi, N.; Knizia, D.; Cullen, M.A.; El Baz, A.; Embaby, M.G.; Liu, S.; Badad, O.; Vuong, T.D.; AbuGhazaleh, A.; et al. Genome-wide identification and analysis of soybean acyl-ACP thioesterase gene family reveals the role of GmFAT to improve fatty acid composition in soybean seed. Theor. Appl. Genet. 2021, 134, 3611–3623. [Google Scholar] [CrossRef] [PubMed]
- Martineau-Côté, D.; Achouri, A.; Karboune, S.; L’Hocine, L. Faba Bean: An Untapped Source of Quality Plant Proteins and Bioactives. Nutrients 2022, 14, 1541. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Abdullah, R.B.; Wan Khadijah, W.E. A review of oxalate poisoning in domestic animals: Tolerance and performance aspects. J. Anim. Physiol. Anim. Nutr. 2012, 97, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Shadi, H.; Rouzbehan, Y.; Rezaei, J.; Fazaeli, H. Yield, chemical composition, fermentation characteristics, in vitro ruminal variables, and degradability of ensiled amaranth (Amaranthus hypochondriacus) cultivars compared with corn (Zea mays) silage. Transl. Anim. Sci. 2020, 4, txaa180. [Google Scholar] [CrossRef]
- Geraldo, R.; Santos, C.S.; Pinto, E.; Vasconcelos, M.W. Widening the Perspectives for Legume Consumption: The Case of Bioactive Non-utrients. Front. Plant Sci. 2022, 13, 772054. [Google Scholar] [CrossRef]
- Avramidou, E.; Irakli, M.; Parisi, Z.; Abraham, E.; Madesis, P. Comparative Assessment of Advanced Lines and Cultivars of Faba Bean (Vicia faba L.) for Antinutritional Factors Using Molecular Markers, Joint Seminar of the FAO CIHEAM, Networks on Pasture and Forage, Crops and on Sheep and Goat, Nutrition; Book of Abstracts; FAO-CIHEAM: Catania, Italy, 2022; p. 30. [Google Scholar]
- Hou, A.; Chen, P.; Shi, A.; Zhang, B.; Wang, Y. Sugar variation in soybean seed assessed with a rapid extraction and quantification method. Int. J. Agron. 2008, 2009, 484571. [Google Scholar] [CrossRef]
- Ali, N.; Paul, S.; Gayen, D.; Sarkar, S.N.; Datta, S.K.; Datta, K. RNAi mediated down regulation of myo-inositol-3-phosphate synthase to generate low phytate rice. Rice 2013, 6, 12. [Google Scholar] [CrossRef]
- Zehring, J.; Walter, S.; Quendt, U.; Zocher, K.; Rohn, S. Phytic Acid Content of Faba Beans (Vicia faba)—Annual and Varietal Effects, and Influence of Organic Cultivation Practices. Agronomy 2022, 12, 889. [Google Scholar] [CrossRef]
- Hanieh, H.; Sakaguchi, E. Effect of D-mannitol on feed digestion and cecotrophic system in rabbits. Anim. Sci. J. 2009, 80, 157–162. [Google Scholar] [CrossRef]
- Min, X.; Li, X.; Hiura, S.; Kawasaki, K.; Xiao, J.; Sakaguchi, E. Effect of D-mannitol on nitrogen retention, fiber digestibility and digesta transit time in adult rabbits. Anim. Sci. J. 2013, 84, 551–555. [Google Scholar] [CrossRef]
- Ghomi, K.; Rabiei, B.; Sabouri, H.; Gholamalipour Alamdari, E. Association analysis, genetic diversity and population structure of barley (Hordeum vulgare L.) under heat stress conditions using SSR and ISSR markers linked to primary and secondary metabolites. Mol. Biol. Rep. 2021, 48, 6673–6694. [Google Scholar] [CrossRef]
- Tong, H.; Küken, A.; Razaghi-Moghadam, Z.; Nikoloski, Z. Characterization of effects of genetic variants via genome-scale metabolic modelling. Cell. Mol. Life Sci. 2021, 78, 5123–5138. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Peakall, R.; Smouse, E.P. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Michalakis, Y.; Excoffier, L. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 1996, 142, 1061–1064. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; WF Freeman & Co.: San Francisco, CA, USA, 1973. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Geneticps Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Diniz-Filho, J.A.; Soares, T.N.; Lima, J.S.; Dobrovolski, R.; Landeiro, V.L.; de Campos Telles, M.P.; Rangel, T.F.; Bini, L.M. Mantel test in population genetics. Genet. Mol. Biol. 2013, 36, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Hood, G.M. Poptools-Software for the Analysis of Ecological Models. Available online: http://www.cse.csiro.au/poptools (accessed on 13 February 2023).
