Regulation of Reactive Oxygen Species during Salt Stress in Plants and Their Crosstalk with Other Signaling Molecules—Current Perspectives and Future Directions
Abstract
1. Introduction
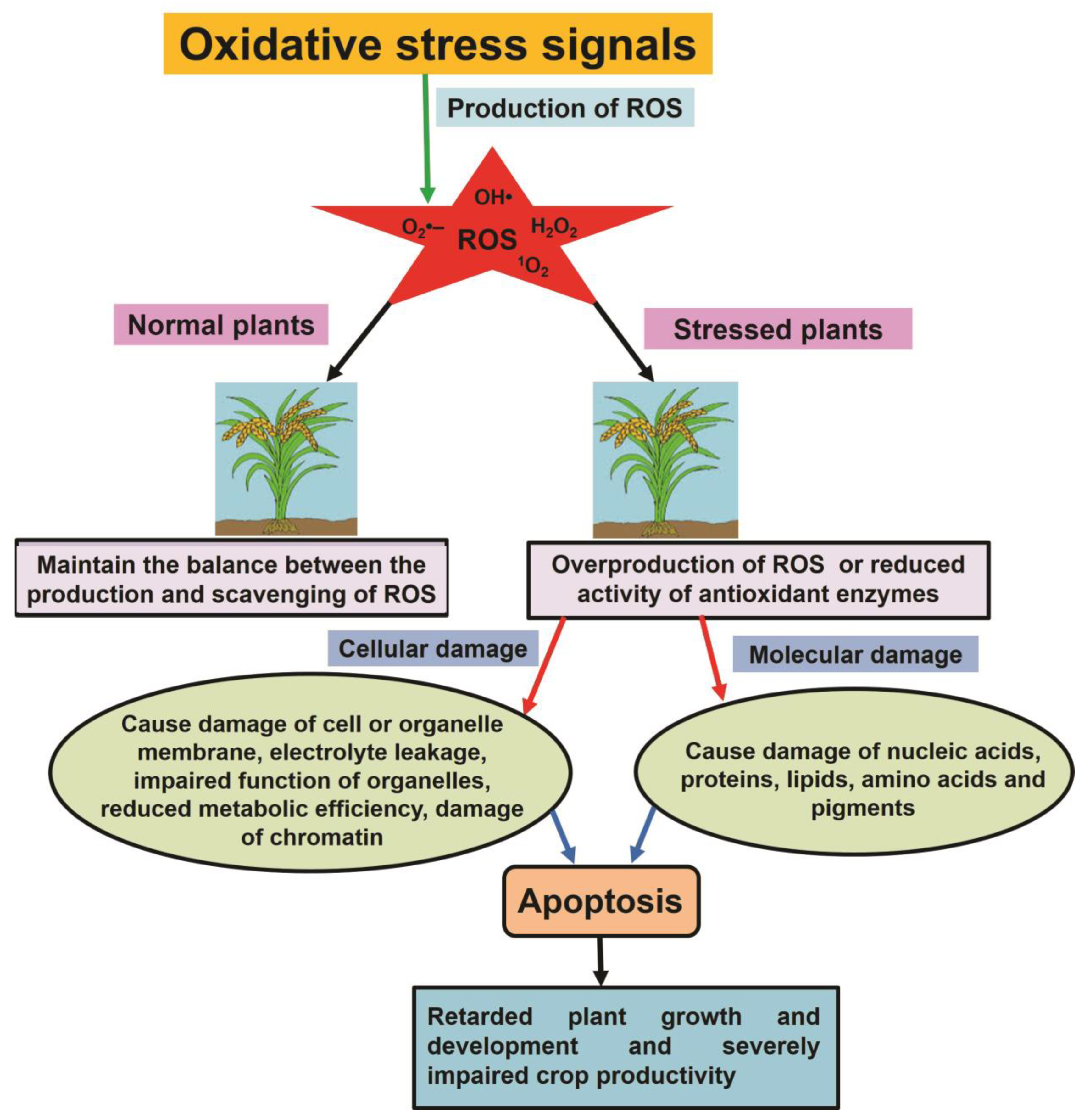
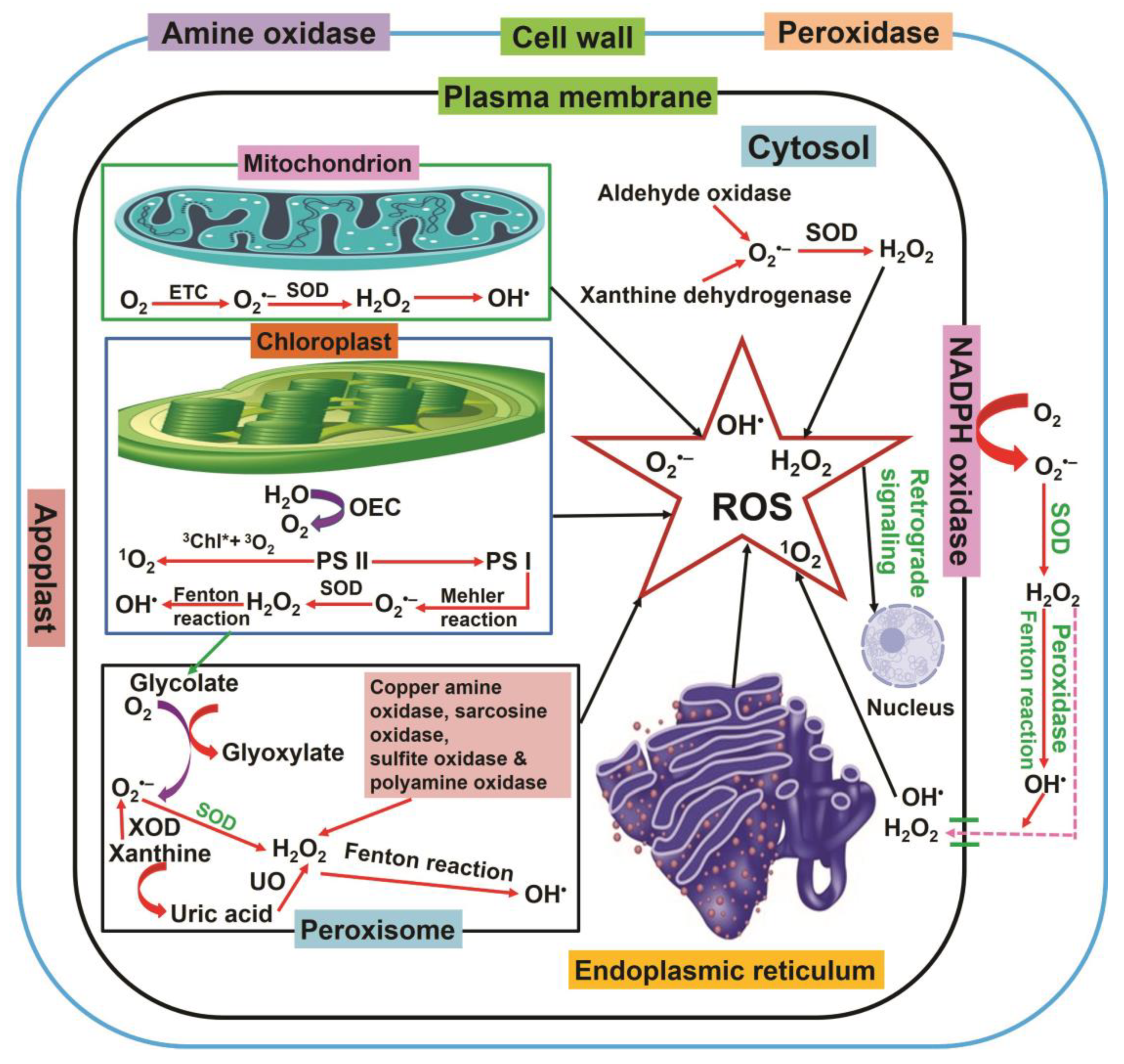
2. Production of ROS in Plant Cells
3. Role of Antioxidants in Oxidative Stress
3.1. Enzymatic Antioxidants
3.1.1. SOD
- O2•−+ SOD − M2+ → O2 + M+--------Step 1
- O2•−+ 2H+-SOD − M → H2O2 + M2+-------Step 2
- O2•−+ O2•−+ 2H+ → 2H2O2 + O2
3.1.2. CAT
- RH2 + H2O2 → R + 2H2O
- CAT-Fe-OOH + C2H5OH → CAT-Fe-OH + H2O + CH3CHO (peroxidatic reaction)
- CAT-Fe-OOH + H2O2 → CAT-Fe-OH + H2O + O2 (catalytic reaction)
3.1.3. APX
- H2O2 + AA→2H2O + DHA
To provide significant protection during oxidative stress, the enzymatic and non-enzymatic antioxidant molecules closely coordinate their actions. By keeping the redox equilibrium under stress, APX connects the two pathways. Furthermore, it has a particular affinity for H2O2 [71]. Plants lacking APX1 have delayed growth and development. In the absence of APX1, H2O2 levels increase and cause aberrant stomatal closure [73]. In APX1-deficient plants, light stress results in H2O2-mediated-enhanced activation of heat shock proteins [51,74]. Furthermore, APX and CAT at least partially complement each other’s shortcomings [75]. Moreover, the activities of SOD, CAT, reduced glutathione (GSH) reductase, and APX typically increase in response to various environmental stresses [72].