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association Mapping in Structured Populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. POPHELPER: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2016, 17, 27–32. [Google Scholar] [CrossRef]
- Papastylianou, P.; Vlachostergios, D.N.; Dordas, C.; Tigka, E.; Papakaloudis, P.; Kargiotidou, A.; Pratsinakis, E.; Koskosidis, A.; Pankou, C.; Kousta, A.; et al. Genotype X Environment Interaction Analysis of Faba Bean (Vicia Faba L.) for Biomass and Seed Yield across Different Environments. Sustainability 2021, 13, 2586. [Google Scholar] [CrossRef]
- Kostopoulou, S.; Ntatsi, G.; Arapis, G.; Aliferis, K.A. Assessment of the Effects of Metribuzin, Glyphosate, and Their Mixtures on the Metabolism of the Model Plant Lemna Minor L. Applying Metabolomics. Chemosphere 2020, 239, 124582. (In English) [Google Scholar] [CrossRef]
- Papadopoulou, E.; Nicolescu, A.; Haug, L.S.; Husøy, T.; Deleanu, C.; Dirven, H.; Lindeman, B. Lipoprotein profiles associated with exposure to poly- and perfluoroalkyl substances (PFASs) in the EuroMix human biomonitoring study. Environ. Pollut. 2022, 308, 119664. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheyst, J.; Fiehn, O.; Arita, M. Ms-Dial: Data-Independent Ms/Ms Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
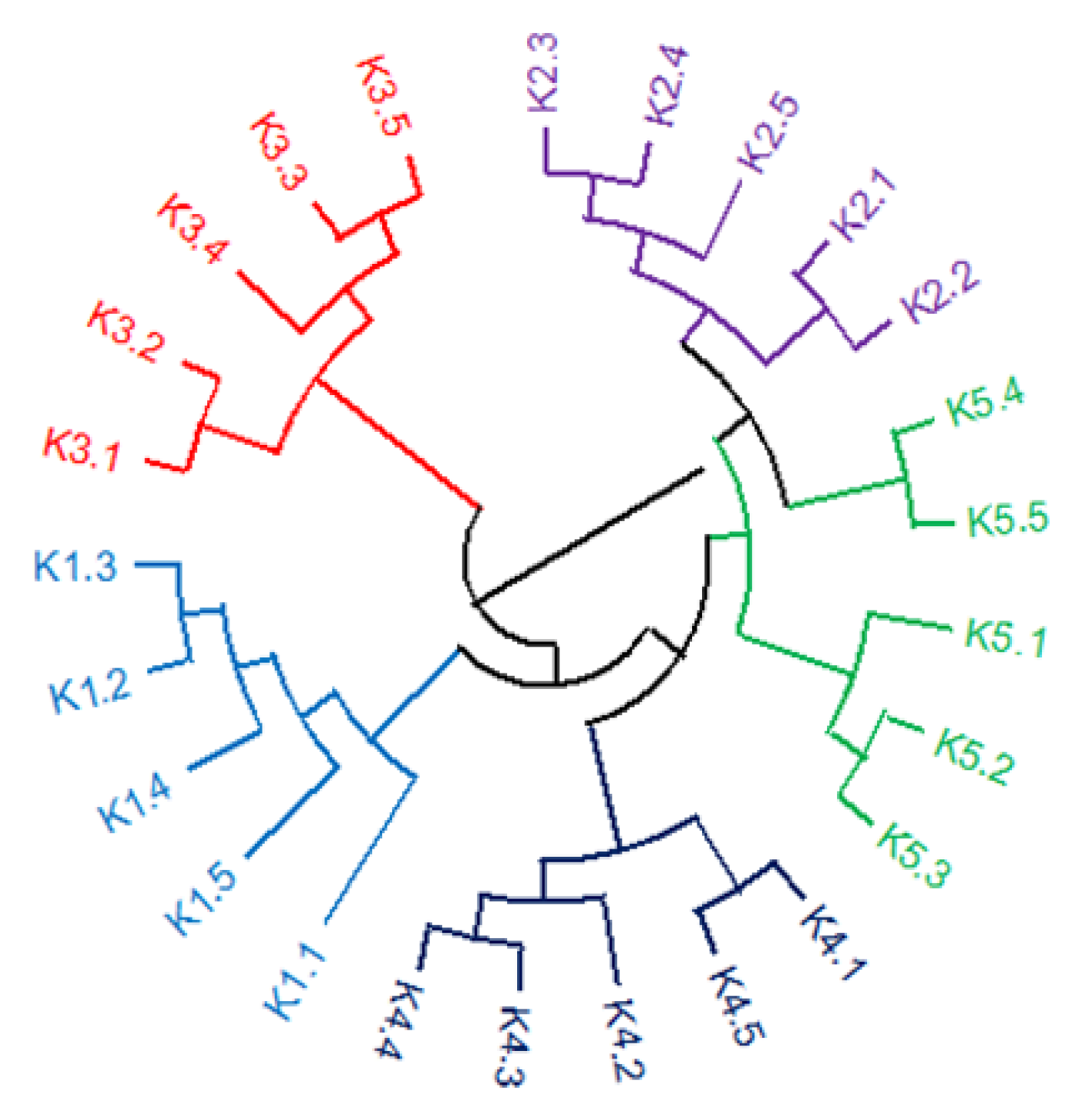

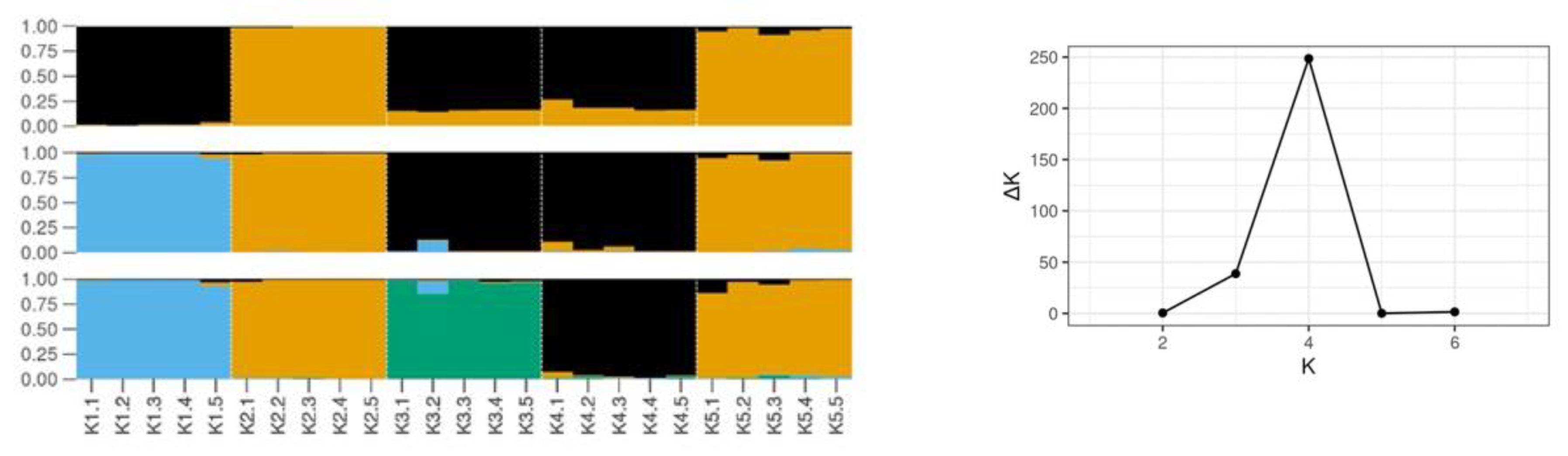
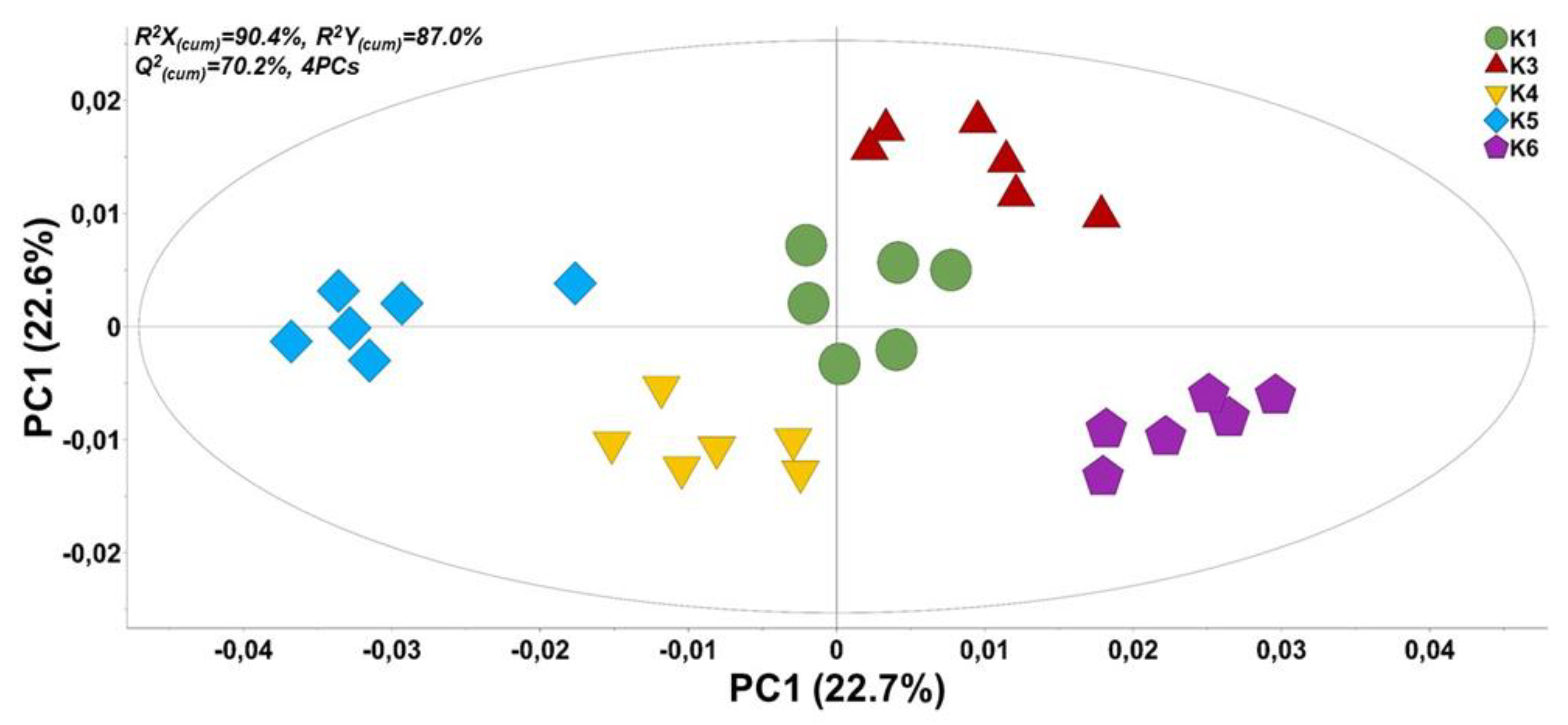
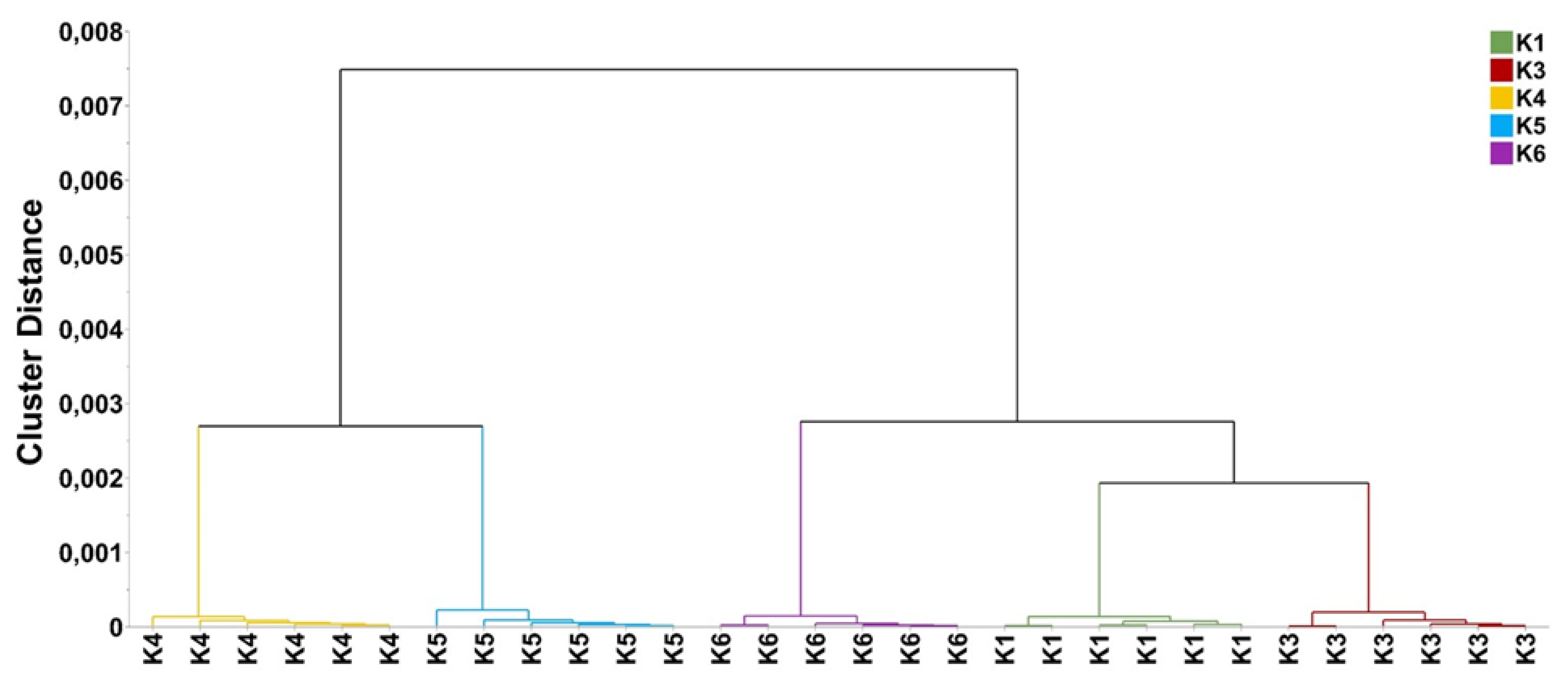
| Populations | N (1) | NPB | No. Private Bands | P (%) | Shannon Index (I) | GD |
|---|---|---|---|---|---|---|
| ΚΚ18 | 5 | 88 | 5 | 51.46 | 0.319 | 0.222 |
| TANAGRA | 5 | 78 | 0 | 50.49 | 0.300 | 0.205 |
| POLIKARPI | 5 | 81 | 4 | 66.02 | 0.392 | 0.269 |
| ΚΚ14 | 5 | 75 | 0 | 62.14 | 0.373 | 0.256 |
| ΚΚ10 | 5 | 83 | 1 | 61.17 | 0.370 | 0.255 |
| Mean | 5 | 81 | 58.25 | 0.351 | 0.241 | |
| Species level | 25 |
| Source | Df (1) | SS (2) | MS (3) | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 4 | 202.080 | 50.520 | 6.996 | 31% |
| Within Pops | 20 | 310.800 | 15.540 | 15.540 | 69% |
| Total | 24 | 512.880 | 22.536 | 100% | |
| Stat | Value | p (rand >= data) | |||
| PhiPT | 0.310 | 0.001 | |||
| KK18 | TANAGRA | POLIKARPI | KK14 | KK10 | |
|---|---|---|---|---|---|
| 0.000 | KK18 | ||||
| 0.389 | 0.000 | TANAGRA | |||
| 0.357 | 0.355 | 0.000 | POLIKARPI | ||
| 0.344 | 0.322 | 0.265 | 0.000 | KK14 | |
| 0.321 | 0.169 | 0.301 | 0.244 | 0.000 | KK10 |
| Metabolites | Relative Concentration (×10−3 mol/L) | ||||
|---|---|---|---|---|---|
| KK18 | KK10 | KK14 | POLIKARPI | TANAGRA | |
| a-a-trehalose | 771.08 a | 719.19 b | 677.44 c | 758.64 a | 721.56 b |
| Palmitic acid | 24.98 c | 29.51 ab | 30.25 a | 28.32 ab | 28.64 ab |