3.1.4. MDHAR
- MDHA + NADPH→AA + NADP+
3.1.5. DHAR
- DHA + 2GSH→AA + GSSG
3.1.6. GR
- GSSG + NADPH → 2GSH + NADP+
3.1.7. GPX
- H2O2 + GSH → H2O + GSSG
3.2. Non-Enzymatic Antioxidants
3.2.1. AA
3.2.2. GSH
3.2.3. α-Tocopherol
3.2.4. Carotenoids
3.2.5. Flavonoids
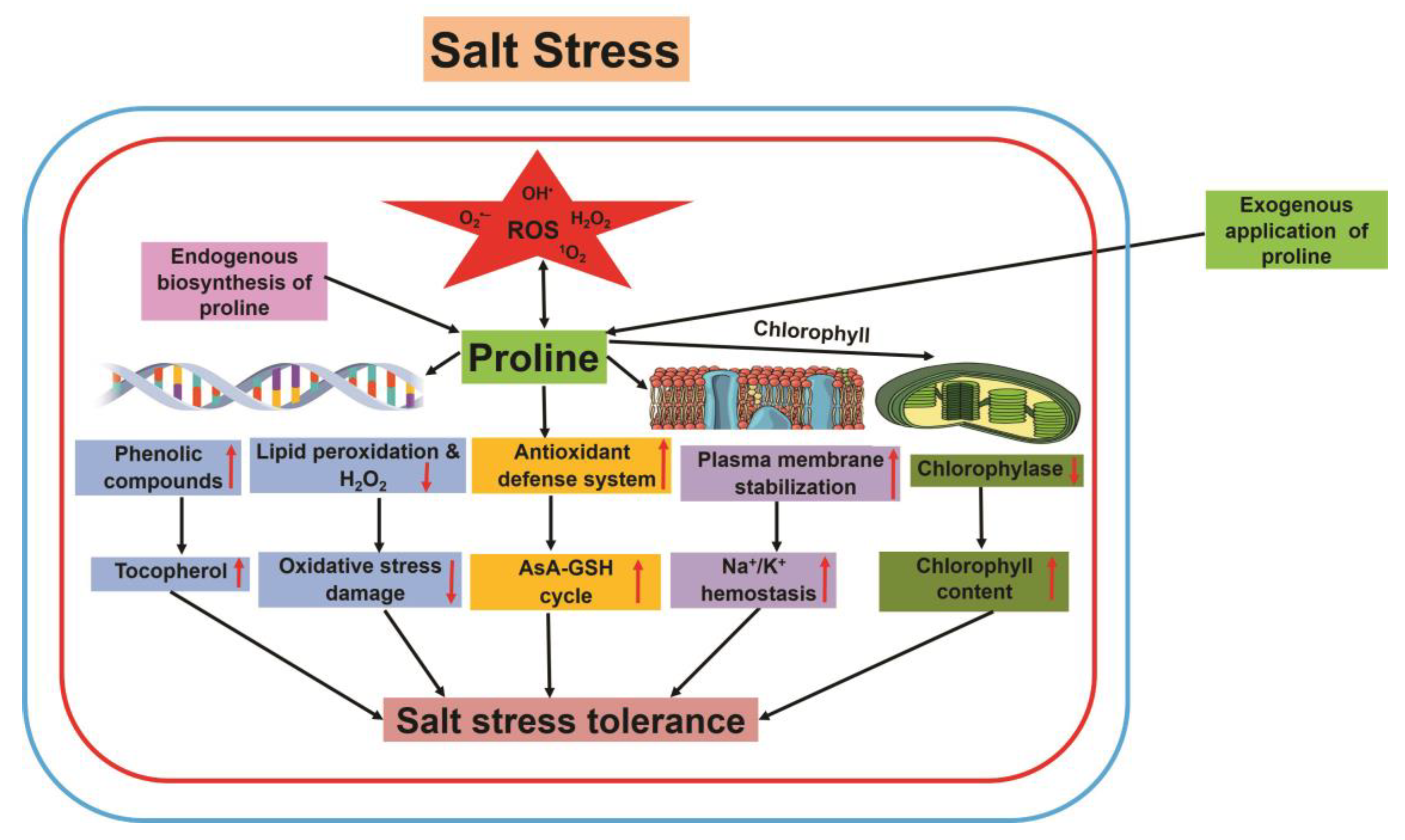
3.2.6. Proline
4. Downstream Redox Signaling during Oxidative Stress in Plants
5. Oxidative Stress under Salt Stress
6. Antioxidant Defense System in Plants under High Salinity Stress
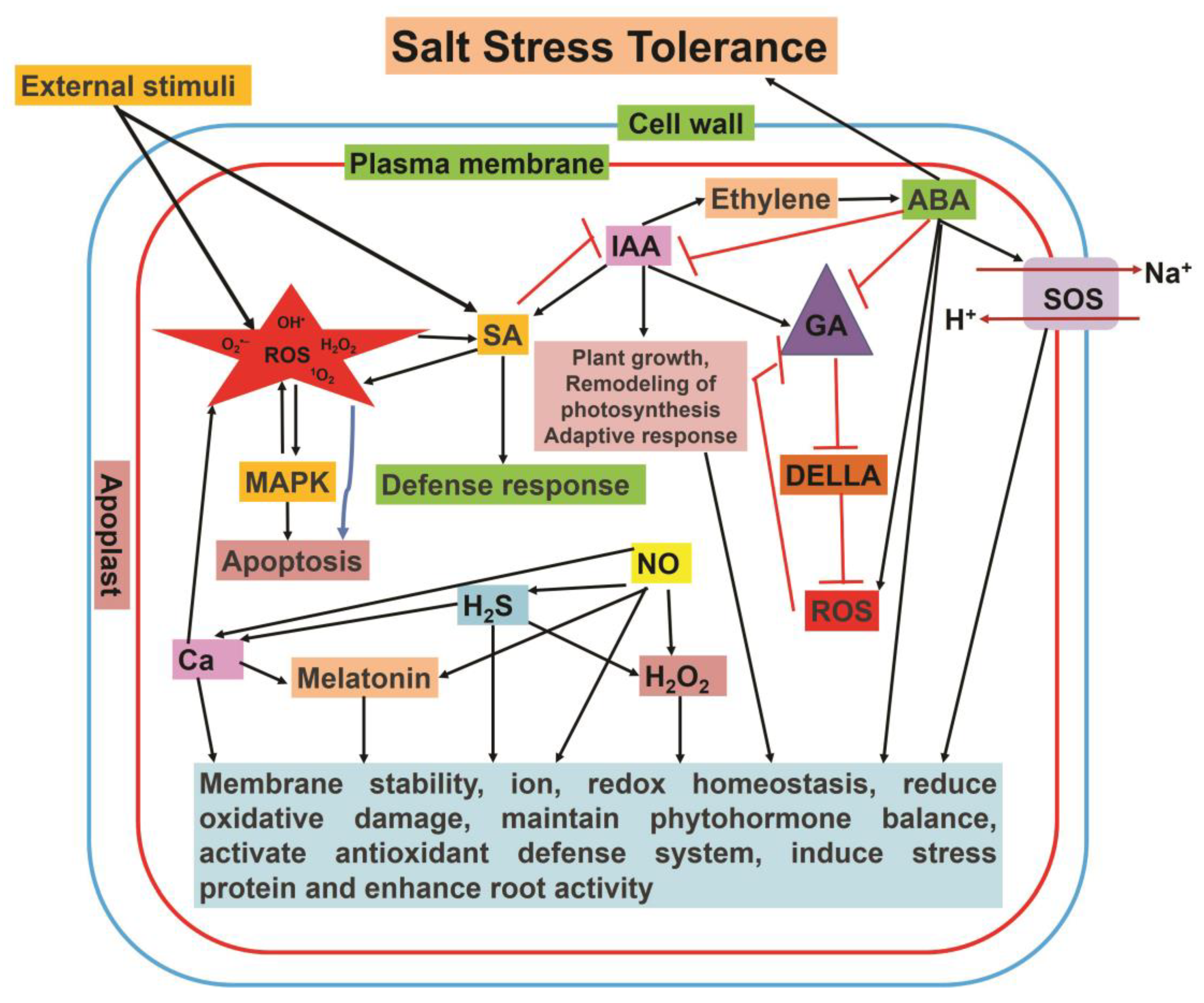
7. Crosstalk between Different Signaling Molecules during Salt Stress
7.1. H2O2
7.2. NO
7.3. ROS
7.4. H2S
7.5. Ca2+
7.6. Phytohormones
8. Transgenic Approach to Improve the Antioxidant System in Plants under Salt Stress
| Gene Name | Source of Genes | Transgenic Plants | Stress Characteristics (Concentration of Salt Solution and Duration of Exposure) | Mode of Action | Reference |
|---|---|---|---|---|---|
| DgNAC1 | Dendronthema grandiflorum | Chrysanthemum | 100, 200, and 400 mM NaCl; 1, 5, 10, 15 days | Reduced levels of O2−•, H2O2, and MDA; increased activities of CAT, POD, and SOD | [268] |
| RaAPX and PaSOD | Rheum australe and Potentilla atrosanguinea | Solanum tuberosum | 0, 50, 100, and 150 mM NaCl; 7 and 15 days | Enhanced activities of SOD and APX in both the transgenic variants of potatoes | [269] |
| OsMYB6 | Oryza sativa | Oryza sativa | 150 mM NaCl; 6 days | Increased concentration of proline, elevated activities of SOD and CAT, decreased content of MDA and REL | [270] |
| StCYS1 in | Solanum tuberosum | Solanum tuberosum | 0.17 mol/L NaCl; 0, 3, 5, and 7 days | Enhanced accumulation of proline and increased scavenging of H2O2 | [271] |
| OsSTAP1 | Oryza sativa | Oryza sativa | 150 mM NaCl; 5 days | Increased activities of SOD, POD, and CAT | [272] |
| DnWRKY11 | Dendrobium nobile | Nicotiana tabacum | 200 mM NaCl; 20 days | Enhanced activities of SOD, CAT, POD; reduced content of MDA | [273] |
| GmMYB84 | Glycine max | Glycine max | 150 and 200 mM NaCl; until the seed germination | Higher activities of antioxidant enzymes POD, CAT, and SOD and accumulation of proline | [274] |
| OsEXPA7 | Oryza sativa | Oryza sativa | 150 mM NaCl; two to three weeks | Increased activity of SOD and POD, reduced accumulation of ROS and MDA, increased accumulation of proline | [275] |
| MsWRKY11 | Medicago sativa | Glycine max | 100 and 200 mM NaCl; 7 days | Increased contents of soluble sugar, chlorophyll, proline; enhanced activities of CAT and SOD; reduced contents of O2−•, H2O2, MDA | [276] |
| nbexo70d1 | Nicotiana benthamiana | Nicotiana benthamiana | 100, 200, and 300 mM NaCl; 5 days | Declined accumulation of ROS and decreased activity of NADPH oxidase | [277] |
| VvIAA18 | Vitis vinifera | Nicotiana tabacum | 200 mM NaCl; every 2 days for 8 weeks | Induced the expression of salt stress-responsive genes LEA5, P5CS, POD, and SOD; increased activities of POD and SOD | [278] |
| VvWRKY30 | Vitis vinifera | Arabidopsis thaliana | 150 mM NaCl; 3, 6, 9, 12, and 24 h | Increased activities of antioxidant enzymes CAT, SOD, and POD | [279] |
| SsMAX2 | Sapium sebiferum | Arabidopsis thaliana | 100 and 150 mM NaCl; 15-day-old seedlings | Enhanced activities of SOD, POD, and APX | [280] |
| MfWRKY70 | Myrothamnus flabellifolia | Arabidopsis thaliana | 50, 100, or 150 mM NaCl; treated to seed | Reduced levels of H2O2 and differential activities of POD, SOD, and CAT | [281] |
| PeHSF | Populus euphratica | Nicotiana tabacum | 150 mM NaCl; 1, 8, 15, and 23 days | Increased activities of APX, GPX, and GSH | [282] |
| ThHSFA1 | Tamarix hispida | Populus trichocarpa | 200 mM NaCl; every 2 days for 10 days | Reduced levels of ROS, increased activities of antioxidant enzymes | [283] |
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, K.; Kumar, M.; Kim, S.-R.; Ryu, H.; Cho, Y.-G. Insights into genomics of salt stress response in rice. Rice 2013, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Ali, A. Genome-wide identification and characterization of the brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Routray, S.; Mohapatra, C.; Saha, D.; Ram, C.; Siddique, K.H.; Kumar, A. Genome-Wide Analysis and Characterization of the Proline-Rich Extensin-like Receptor Kinases (PERKs) Gene Family Reveals Their Role in Different Developmental Stages and Stress Conditions in Wheat (Triticum aestivum L.). Plants 2022, 11, 496. [Google Scholar] [CrossRef]
- Kumar, M.; Kesawat, M.S.; Ali, A.; Lee, S.-C.; Gill, S.S.; Kim, H.U. Integration of abscisic acid signaling with other signaling pathways in plant stress responses and development. Plants 2019, 8, 592. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651. [Google Scholar] [CrossRef]
- Volkov, V.; Beilby, M.J. Salinity Tolerance in Plants: Mechanisms and Regulation of Ion Transport. Front. Plant Sci. 2017, 8, 1795. [Google Scholar]
- Ashraf, M. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 2009, 27, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Singhal, R.K.; Saha, D.; Skalicky, M.; Mishra, U.N.; Chauhan, J.; Behera, L.P.; Lenka, D.; Chand, S.; Kumar, V.; Dey, P. Crucial cell signaling compounds crosstalk and integrative multi-omics techniques for salinity stress tolerance in plants. Front. Plant Sci. 2021, 12, 670369. [Google Scholar] [CrossRef] [PubMed]
- Mehla, N.; Sindhi, V.; Josula, D.; Bisht, P.; Wani, S.H. An introduction to antioxidants and their roles in plant stress tolerance. In Reactive Oxygen Species and Antioxidant Systems in Plants: Role and Regulation under Abiotic Stress; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–23. [Google Scholar]
- Hasanuzzaman, M.; Bhuyan, M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, A.; Yadav, S.; Singh, I.K. Reactive oxygen species-mediated signaling during abiotic stress. Plant Gene 2019, 18, 100173. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Hossain, M.A.; Silva, J.A.; Fujita, M. Plant response and tolerance to abiotic oxidative stress: Antioxidant defense is a key factor. In Crop Stress and Its Management: Perspectives and Strategies; Springer: Berlin/Heidelberg, Germany, 2012; pp. 261–315. [Google Scholar]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Kaur, N.; Kaur, J.; Grewal, S.K.; Singh, I. Effect of heat stress on antioxidative defense system and its amelioration by heat acclimation and salicylic acid pre-treatments in three pigeonpea genotypes. Indian J. Agric. Biochem. 2019, 32, 106–110. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Antoniou, C.; Savvides, A.; Christou, A.; Fotopoulos, V. Unravelling chemical priming machinery in plants: The role of reactive oxygen–nitrogen–sulfur species in abiotic stress tolerance enhancement. Curr. Opin. Plant Biol. 2016, 33, 101–107. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016, 171, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Kohli, S.K.; Khanna, K.; Bhardwaj, R.; Abd_Allah, E.F.; Ahmad, P.; Corpas, F.J. Assessment of subcellular ROS and NO metabolism in higher plants: Multifunctional signaling molecules. Antioxidants 2019, 8, 641. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J. Thiol-based peroxidases and ascorbate peroxidases: Why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol. Cells 2016, 39, 20. [Google Scholar] [PubMed]
- Dogra, V.; Kim, C. Singlet oxygen metabolism: From genesis to signaling. Front. Plant Sci. 2020, 10, 1640. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.; Li, X.-P.; Niyogi, K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K.K. Sensing and responding to excess light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef] [PubMed]
- Krieger-Liszkay, A. Singlet oxygen production in photosynthesis. J. Exp. Bot. 2005, 56, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Krieger-Liszkay, A.; Fufezan, C.; Trebst, A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 2008, 98, 551–564. [Google Scholar] [CrossRef]
- Flors, C.; Fryer, M.J.; Waring, J.; Reeder, B.; Bechtold, U.; Mullineaux, P.M.; Nonell, S.; Wilson, M.T.; Baker, N.R. Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green®. J. Exp. Bot. 2006, 57, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Triantaphylidès, C.; Havaux, M. Singlet oxygen in plants: Production, detoxification and signaling. Trends Plant Sci. 2009, 14, 219–228. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef]
- Kerchev, P.; Waszczak, C.; Lewandowska, A.; Willems, P.; Shapiguzov, A.; Li, Z.; Alseekh, S.; Mühlenbock, P.; Hoeberichts, F.A.; Huang, J. Lack of GLYCOLATE OXIDASE1, but not GLYCOLATE OXIDASE2, attenuates the photorespiratory phenotype of CATALASE2-deficient Arabidopsis. Plant Physiol. 2016, 171, 1704–1719. [Google Scholar] [CrossRef]
- Del Río, L.A.; López-Huertas, E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016, 57, 1364–1376. [Google Scholar] [CrossRef]
- Reumann, S.; Chowdhary, G.; Lingner, T. Characterization, prediction and evolution of plant peroxisomal targeting signals type 1 (PTS1s). Biochim. Biophys. Acta BBA-Mol. Cell Res. 2016, 1863, 790–803. [Google Scholar] [CrossRef]
- Corpas, F.J.; Del Río, L.A.; Palma, J.M. Plant peroxisomes at the crossroad of NO and H2O2 metabolism. J. Integr. Plant Biol. 2019, 61, 803–816. [Google Scholar]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, calcium, and electric signals: Key mediators of rapid systemic signaling in plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant peroxisomes: A factory of reactive species. Front. Plant Sci. 2020, 11, 853. [Google Scholar] [CrossRef]
- Lisenbee, C.S.; Lingard, M.J.; Trelease, R.N. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 2005, 43, 900–914. [Google Scholar] [CrossRef]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; del Río, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef]
- Choudhary, A.; Kumar, A.; Kaur, N. ROS and oxidative burst: Roots in plant development. Plant Divers. 2020, 42, 33–43. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Heyno, E.; Mary, V.; Schopfer, P.; Krieger-Liszkay, A. Oxygen activation at the plasma membrane: Relation between superoxide and hydroxyl radical production by isolated membranes. Planta 2011, 234, 35–45. [Google Scholar] [CrossRef]
- Jeevan Kumar, S.; Rajendra Prasad, S.; Banerjee, R.; Thammineni, C. Seed birth to death: Dual functions of reactive oxygen species in seed physiology. Ann. Bot. 2015, 116, 663–668. [Google Scholar] [CrossRef]
- Janků, M.; Luhová, L.; Petřivalský, M. On the origin and fate of reactive oxygen species in plant cell compartments. Antioxidants 2019, 8, 105. [Google Scholar] [CrossRef]
- Maurya, A.K. Oxidative stress in crop plants. In Agronomic Crops; Springer: Berlin/Heidelberg, Germany, 2020; pp. 349–380. [Google Scholar]
- Camejo, D.; Guzmán-Cedeño, Á.; Moreno, A. Reactive oxygen species, essential molecules, during plant–pathogen interactions. Plant Physiol. Biochem. 2016, 103, 10–23. [Google Scholar] [CrossRef]
- Dumont, S.; Rivoal, J. Consequences of oxidative stress on plant glycolytic and respiratory metabolism. Front. Plant Sci. 2019, 10, 166. [Google Scholar] [CrossRef]
- Martinez, V.; Mestre, T.C.; Rubio, F.; Girones-Vilaplana, A.; Moreno, D.A.; Mittler, R.; Rivero, R.M. Accumulation of flavonols over hydroxycinnamic acids favors oxidative damage protection under abiotic stress. Front. Plant Sci. 2016, 7, 838. [Google Scholar] [CrossRef]
- Karuppanapandian, T.; Kim, W. Cobalt-induced oxidative stress causes growth inhibition associated with enhanced lipid peroxidation and activates antioxidant responses in Indian mustard (Brassica juncea L.) leaves. Acta Physiol. Plant 2013, 35, 2429–2443. [Google Scholar] [CrossRef]
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Dat, J.F.; Foyer, C.H.; Scott, I.M. Changes in salicylic acid and antioxidants during induced thermotolerance in mustard seedlings. Plant Physiol. 1998, 118, 1455–1461. [Google Scholar] [CrossRef]
- Gepstein, S.; Glick, B.R. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 2013, 82, 623–633. [Google Scholar] [CrossRef]
- Corpas, F.J.; Fernandez-Ocana, A.; Carreras, A.; Valderrama, R.; Luque, F.; Esteban, F.J.; Rodríguez-Serrano, M.; Chaki, M.; Pedrajas, J.R.; Sandalio, L.M. The expression of different superoxide dismutase forms is cell-type dependent in olive (Olea europaea L.) leaves. Plant Cell Physiol. 2006, 47, 984–994. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Pazmino, D.M.; Testillano, P.S.; Risueño, M.C.; Del Río, L.A.; Sandalio, L.M. Cellular response of pea plants to cadmium toxicity: Cross talk between reactive oxygen species, nitric oxide, and calcium. Plant Physiol. 2009, 150, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Boguszewska, D.; Grudkowska, M.; Zagdańska, B. Drought-responsive antioxidant enzymes in potato (Solanum tuberosum L.). Potato Res. 2010, 53, 373–382. [Google Scholar] [CrossRef]
- Nicholls, P.; Fita, I.; Loewen, P.C. Enzymology and Structure of Catalases. Adv Inorg Chem 2000, 51, 51–106. [Google Scholar]
- Zámocký, M.; Gasselhuber, B.; Furtmüller, P.G.; Obinger, C. Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys. 2012, 525, 131–144. [Google Scholar] [CrossRef]
- Loewen, P.C.; Klotz, M.G.; Hassett, D.J. Catalase—An “old” enzyme that continues to surprise us. ASM News 2000, 66, 76–82. [Google Scholar]
- Mhamdi, A.; Hager, J.; Chaouch, S.; Queval, G.; Han, Y.; Taconnat, L.; Saindrenan, P.; Gouia, H.; Issakidis-Bourguet, E.; Renou, J.-P. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010, 153, 1144–1160. [Google Scholar] [CrossRef]
- Ogren, W.L. Photorespiration: Pathways, regulation, and modification. Annu. Rev. Plant Physiol. 1984, 35, 415–442. [Google Scholar] [CrossRef]
- Andre, C.; Kim, S.W.; Yu, X.-H.; Shanklin, J. Fusing catalase to an alkane-producing enzyme maintains enzymatic activity by converting the inhibitory byproduct H2O2 to the cosubstrate O2. Proc. Natl. Acad. Sci. USA 2013, 110, 3191–3196. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Braus-Stromeyer, S.A.; Timpner, C.; Valerius, O.; von Tiedemann, A.; Karlovsky, P.; Druebert, C.; Polle, A.; Braus, G.H. The plant host Brassica napus induces in the pathogen Verticillium longisporum the expression of functional catalase peroxidase which is required for the late phase of disease. Mol. Plant Microbe Interact. 2012, 25, 569–581. [Google Scholar] [CrossRef]
- Scandalios, J.G. Regulation and properties of plant catalases. In Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants; CRC Press: Boca Raton, FL, USA, 2019; pp. 275–316. [Google Scholar]
- Sharma, I.; Ahmad, P. Catalase: A versatile antioxidant in plants. In Oxidative Damage to Plants; Elsevier: Amsterdam, The Netherlands, 2014; pp. 131–148. [Google Scholar]
- Gondim, F.A.; Gomes-Filho, E.; Costa, J.H.; Alencar, N.L.M.; Prisco, J.T. Catalase plays a key role in salt stress acclimation induced by hydrogen peroxide pretreatment in maize. Plant Physiol. Biochem. 2012, 56, 62–71. [Google Scholar] [CrossRef]
- Kerdnaimongkol, K.; Woodson, W.R. Inhibition of catalase by antisense RNA increases susceptibility to oxidative stress and chilling injury in transgenic tomato plants. J. Am. Soc. Hortic. Sci. 1999, 124, 330–336. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic stress tolerance in plants: Myriad roles of ascorbate peroxidase. Front. Plant Sci. 2017, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Liang, H.; Rozenberg, M.; Mittler, R. Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J. 2003, 34, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.; Desikan, R.; Hancock, J. Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 2002, 5, 388–395. [Google Scholar] [CrossRef]
- Vandenabeele, S.; Vanderauwera, S.; Vuylsteke, M.; Rombauts, S.; Langebartels, C.; Seidlitz, H.K.; Zabeau, M.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 2004, 39, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Young, T.E.; Ling, J.; Chang, S.-C.; Gallie, D.R. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc. Natl. Acad. Sci. USA 2003, 100, 3525–3530. [Google Scholar] [CrossRef]
- Mittova, V.; Volokita, M.; Guy, M.; Tal, M. Activities of SOD and the ascorbate-glutathione cycle enzymes in subcellular compartments in leaves and roots of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol. Plant. 2000, 110, 42–51. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Shibahara, T.; Inanaga, S.; Tanaka, K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef]
- Hossain, M.A.; Fujita, M. Evidence for a role of exogenous glycinebetaine and proline in antioxidant defense and methylglyoxal detoxification systems in mung bean seedlings under salt stress. Physiol. Mol. Biol. Plants 2010, 16, 19–29. [Google Scholar] [CrossRef]
- Chen, Z.; Gallie, D.R. The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 2004, 16, 1143–1162. [Google Scholar] [CrossRef]
- Noshi, M.; Hatanaka, R.; Tanabe, N.; Terai, Y.; Maruta, T.; Shigeoka, S. Redox regulation of ascorbate and glutathione by a chloroplastic dehydroascorbate reductase is required for high-light stress tolerance in Arabidopsis. Biosci. Biotechnol. Biochem. 2016, 80, 870–877. [Google Scholar] [CrossRef]
- Rahantaniaina, M.-S.; Li, S.; Chatel-Innocenti, G.; Tuzet, A.; Issakidis-Bourguet, E.; Mhamdi, A.; Noctor, G. Cytosolic and chloroplastic DHARs cooperate in oxidative stress-driven activation of the salicylic acid pathway. Plant Physiol. 2017, 174, 956–971. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Wang, B.; Han, Y.; Li, S. The pivotal function of dehydroascorbate reductase in glutathione homeostasis in plants. J. Exp. Bot. 2020, 71, 3405–3416. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, T.; Yokota, A.; Miyake, C. Purification and characterization of chloroplast dehydroascorbate reductase from spinach leaves. Plant Cell Physiol. 2000, 41, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.; Shi, Q.; Yu, X. Ascorbic acid contents in transgenic potato plants overexpressing two dehydroascorbate reductase genes. Mol. Biol. Rep. 2011, 38, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Sanmartin, M.; Drogoudi, P.D.; Lyons, T.; Pateraki, I.; Barnes, J.; Kanellis, A.K. Over-expression of ascorbate oxidase in the apoplast of transgenic tobacco results in altered ascorbate and glutathione redox states and increased sensitivity to ozone. Planta 2003, 216, 918–928. [Google Scholar] [CrossRef]