| Oxalic acid | 18.51 b | 24.79 ab | 28.57 a | 25.66 ab | 18.99 b |
| D-myo-inositol phosphate | 15.21 b | 17.80 ab | 16.80 ab | 16.06 ab | 18.83 a |
| Stearic acid | 12.08 c | 16.41 a | 14.22 b | 15.07 ab | 13.50 bc |
| L-lactic acid | 5.42 a | 3.77 bc | 3.20 c | 2.98 c | 4.75 ab |
| Myo-inositol | 4.06 b | 4.72 a | 3.46 bc | 3.81 b | 2.95 c |
| D-Gluconic acid | 3.76 b | 5.18 a | 3.45 bc | 2.44 c | 3.44 bc |
| Malic acid | 1.93 c | 1.32 c | 3.11 b | 1.53 c | 4.48 a |
| D-Mannitol | 1.91 b | 3.17 a | 2.86 a | 1.52 b | 3.25 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avramidou, E.; Sarri, E.; Ganopoulos, I.; Madesis, P.; Kougiteas, L.; Papadopoulou, E.-A.; Aliferis, K.A.; Abraham, E.M.; Tani, E. Genetic and Metabolite Variability among Commercial Varieties and Advanced Lines of Vicia faba L. Plants 2023, 12, 908. https://doi.org/10.3390/plants12040908
Avramidou E, Sarri E, Ganopoulos I, Madesis P, Kougiteas L, Papadopoulou E-A, Aliferis KA, Abraham EM, Tani E. Genetic and Metabolite Variability among Commercial Varieties and Advanced Lines of Vicia faba L. Plants. 2023; 12(4):908. https://doi.org/10.3390/plants12040908
Chicago/Turabian StyleAvramidou, Eleni, Efi Sarri, Ioannis Ganopoulos, Panagiotis Madesis, Leonidas Kougiteas, Evgenia-Anna Papadopoulou, Konstantinos A. Aliferis, Eleni M. Abraham, and Eleni Tani. 2023. "Genetic and Metabolite Variability among Commercial Varieties and Advanced Lines of Vicia faba L." Plants 12, no. 4: 908. https://doi.org/10.3390/plants12040908
APA StyleAvramidou, E., Sarri, E., Ganopoulos, I., Madesis, P., Kougiteas, L., Papadopoulou, E.-A., Aliferis, K. A., Abraham, E. M., & Tani, E. (2023). Genetic and Metabolite Variability among Commercial Varieties and Advanced Lines of Vicia faba L. Plants, 12(4), 908. https://doi.org/10.3390/plants12040908