- Dardalhon, M.; Kumar, C.; Iraqui, I.; Vernis, L.; Kienda, G.; Banach-Latapy, A.; He, T.; Chanet, R.; Faye, G.; Outten, C.E. Redox-sensitive YFP sensors monitor dynamic nuclear and cytosolic glutathione redox changes. Free Radic. Biol. Med. 2012, 52, 2254–2265. [Google Scholar] [CrossRef]
- Rao, A.; Reddy, A.R. Glutathione reductase: A putative redox regulatory system in plant cells. In Sulfur Assimilation and Abiotic Stress in Plants; Springer: Berlin/Heidelberg, Germany, 2008; pp. 111–147. [Google Scholar]
- Bass, R.; Ruddock, L.W.; Klappa, P.; Freedman, R.B. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J. Biol. Chem. 2004, 279, 5257–5262. [Google Scholar] [CrossRef]
- Raturi, A.; Mutus, B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic. Biol. Med. 2007, 43, 62–70. [Google Scholar] [CrossRef]
- Kubo, A.; Sano, T.; Saji, H.; Tanaka, K.; Kondo, N.; Tanaka, K. Primary structure and properties of glutathione reductase from Arabidopsis thaliana. Plant Cell Physiol. 1993, 34, 1259–1266. [Google Scholar]
- Creissen, G.P.; Mullineaux, P.M. Cloning and characterisation of glutathione reductase cDNAs and identification of two genes encoding the tobacco enzyme. Planta 1995, 197, 422–425. [Google Scholar] [CrossRef]
- Kaminaka, H.; Morita, S.; Nakajima, M.; Masumura, T.; Tanaka, K. Gene cloning and expression of cytosolic glutathione reductase in rice (Oryza sativa L.). Plant Cell Physiol. 1998, 39, 1269–1280. [Google Scholar] [CrossRef]
- Contour-Ansel, D.; Torres-Franklin, M.L.; Cruz De Carvalho, M.H.; D’Arcy-Lameta, A.; Zuily-Fodil, Y. Glutathione reductase in leaves of cowpea: Cloning of two cDNAs, expression and enzymatic activity under progressive drought stress, desiccation and abscisic acid treatment. Ann. Bot. 2006, 98, 1279–1287. [Google Scholar] [CrossRef]
- Yousuf, P.Y.; Hakeem, K.U.R.; Chandna, R.; Ahmad, P. Role of glutathione reductase in plant abiotic stress. In Abiotic Stress Responses in Plants; Springer: Berlin/Heidelberg, Germany, 2012; pp. 149–158. [Google Scholar]
- Ghisla, S.; Massey, V. Mechanisms of flavoprotein-catalyzed reactions. Eur. J. Biochem. 1989, 181, 1–17. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Hasanuzzaman, M.; Gill, R.; Trivedi, D.K.; Ahmad, I.; Pereira, E.; Tuteja, N. Glutathione and glutathione reductase: A boon in disguise for plant abiotic stress defense operations. Plant Physiol. Biochem. 2013, 70, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Luthje, S. Properties of guaiacol peroxidase activities isolated from corn root plasma membranes. Plant Physiol. 2003, 132, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.; Sasaki, K.; Ito, H.; Ohashi, Y.; Matsui, H. A large family of class III plant peroxidases. Plant Cell Physiol. 2001, 42, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Xiao, W.; Hao, H.; Xiaoqing, L.; Liang, C.; Chao, L.; Mingyu, S.; Fashui, H. Oxidative stress induced by lead in chloroplast of spinach. Biol. Trace Elem. Res. 2008, 126, 257–268. [Google Scholar] [CrossRef]
- Castro, D.; Contreras, L.M.; Kurz, L.; Wilkesman, J. Detection of Guaiacol Peroxidase on Electrophoretic Gels. In Zymography; Springer: Berlin/Heidelberg, Germany, 2017; pp. 199–204. [Google Scholar]
- Parvanova, D.; Ivanov, S.; Konstantinova, T.; Karanov, E.; Atanassov, A.; Tsvetkov, T.; Alexieva, V.; Djilianov, D. Transgenic tobacco plants accumulating osmolytes show reduced oxidative damage under freezing stress. Plant Physiol. Biochem. 2004, 42, 57–63. [Google Scholar] [CrossRef] [PubMed]
- de Pinto, M.C.; De Gara, L. Changes in the ascorbate metabolism of apoplastic and symplastic spaces are associated with cell differentiation. J. Exp. Bot. 2004, 55, 2559–2569. [Google Scholar] [CrossRef]
- Cruz-Rus, E.; Amaya, I.; Valpuesta, V. The challenge of increasing vitamin C content in plant foods. Biotechnol. J. 2012, 7, 1110–1121. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.; Zheng, Y.; Lyons, T. Plant resistance to ozone: The role of ascorbate. In Air Pollution and Plant Biotechnology; Springer: Berlin/Heidelberg, Germany, 2002; pp. 235–252. [Google Scholar]
- Barata-Soares, A.D.; Gomez, M.L.; Mesquita, C.H.d.; Lajolo, F.M. Ascorbic acid biosynthesis: A precursor study on plants. Braz. J. Plant Physiol. 2004, 16, 147–154. [Google Scholar] [CrossRef]
- Horemans, N.; Foyer, C.H.; Asard, H. Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci. 2000, 5, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. L-ascorbic acid: A multifunctional molecule supporting plant growth and development. Scientifica 2013, 2013, 795964. [Google Scholar] [CrossRef] [PubMed]
- HongBo, S.; ZongSuo, L.; MingAn, S.; ShiMeng, S.; ZanMin, H. Investigation on dynamic changes of photosynthetic characteristics of 10 wheat (Triticum aestivum L.) genotypes during two vegetative-growth stages at water deficits. Colloids Surf. B 2005, 43, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Smirnof, N. Therole ofactive oxygenin theresponse of plants to water deficit and desiccation. New Phytol. 1993, 125, 27–58. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S. Increased antioxidant activity in Cassia seedlings under UV-B radiation. Biol. Plant. 2007, 51, 157–160. [Google Scholar] [CrossRef]
- Ramírez, L.; Bartoli, C.G.; Lamattina, L. Glutathione and ascorbic acid protect Arabidopsis plants against detrimental effects of iron deficiency. J. Exp. Bot. 2013, 64, 3169–3178. [Google Scholar] [CrossRef]
- Szarka, A.; Tomasskovics, B.; Bánhegyi, G. The ascorbate-glutathione-α-tocopherol triad in abiotic stress response. Int. J. Mol. Sci. 2012, 13, 4458–4483. [Google Scholar] [CrossRef]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef]
- Larson, R.A. The antioxidants of higher plants. Phytochemistry 1988, 27, 969–978. [Google Scholar] [CrossRef]
- Mullineaux, P.M.; Rausch, T. Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth. Res. 2005, 86, 459–474. [Google Scholar] [CrossRef]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef]
- Millar, A.H.; Mittova, V.; Kiddle, G.; Heazlewood, J.L.; Bartoli, C.G.; Theodoulou, F.L.; Foyer, C.H. Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol. 2003, 133, 443–447. [Google Scholar] [CrossRef]
- May, M.J.; Vernoux, T.; Leaver, C.; Montagu, M.V.; Inzé, D. Glutathione homeostasis in plants: Implications for environmental sensing and plant development. J. Exp. Bot. 1998, 49, 649–667. [Google Scholar] [CrossRef]
- Roychoudhury, A.; Basu, S. Ascorbate-glutathione and plant tolerance to various abiotic stresses. In Oxidative Stress in Plants: Causes, Consequences and Tolerance; IK International Publishers: New Delhi, India, 2012; pp. 177–258. [Google Scholar]
- Kamal-Eldin, A.; Appelqvist, L.Å. The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids 1996, 31, 671–701. [Google Scholar] [CrossRef]
- Holländer-Czytko, H.; Grabowski, J.; Sandorf, I.; Weckermann, K.; Weiler, E.W. Tocopherol content and activities of tyrosine aminotransferase and cystine lyase in Arabidopsis under stress conditions. J. Plant Physiol. 2005, 162, 767–770. [Google Scholar] [CrossRef]
- Kiffin, R.; Bandyopadhyay, U.; Cuervo, A.M. Oxidative stress and autophagy. Antioxid. Redox Signal. 2006, 8, 152–162. [Google Scholar] [CrossRef]
- Kruk, J.; Holländer-Czytko, H.; Oettmeier, W.; Trebst, A. Tocopherol as singlet oxygen scavenger in photosystem II. J. Plant Physiol. 2005, 162, 749–757. [Google Scholar] [CrossRef]
- Noctor, G. Metabolic signalling in defence and stress: The central roles of soluble redox couples. Plant Cell Environ. 2006, 29, 409–425. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Hill, R.D. Nitrate, NO and haemoglobin in plant adaptation to hypoxia: An alternative to classic fermentation pathways. J. Exp. Bot. 2004, 55, 2473–2482. [Google Scholar] [CrossRef]
- Yu, S.-W.; Tang, K.-X. MAP kinase cascades responding to environmental stress in plants. Acta Bot. Sin. 2004, 46, 127–136. [Google Scholar]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- Stahl, W.; Sies, H. Antioxidant activity of carotenoids. Mol. Asp. Med. 2003, 24, 345–351. [Google Scholar] [CrossRef]
- Muller, L.; Bohm, V. Antioxidant activity of β-carotene compounds in different in vitro assays. Molecules 2011, 16, 1055–1069. [Google Scholar] [CrossRef]
- Mortensen, A.; Skibsted, L.; Truscott, T. The interaction of dietary carotenoids with radical species. Arch. Biochem. Biophys. 2001, 385, 13–19. [Google Scholar] [CrossRef]
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach; John Wiley & Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Gruszecki, W.I.; Strzałka, K. Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta Mol. Basis Dis. 2005, 1740, 108–115. [Google Scholar] [CrossRef]
- Heber, D.; Lu, Q.-Y. Overview of mechanisms of action of lycopene. Exp. Biol. Med. 2002, 227, 920–923. [Google Scholar] [CrossRef]
- Woodall, A.A.; Lee, S.W.-M.; Weesie, R.J.; Jackson, M.J.; Britton, G. Oxidation of carotenoids by free radicals: Relationship between structure and reactivity. Biochim. Biophys. Acta Gen. Subj. 1997, 1336, 33–42. [Google Scholar] [CrossRef]
- Ramel, F.; Birtic, S.; Cuiné, S.; Triantaphylidès, C.; Ravanat, J.-L.; Havaux, M. Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 2012, 158, 1267–1278. [Google Scholar] [CrossRef]
- Ozhogina, O.A.; Kasaikina, O.T. β-Carotene as an interceptor of free radicals. Free Radic. Biol. Med. 1995, 19, 575–581. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Polovka, M.; Brezová, V.; Staško, A. Antioxidant properties of tea investigated by EPR spectroscopy. Biophys. Chem. 2003, 106, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Bravo, L. Polyphenols: Chemistry, dietary sources, metabolism, and nutritional significance. Nutr. Rev. 1998, 56, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.M.; Markham, K.R. Flavonoids: Chemistry, Biochemistry and Applications; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Chen, Z.; Chan, P.; Ho, K.; Fung, K.; Wang, J. Antioxidant activity of natural flavonoids is governed by number and location of their aromatic hydroxyl groups. Chem. Phys. Lipids 1996, 79, 157–163. [Google Scholar] [CrossRef]
- Morel, I.; Lescoat, G.; Cogrel, P.; Sergent, O.; Pasdeloup, N.; Brissot, P.; Cillard, P.; Cillard, J. Antioxidant and iron-chelating activities of the flavonoids catechin, quercetin and diosmetin on iron-loaded rat hepatocyte cultures. Biochem. Pharmacol. 1993, 45, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Yordi, E.G.; Pérez, E.M.; Matos, M.J.; Villares, E.U. Antioxidant and pro-oxidant effects of polyphenolic compounds and structure-activity relationship evidence. Nutr. Well-Being Health 2012, 2, 23–48. [Google Scholar]
- Nijveldt, R.J.; Van Nood, E.; Van Hoorn, D.E.; Boelens, P.G.; Van Norren, K.; Van Leeuwen, P.A. Flavonoids: A review of probable mechanisms of action and potential applications. Am. J. Clin. Nutr. 2001, 74, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Qamar, A.; Mysore, K.S.; Senthil-Kumar, M. Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front. Plant Sci. 2015, 6, 503. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Decros, G.; Baldet, P.; Beauvoit, B.; Stevens, R.; Flandin, A.; Colombié, S.; Gibon, Y.; Pétriacq, P. Get the balance right: ROS homeostasis and redox signalling in fruit. Front. Plant Sci. 2019, 10, 1091. [Google Scholar] [CrossRef] [PubMed]
- Paciolla, C.; Paradiso, A.; De Pinto, M. Cellular redox homeostasis as central modulator in plant stress response. In Redox State as a Central Regulator of Plant-Cell Stress Responses; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–23. [Google Scholar]
- Finkel, T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011, 194, 7–15. [Google Scholar] [CrossRef]
- Rehman, S.; Abbas, G.; Shahid, M.; Saqib, M.; Farooq, A.B.U.; Hussain, M.; Murtaza, B.; Amjad, M.; Naeem, M.A.; Farooq, A. Effect of salinity on cadmium tolerance, ionic homeostasis and oxidative stress responses in conocarpus exposed to cadmium stress: Implications for phytoremediation. Ecotoxicol. Environ. Saf. 2019, 171, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-W.; Kong, X.-W.; Wang, N.; Wang, T.-T.; Chen, J.; Shi, Z.Q. Thymol confers tolerance to salt stress by activating anti-oxidative defense and modulating Na+ homeostasis in rice root. Ecotoxicol. Environ. Saf. 2020, 188, 109894. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Mir, R.A.; Alyemeni, M.N.; Ahmad, P. Combined effects of brassinosteroid and kinetin mitigates salinity stress in tomato through the modulation of antioxidant and osmolyte metabolism. Plant Physiol. Biochem. 2020, 147, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Ahanger, M.A.; Alam, P.; Alyemeni, M.N.; Wijaya, L.; Ali, S.; Ashraf, M. Silicon (Si) supplementation alleviates NaCl toxicity in mung bean [Vigna radiata (L.) Wilczek] through the modifications of physio-biochemical attributes and key antioxidant enzymes. J. Plant Growth Regul. 2019, 38, 70–82. [Google Scholar] [CrossRef]
- Arora, M.; Saxena, P.; Abdin, M.; Varma, A. Interaction between Piriformospora indica and Azotobacter chroococcum diminish the effect of salt stress in Artemisia annua L. by enhancing enzymatic and non-enzymatic antioxidants. Symbiosis 2020, 80, 61–73. [Google Scholar] [CrossRef]
- Lalarukh, I.; Shahbaz, M. Response of antioxidants and lipid peroxidation to exogenous application of alpha-tocopherol in sunflower (Helianthus annuus L.) under salt stress. Pak. J. Bot. 2020, 52, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Shahbaz, M. Glycinebetaine induced modulation in oxidative defense system and mineral nutrients sesame (Sesamum indicum L.) under saline regimes. Pak. J. Bot. 2020, 52, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Mhadhbi, H.; Fotopoulos, V.; Mylona, P.V.; Jebara, M.; Aouani, M.E.; Polidoros, A.N. Alternative oxidase 1 (Aox1) gene expression in roots of Medicago truncatula is a genotype-specific component of salt stress tolerance. J. Plant Physiol. 2013, 170, 111–114. [Google Scholar] [CrossRef]
- Filippou, P.; Bouchagier, P.; Skotti, E.; Fotopoulos, V. Proline and reactive oxygen/nitrogen species metabolism is involved in the tolerant response of the invasive plant species Ailanthus altissima to drought and salinity. Environ. Exp. Bot. 2014, 97, 1–10. [Google Scholar] [CrossRef]
- Cunha, J.R.; Neto, M.C.L.; Carvalho, F.E.; Martins, M.O.; Jardim-Messeder, D.; Margis-Pinheiro, M.; Silveira, J.A. Salinity and osmotic stress trigger different antioxidant responses related to cytosolic ascorbate peroxidase knockdown in rice roots. Environ. Exp. Bot. 2016, 131, 58–67. [Google Scholar] [CrossRef]
- Vighi, I.; Benitez, L.; Amaral, M.; Moraes, G.; Auler, P.; Rodrigues, G.; Deuner, S.; Maia, L.; Braga, E. Functional characterization of the antioxidant enzymes in rice plants exposed to salinity stress. Biol. Plant. 2017, 61, 540–550. [Google Scholar] [CrossRef]
- Zeeshan, M.; Lu, M.; Sehar, S.; Holford, P.; Wu, F. Comparison of biochemical, anatomical, morphological, and physiological responses to salinity stress in wheat and barley genotypes deferring in salinity tolerance. Agronomy 2020, 10, 127. [Google Scholar] [CrossRef]
- Alzahrani, S.M.; Alaraidh, I.A.; Migdadi, H.; Alghamdi, S.; Khan, M.A.; Ahmad, P. Physiological, biochemical, and antioxidant properties of two genotypes of Vicia faba grown under salinity stress. Pak. J. Bot. 2019, 51, 786–798. [Google Scholar] [CrossRef]
- Alsahli, A.; Mohamed, A.-K.; Alaraidh, I.; Al-Ghamdi, A.; Al-Watban, A.; El-Zaidy, M.; Alzahrani, S.M. Salicylic acid alleviates salinity stress through the modulation of biochemical attributes and some key antioxidants in wheat seedlings. Pak. J. Bot. 2019, 51, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.Y.A.; Ibrahim, M.E.H.; Zhou, G.; Nimir, N.E.A.; Jiao, X.; Zhu, G.; Elsiddig, A.M.I.; Suliman, M.S.E.; Elradi, S.B.M.; Yue, W. Exogenous jasmonic acid and humic acid increased salinity tolerance of sorghum. Agronomy 2020, 112, 871–884. [Google Scholar] [CrossRef]
- Tanou, G.; Ziogas, V.; Belghazi, M.; Christou, A.; Filippou, P.; Job, D.; Fotopoulos, V.; Molassiotis, A. Polyamines reprogram oxidative and nitrosative status and the proteome of citrus plants exposed to salinity stress. Plant Cell Environ. 2014, 37, 864–885. [Google Scholar] [CrossRef] [PubMed]
- Ahanger, M.A.; Qin, C.; Begum, N.; Maodong, Q.; Dong, X.X.; El-Esawi, M.; El-Sheikh, M.A.; Alatar, A.A.; Zhang, L. Nitrogen availability prevents oxidative effects of salinity on wheat growth and photosynthesis by up-regulating the antioxidants and osmolytes metabolism, and secondary metabolite accumulation. BMC Plant Biol. 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.S.; Kim, K.-S.; Hamayun, M.; Kim, Y. Silicon confers soybean resistance to salinity stress through regulation of reactive oxygen and reactive nitrogen species. Front. Plant Sci. 2020, 10, 1725. [Google Scholar] [CrossRef] [PubMed]
- Christou, A.; Manganaris, G.A.; Papadopoulos, I.; Fotopoulos, V. Hydrogen sulfide induces systemic tolerance to salinity and non-ionic osmotic stress in strawberry plants through modification of reactive species biosynthesis and transcriptional regulation of multiple defence pathways. J. Exp. Bot. 2013, 64, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Santander, C.; Ruiz, A.; García, S.; Aroca, R.; Cumming, J.; Cornejo, P. Efficiency of two arbuscular mycorrhizal fungal inocula to improve saline stress tolerance in lettuce plants by changes of antioxidant defense mechanisms. J. Sci. Food Agric. 2020, 100, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Rady, M.M.; Kuşvuran, A.; Alharby, H.F.; Alzahrani, Y.; Kuşvuran, S. Pretreatment with proline or an organic bio-stimulant induces salt tolerance in wheat plants by improving antioxidant redox state and enzymatic activities and reducing the oxidative stress. J. Plant Growth Regul. 2019, 38, 449–462. [Google Scholar] [CrossRef]
- Rady, M.M.; Elrys, A.S.; El-Maati, M.F.A.; Desoky, E.-S.M. Interplaying roles of silicon and proline effectively improve salt and cadmium stress tolerance in Phaseolus vulgaris plant. Plant Physiol. Biochem. 2019, 139, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Heydari, H.; Rezayian, M.; Niknam, V.; Ebrahimzadeh, H. Role of Penconazole in salt stress amelioration in Sesamum indicum L. Soil Sci. Plant Nutr. 2019, 65, 243–250. [Google Scholar] [CrossRef]
- Aubert, S.; Assard, N.; Boutin, J.P.; Frenot, Y.; Dorne, A.J. Carbon metabolism in the subantarctic Kerguelen cabbage Pringlea antiscorbutica R. Br.: Environmental controls over carbohydrates and proline contents and relation to phenology. Plant Cell Environ. 1999, 22, 243–254. [Google Scholar] [CrossRef]
- Yan, H. Effects of exogenous proline on the physiology of soyabean plantlets regenerated from embryos in vitro and on the ultrastructure of their mitochondria under NaCl stress. Soybean Sci. 2000, 19, 314–319. [Google Scholar]
- Djilianov, D.; Georgieva, T.; Moyankova, D.; Atanassov, A.; Shinozaki, K.; Smeeken, S.; Verma, D.; Murata, N. Improved abiotic stress tolerance in plants by accumulation of osmoprotectants—Gene transfer approach. Biotechnol. Biotechnol. Equip. 2005, 19 (Suppl. 3), 63–71. [Google Scholar] [CrossRef]
- Jaarsma, R.; de Vries, R.S.; de Boer, A.H. Effect of salt stress on growth, Na+ accumulation and proline metabolism in potato (Solanum tuberosum) cultivars. PLoS ONE 2013, 8, e60183. [Google Scholar] [CrossRef] [PubMed]
- El-Shawa, G.M.; Rashwan, E.M.; Abdelaal, K.A. Mitigating salt stress effects by exogenous application of proline and yeast extract on morpho-physiological, biochemical and anatomical characters of calendula plants. Sci. J. Flowers Ornam. Plants 2020, 7, 461–482. [Google Scholar] [CrossRef]
- Abdelaal, K.; El-Afry, M.; Metwaly, M.; Zidan, M.; Rashwan, E. Salt tolerance activation in faba bean plants using proline and salicylic acid associated with physio-biochemical and yield characters improvement. Fresenius Environ. Bull. 2021, 30, 3175–3186. [Google Scholar]
- De la Torre-González, A.; Montesinos-Pereira, D.; Blasco, B.; Ruiz, J. Influence of the proline metabolism and glycine betaine on tolerance to salt stress in tomato (Solanum lycopersicum L.) commercial genotypes. J. Plant Physiol. 2018, 231, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Hannachi, S.; Van Labeke, M.-C. Salt stress affects germination, seedling growth and physiological responses differentially in eggplant cultivars (Solanum melongena L.). Sci. Hortic. 2018, 228, 56–65. [Google Scholar] [CrossRef]
- Abdelaal, K.; Attia, K.A.; Niedbała, G.; Wojciechowski, T.; Hafez, Y.; Alamery, S.; Alateeq, T.K.; Arafa, S.A. Mitigation of Drought Damages by Exogenous Chitosan and Yeast Extract with Modulating the Photosynthetic Pigments, Antioxidant Defense System and Improving the Productivity of Garlic Plants. Horticulturae 2021, 7, 510. [Google Scholar] [CrossRef]
- Hare, P.; Cress, W. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997, 21, 79–102. [Google Scholar] [CrossRef]
- Székely, G.; Abrahám, E.; Cséplo, A.; Rigó, G.; Zsigmond, L.; Csiszár, J.; Ayaydin, F.; Strizhov, N.; Jásik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef]
- Chen, T.H.; Murata, N. Glycinebetaine: An effective protectant against abiotic stress in plants. Trends Plant Sci. 2008, 13, 499–505. [Google Scholar] [CrossRef]
- Hoque, M.A.; Banu, M.N.A.; Okuma, E.; Amako, K.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Exogenous proline and glycinebetaine increase NaCl-induced ascorbate–glutathione cycle enzyme activities, and proline improves salt tolerance more than glycinebetaine in tobacco Bright Yellow-2 suspension-cultured cells. J. Plant Physiol. 2007, 164, 1457–1468. [Google Scholar] [CrossRef]
- Banu, M.N.A.; Hoque, M.A.; Watanabe-Sugimoto, M.; Matsuoka, K.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Proline and glycinebetaine induce antioxidant defense gene expression and suppress cell death in cultured tobacco cells under salt stress. J. Plant Physiol. 2009, 166, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Fotopoulos, V.; Sanmartin, M.; Kanellis, A.K. Effect of ascorbate oxidase over-expression on ascorbate recycling gene expression in response to agents imposing oxidative stress. J. Exp. Bot. 2006, 57, 3933–3943. [Google Scholar] [CrossRef]
- Neill, S.J.; Desikan, R.; Clarke, A.; Hurst, R.D.; Hancock, J.T. Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 2002, 53, 1237–1247. [Google Scholar] [CrossRef]
- Molassiotis, A.; Fotopoulos, V. Oxidative and nitrosative signaling in plants: Two branches in the same tree? Plant Signal. Behav. 2011, 6, 210–214. [Google Scholar] [CrossRef]
- Zhao, M.-G.; Tian, Q.-Y.; Zhang, W.-H. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 2007, 144, 206–217. [Google Scholar] [CrossRef]
- Qiao, W.; Xiao, S.; Yu, L.; Fan, L.-M. Expression of a rice gene OsNOA1 re-establishes nitric oxide synthesis and stress-related gene expression for salt tolerance in Arabidopsis nitric oxide-associated 1 mutant Atnoa1. Environ. Exp. Bot. 2009, 65, 90–98. [Google Scholar] [CrossRef]
- Wahid, A.; Perveen, M.; Gelani, S.; Basra, S.M. Pretreatment of seed with H2O2 improves salt tolerance of wheat seedlings by alleviation of oxidative damage and expression of stress proteins. J. Plant Physiol. 2007, 164, 283–294. [Google Scholar] [CrossRef]
- Lu, S.; Su, W.; Li, H.; Guo, Z. Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2-and NO-induced antioxidant enzyme activities. Plant Physiol. Biochem. 2009, 47, 132–138. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, J.; Guo, Z.; Lu, S.; He, S.; Shu, W.; Zhou, B. Increased abscisic acid levels in transgenic tobacco over-expressing 9 cis-epoxycarotenoid dioxygenase influence H2O2 and NO production and antioxidant defences. Plant Cell Environ. 2009, 32, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ling, T.; Han, Y.; Liu, K.; Zheng, Q.; Huang, L.; Yuan, X.; He, Z.; Hu, B.; Fang, L. Carbon monoxide enhances salt tolerance by nitric oxide-mediated maintenance of ion homeostasis and up-regulation of antioxidant defence in wheat seedling roots. Plant Cell Environ. 2008, 31, 1864–1881. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Zhao, Y.; Yu, X.; Kiprotich, F.; Han, H.; Guan, R.; Wang, R.; Shen, W. Nitric oxide is required for melatonin-enhanced tolerance against salinity stress in rapeseed (Brassica napus L.) seedlings. Int. J. Mol. Sci. 2018, 19, 1912. [Google Scholar] [CrossRef]
- Poór, P.; Tari, I. Ethylene-regulated reactive oxygen species and nitric oxide under salt stress in tomato cell suspension culture. Acta Biol. Szeged. 2011, 55, 143–146. [Google Scholar]
- Molassiotis, A.; Tanou, G.; Diamantidis, G. NO says more than ‘YES’ to salt tolerance: Salt priming and systemic nitric oxide signaling in plants. Plant Signal. Behav. 2010, 5, 209–212. [Google Scholar] [CrossRef]
- Xu, J.; Wang, W.; Sun, J.; Zhang, Y.; Ge, Q.; Du, L.; Yin, H.; Liu, X. Involvement of auxin and nitric oxide in plant Cd-stress responses. Plant Soil 2011, 346, 107–119. [Google Scholar] [CrossRef]
- Kong, X.; Wang, T.; Li, W.; Tang, W.; Zhang, D.; Dong, H. Exogenous nitric oxide delays salt-induced leaf senescence in cotton (Gossypium hirsutum L.). Acta Physiol. Plant 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Wang, H.; Liang, X.; Wan, Q.; Wang, X.; Bi, Y. Ethylene and nitric oxide are involved in maintaining ion homeostasis in Arabidopsis callus under salt stress. Planta 2009, 230, 293–307. [Google Scholar] [CrossRef]
- Simaei, M.; Khavarinejad, R.; Saadatmand, S.; Bernard, F.; Fahimi, H. Interactive effects of salicylic acid and nitric oxide on soybean plants under NaCl salinity. Russian J. Plant Physiol. 2011, 58, 783–790. [Google Scholar] [CrossRef]
- Dong, F.; Simon, J.; Rienks, M.; Lindermayr, C.; Rennenberg, H. Effects of rhizopheric nitric oxide (NO) on N uptake in Fagus sylvatica seedlings depend on soil CO2 concentration, soil N availability and N source. Tree Physiol. 2015, 35, 910–920. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Das, B.K.; Bhaganagare, G.R. Genome-wide identification, evolutionary and expression analyses of putative Fe–S biogenesis genes in rice (Oryza sativa). Genome 2012, 55, 571–583. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Das, B.K.; Kumar, M.; Bhaganagare, G.R. An overview on Fe–S protein biogenesis from prokaryotes to eukaryotes. In Biological Nitrogen Fixation; Wiley Online Library: Hoboken, NJ, USA, 2015; pp. 57–74. [Google Scholar]
- Fatma, M.; Masood, A.; Per, T.S.; Khan, N.A. Nitric oxide alleviates salt stress inhibited photosynthetic performance by interacting with sulfur assimilation in mustard. Front. Plant Sci. 2016, 7, 521. [Google Scholar] [CrossRef]
- Fan, H.-F.; Du, C.-X.; Guo, S.-R. Effect of nitric oxide on proline metabolism in cucumber seedlings under salinity stress. J. Am. Soc. Hortic. Sci. 2012, 137, 127–133. [Google Scholar] [CrossRef]
- Khan, M.N.; Siddiqui, M.H.; Mohammad, F.; Naeem, M. Interactive role of nitric oxide and calcium chloride in enhancing tolerance to salt stress. Nitric Oxide 2012, 27, 210–218. [Google Scholar] [CrossRef]
- Campos, F.V.; Oliveira, J.A.; Pereira, M.G.; Farnese, F.S. Nitric oxide and phytohormone interactions in the response of Lactuca sativa to salinity stress. Planta 2019, 250, 1475–1489. [Google Scholar] [CrossRef]
- Babaei, S.; Niknam, V.; Behmanesh, M. Comparative effects of nitric oxide and salicylic acid on salinity tolerance in saffron (Crocus sativus). Plant Biosyst. 2021, 155, 73–82. [Google Scholar] [CrossRef]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.-K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Kurusu, T.; Kuchitsu, K.; Tada, Y. Plant signaling networks involving Ca2+ and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 2015, 6, 427. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef]
- Kwak, J.M.; Mori, I.C.; Pei, Z.-M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Szepesi, Á.; Csiszár, J.; Gémes, K.; Horváth, E.; Horváth, F.; Simon, M.L.; Tari, I. Salicylic acid improves acclimation to salt stress by stimulating abscisic aldehyde oxidase activity and abscisic acid accumulation, and increases Na+ content in leaves without toxicity symptoms in Solanum lycopersicum L. J. Plant Physiol. 2009, 166, 914–925. [Google Scholar] [CrossRef]
- Takeda, S.; Gapper, C.; Kaya, H.; Bell, E.; Kuchitsu, K.; Dolan, L. Local positive feedback regulation determines cell shape in root hair cells. Science 2008, 319, 1241–1244. [Google Scholar] [CrossRef]
- Pottosin, I.; Velarde-Buendía, A.M.; Bose, J.; Zepeda-Jazo, I.; Shabala, S.; Dobrovinskaya, O. Cross-talk between reactive oxygen species and polyamines in regulation of ion transport across the plasma membrane: Implications for plant adaptive responses. J. Exp. Bot. 2014, 65, 1271–1283. [Google Scholar] [CrossRef]
- Xu, P.; Liu, Z.; Fan, X.; Gao, J.; Zhang, X.; Zhang, X.; Shen, X. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in Gossypium aridum under salt stress. Gene 2013, 525, 26–34. [Google Scholar] [CrossRef]
- Choi, W.-G.; Toyota, M.; Kim, S.-H.; Hilleary, R.; Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef] [PubMed]
- Mostofa, M.G.; Rahman, A.; Ansary, M.; Uddin, M.; Watanabe, A.; Fujita, M.; Tran, L.-S.P. Hydrogen sulfide modulates cadmium-induced physiological and biochemical responses to alleviate cadmium toxicity in rice. Sci. Rep. 2015, 5, 1–17. [Google Scholar] [CrossRef]
- Kodela, R.; Chattopadhyay, M.; Kashfi, K. NOSH-Aspirin: A novel nitric oxide–hydrogen sulfide-releasing hybrid: A new class of anti-inflammatory pharmaceuticals. ACS Med. Chem. Lett. 2012, 3, 257–262. [Google Scholar] [CrossRef]
- Zhang, L.; Pei, Y.; Wang, H.; Jin, Z.; Liu, Z.; Qiao, Z.; Fang, H.; Zhang, Y. Hydrogen sulfide alleviates cadmium-induced cell death through restraining ROS accumulation in roots of Brassica rapa L. ssp. pekinensis. Oxid. Med. Cell. Longev. 2015, 2015, 804603. [Google Scholar] [CrossRef]
- Hancock, J.T.; Whiteman, M. Hydrogen sulfide signaling: Interactions with nitric oxide and reactive oxygen species. Ann. N. Y. Acad. Sci. 2016, 1365, 5–14. [Google Scholar] [CrossRef]
- Paranhos, A. Interplay of calcium, cAMP and PKA in flavonoid accumulation by cell cultures of Hypericum androsaemum L. Planta Med. 2014, 80, P2O63. [Google Scholar] [CrossRef]
- Vafadar, F.; Amooaghaie, R.; Ehsanzadeh, P.; Ghanati, F.; Sajedi, R.H. Crosstalk between melatonin and Ca2+/CaM evokes systemic salt tolerance in Dracocephalum kotschyi. J. Plant Physiol. 2020, 252, 153237. [Google Scholar] [CrossRef]
- Hajihashemi, S.; Skalicky, M.; Brestic, M.; Pavla, V. Cross-talk between nitric oxide, hydrogen peroxide and calcium in salt-stressed Chenopodium quinoa Willd. At seed germination stage. Plant Physiol. Biochem. 2020, 154, 657–664. [Google Scholar] [CrossRef]
- Bouallègue, A.; Souissi, F.; Nouairi, I.; Souibgui, M.; Abbes, Z.; Mhadhbi, H. Salicylic acid and hydrogen peroxide pretreatments alleviate salt stress in faba bean (Vicia faba) seeds during germination. Seed Sci. Technol. 2017, 45, 675–690. [Google Scholar] [CrossRef]
- Pathak, M.R.; Teixeira da Silva, J.A.; Wani, S.H. Polyamines in response to abiotic stress tolerance through transgenic approaches. GM Crops Food 2014, 5, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chan, Z. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J. Integr. Plant Biol. 2014, 56, 114–121. [Google Scholar] [CrossRef]
- Do, P.T.; Drechsel, O.; Heyer, A.G.; Hincha, D.K.; Zuther, E. Changes in free polyamine levels, expression of polyamine biosynthesis genes, and performance of rice cultivars under salt stress: A comparison with responses to drought. Front. Plant Sci. 2014, 5, 182. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Su, S.; Wang, Z.; Wang, Y.; Xu, X. Comprehensive Genome-Wide Identification and Transcript Profiling of GABA Pathway Gene Family in Apple (Malus domestica). Genes 2021, 12, 1973. [Google Scholar] [CrossRef] [PubMed]
- Velarde-Buendía, A.M.; Shabala, S.; Cvikrova, M.; Dobrovinskaya, O.; Pottosin, I. Salt-sensitive and salt-tolerant barley varieties differ in the extent of potentiation of the ROS-induced K+ efflux by polyamines. Plant Physiol. Biochem. 2012, 61, 18–23. [Google Scholar] [CrossRef]
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: Where do we stand? Nitric Oxide 2017, 63, 30–38. [Google Scholar] [CrossRef]
- Gupta, P.; Srivastava, S.; Seth, C.S. 24-Epibrassinolide and sodium nitroprusside alleviate the salinity stress in Brassica juncea L. cv. Varuna through cross talk among proline, nitrogen metabolism and abscisic acid. Plant Soil 2017, 411, 483–498. [Google Scholar] [CrossRef]
- Siddiqui, M.H.; Mohammad, F.; Khan, M.; Al-Whaibi, M.H. Cumulative effect of nitrogen and sulphur on Brassica juncea L. genotypes under NaCl stress. Protoplasma 2012, 249, 139–153. [Google Scholar] [CrossRef]
- Prakash, V.; Singh, V.P.; Tripathi, D.K.; Sharma, S.; Corpas, F.J. Crosstalk between nitric oxide (NO) and abscisic acid (ABA) signalling molecules in higher plants. Environ. Exp. Bot. 2019, 161, 41–49. [Google Scholar] [CrossRef]
- Kumar, M.; Kherawat, B.S.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.K.; Ghodake, G.S.; Kadam, A.A.; Kim, H.-U.; Chung, S.-M. Genome-wide identification and characterization of PIN-FORMED (PIN) gene family reveals role in developmental and various stress conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef]
- Iqbal, N.; Masood, A.; Khan, N.A. Phytohormones in salinity tolerance: Ethylene and gibberellins cross talk. In Phytohormones and Abiotic Stress Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2012; pp. 77–98. [Google Scholar]
- Bialecka, B.; Kepczynski, J. Effect of ethephon and gibberellin A3 on Amaranthus caudatus seed germination and alpha-and beta-amylase activity under salinity stress. Acta Biol. Crac. Ser. Bot. 2009, 2, 119–125. [Google Scholar]
- Foo, E.; Ross, J.J.; Davies, N.W.; Reid, J.B.; Weller, J.L. A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J. 2006, 46, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yang, L.; Paul, M.; Zu, Y.; Tang, Z. Ethylene promotes germination of Arabidopsis seed under salinity by decreasing reactive oxygen species: Evidence for the involvement of nitric oxide simulated by sodium nitroprusside. Plant Physiol. Biochem. 2013, 73, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Yalpani, N.; Enyedi, A.J.; León, J.; Raskin, I. Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta 1994, 193, 372–376. [Google Scholar] [CrossRef]
- Durner, J.; Klessig, D.F. Salicylic acid is a modulator of tobacco and mammalian catalases. J. Biol. Chem. 1996, 271, 28492–28501. [Google Scholar] [CrossRef]
- Szalai, G.; Tari, I.; Janda, T.; Pestenacz, A.; Páldi, E. Effects of cold acclimation and salicylic acid on changes in ACC and MACC contents in maize during chilling. Biol. Plant. 2000, 43, 637–640. [Google Scholar] [CrossRef]
- Klessig, D.F.; Malamy, J. The salicylic acid signal in plants. Plant Mol. Biol. 1994, 26, 1439–1458. [Google Scholar] [CrossRef]
- Kováčik, J.; Grúz, J.; Bačkor, M.; Strnad, M.; Repčák, M. Salicylic acid-induced changes to growth and phenolic metabolism in Matricaria chamomilla plants. Plant Cell Rep. 2009, 28, 135–143. [Google Scholar] [CrossRef]
- Li, T.; Hu, Y.; Du, X.; Tang, H.; Shen, C.; Wu, J. Salicylic acid alleviates the adverse effects of salt stress in Torreya grandis cv. Merrillii seedlings by activating photosynthesis and enhancing antioxidant systems. PLoS ONE 2014, 9, e109492. [Google Scholar] [CrossRef]
- Arfan, M.; Athar, H.R.; Ashraf, M. Does exogenous application of salicylic acid through the rooting medium modulate growth and photosynthetic capacity in two differently adapted spring wheat cultivars under salt stress? J. Plant Physiol. 2007, 164, 685–694. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Z.-W.; Xue, L.-W.; Du, J.-B.; Shang, J.; Xu, F.; Yuan, S.; Lin, H.-H. Lack of salicylic acid in Arabidopsis protects plants against moderate salt stress. Z. Naturforsch. C J. Biosci. 2009, 64, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Friml, J. Auxin transport—Shaping the plant. Curr. Opin. Plant Biol. 2003, 6, 7–12. [Google Scholar] [CrossRef]
- Ganguly, A.; Park, M.; Kesawat, M.S.; Cho, H.-T. Functional analysis of the hydrophilic loop in intracellular trafficking of Arabidopsis PIN-FORMED proteins. Plant Cell 2014, 26, 1570–1585. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Niu, H.; Xin, D.; Long, Y.; Wang, G.; Liu, Z.; Li, G.; Zhang, F.; Qi, M.; Ye, Y. OsIAA18, an aux/IAA transcription factor gene, is involved in salt and drought tolerance in rice. Front. Plant Sci. 2021, 12, 2571. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, W.; Hu, H.; Li, B.; Wang, Y.; Zhao, Y.; Li, K.; Liu, M.; Li, X. Salt modulates gravity signaling pathway to regulate growth direction of primary roots in Arabidopsis. Plant Physiol. 2008, 146, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Bai, Y.; Wang, S.; Zhang, S.; Wu, Y.; Chen, M.; Jiang, D.; Qi, Y. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 2010, 277, 2954–2969. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Xiong, L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 2009, 229, 577–591. [Google Scholar] [CrossRef]
- Li, G.; Ye, Y.; Ren, X.; Qi, M.; Zhao, H.; Zhou, Q.; Chen, X.; Wang, J.; Yuan, C.; Wang, F. The rice Aux/IAA transcription factor gene OsIAA18 enhances salt and osmotic tolerance in Arabidopsis. Biol. Plant 2020, 64, 454–464. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.-K.; Do Choi, Y.; Kim, J.-K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Dang, C.; Ye, Y.; Wang, Z.; Hu, L.; Zhang, F.; Zhang, Y.; Qian, X.; Shi, J.; Guo, Y. Overexpression of grapevine VvIAA18 gene enhanced salt tolerance in tobacco. Int. J. Mol. Sci. 2020, 21, 1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhong, M.; Wu, Y.-H.; Bai, Z.-Y.; Liang, Q.-Y.; Liu, Q.-L.; Pan, Y.-Z.; Zhang, L.; Jiang, B.-B.; Jia, Y. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 571–581. [Google Scholar] [CrossRef]
- Shafi, A.; Pal, A.K.; Sharma, V.; Kalia, S.; Kumar, S.; Ahuja, P.S.; Singh, A.K. Transgenic potato plants overexpressing SOD and APX exhibit enhanced lignification and starch biosynthesis with improved salt stress tolerance. Plant Mol. Biol. Rep. 2017, 35, 504–518. [Google Scholar] [CrossRef]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef]
- Liu, M.M.; Li, Y.L.; Li, G.C.; Dong, T.T.; Liu, S.Y.; Liu, P.; Wang, Q.G. Overexpression of StCYS1 gene enhances tolerance to salt stress in the transgenic potato (Solanum tuberosum L.) plant. J. Integr. Agric. 2020, 19, 2239–2246. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Zhao, X.; Yang, S.; Huang, L.; Du, F.; Li, Z.; Zhao, X.; Fu, B.; Wang, W. Overexpression of the transcription factor gene OsSTAP1 increases salt tolerance in rice. Rice 2020, 13, 1–12. [Google Scholar] [CrossRef]
- Xu, X.-B.; Pan, Y.-Y.; Wang, C.-L.; Ying, Q.-C.; Song, H.-M.; Wang, H.-Z. Overexpression of DnWRKY11 enhanced salt and drought stress tolerance of transgenic tobacco. Biologia 2014, 69, 994–1000. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, N.; Yang, J.; Guo, H.; Liu, Z.; Zheng, X.; Li, S.; Xiang, F. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020, 291, 110326. [Google Scholar] [CrossRef]
- Jadamba, C.; Kang, K.; Paek, N.-C.; Lee, S.I.; Yoo, S.-C. Overexpression of rice expansin7 (Osexpa7) confers enhanced tolerance to salt stress in rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef]
- Wen, W.; Wang, R.; Su, L.; Lv, A.; Zhou, P.; An, Y. MsWRKY11, activated by MsWRKY22, functions in drought tolerance and modulates lignin biosynthesis in alfalfa (Medicago sativa L.). Environ. Exp. Bot. 2021, 184, 104373. [Google Scholar] [CrossRef]
- Trinh, N.; Le, H.; Nguyen, T. Overexpression of the dominant negative nbexo70d1 mutantion confers tolerance to salt stress in transgenic tobacco. Biol. Plant. 2019, 63, 484–495. [Google Scholar]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019, 280, 132–142. [Google Scholar] [CrossRef]
- Wang, Q.; Ni, J.; Shah, F.; Liu, W.; Wang, D.; Yao, Y.; Hu, H.; Huang, S.; Hou, J.; Fu, S. Overexpression of the stress-inducible SsMAX2 promotes drought and salt resistance via the regulation of redox homeostasis in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 837. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.-Y.; Chen, J.; Xu, W.-X.; Qiu, J.-R.; Song, L.; Wang, J.-T.; Tang, R.; Chen, D.; Jiang, C.-Z.; Huang, Z. Dehydration-induced WRKY transcriptional factor MfWRKY70 of Myrothamnus flabellifolia enhanced drought and salinity tolerance in Arabidopsis. Biomolecules 2021, 11, 327. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Ding, M.; Sun, J.; Deng, S.; Zhao, R.; Wang, M.; Ma, X.; Wang, F.; Zhang, H.; Qian, Z. Overexpression of PeHSF mediates leaf ROS homeostasis in transgenic tobacco lines grown under salt stress conditions. Plant Cell Tissue Organ Cult. 2013, 115, 299–308. [Google Scholar] [CrossRef]
- Sun, T.-T.; Wang, C.; Liu, R.; Zhang, Y.; Wang, Y.-C.; Wang, L.-Q. ThHSFA1 confers salt stress tolerance through modulation of reactive oxygen species scavenging by directly regulating ThWRKY4. Int. J. Mol. Sci. 2021, 22, 5048. [Google Scholar] [CrossRef]
- Yang, D.-Y.; Zhuang, K.-Y.; Ma, N.-N. Overexpression of SlGGP-LIKE gene enhanced the resistance of tomato to salt stress. In Protoplasma; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–11. [Google Scholar]

| Types of ROS | Sites of Production | Mechanisms of Action | Radical Type | Scavenging System |
|---|---|---|---|---|
| Hydroxyl radical (HO•) | Mitochondria, chloroplasts, and plasma membranes | Reacts with all biomolecules: proteins, lipids, RNA, and DNA. | Free radical | Sugars, flavonoids, proline, and ascorbate |
| Singlet oxygen (1O2) | Nucleus, chloroplasts, mitochondria, and plasma membranes | Oxidizes proteins containing cysteine, methionine, tryptophan, tyrosine, and histidine residues; polyunsaturated fatty acids such as methylene-interrupted polyenes and others; and guanine residues of DNA. | Non-radical | α-tocopherol and carotenoids |
| Superoxide (O2•−) | Mitochondria, peroxisomes, chloroplasts, electron transfer chains, and apoplast | Dismutates to H2O2 and reacts with double-bond-containing proteins such as iron-sulfur proteins via the iron atom. | Free radical | Flavonoids, ascorbate, and superoxide dismutase |
| Hydrogen peroxide (H2O2) | Mitochondria, peroxisomes, chloroplasts, cytosol and apoplast | Reacts with DNA. Oxidizes proteins and forms HO• and O2−•. It further reacts with proteins by attacking methionine and cysteine residues. Reacts with heme proteins. | Non-radical | Catalase, ascorbate peroxidase, guaiacol peroxidase, peroxiredoxins, glutathione, and ascorbate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kesawat, M.S.; Satheesh, N.; Kherawat, B.S.; Kumar, A.; Kim, H.-U.; Chung, S.-M.; Kumar, M. Regulation of Reactive Oxygen Species during Salt Stress in Plants and Their Crosstalk with Other Signaling Molecules—Current Perspectives and Future Directions. Plants 2023, 12, 864. https://doi.org/10.3390/plants12040864
Kesawat MS, Satheesh N, Kherawat BS, Kumar A, Kim H-U, Chung S-M, Kumar M. Regulation of Reactive Oxygen Species during Salt Stress in Plants and Their Crosstalk with Other Signaling Molecules—Current Perspectives and Future Directions. Plants. 2023; 12(4):864. https://doi.org/10.3390/plants12040864
Chicago/Turabian StyleKesawat, Mahipal Singh, Neela Satheesh, Bhagwat Singh Kherawat, Ajay Kumar, Hyun-Uk Kim, Sang-Min Chung, and Manu Kumar. 2023. "Regulation of Reactive Oxygen Species during Salt Stress in Plants and Their Crosstalk with Other Signaling Molecules—Current Perspectives and Future Directions" Plants 12, no. 4: 864. https://doi.org/10.3390/plants12040864
APA StyleKesawat, M. S., Satheesh, N., Kherawat, B. S., Kumar, A., Kim, H.-U., Chung, S.-M., & Kumar, M. (2023). Regulation of Reactive Oxygen Species during Salt Stress in Plants and Their Crosstalk with Other Signaling Molecules—Current Perspectives and Future Directions. Plants, 12(4), 864. https://doi.org/10.3390/plants12040864










