Overview of Repressive miRNA Regulation by Short Tandem Target Mimic (STTM): Applications and Impact on Plant Biology
Abstract
1. Introduction
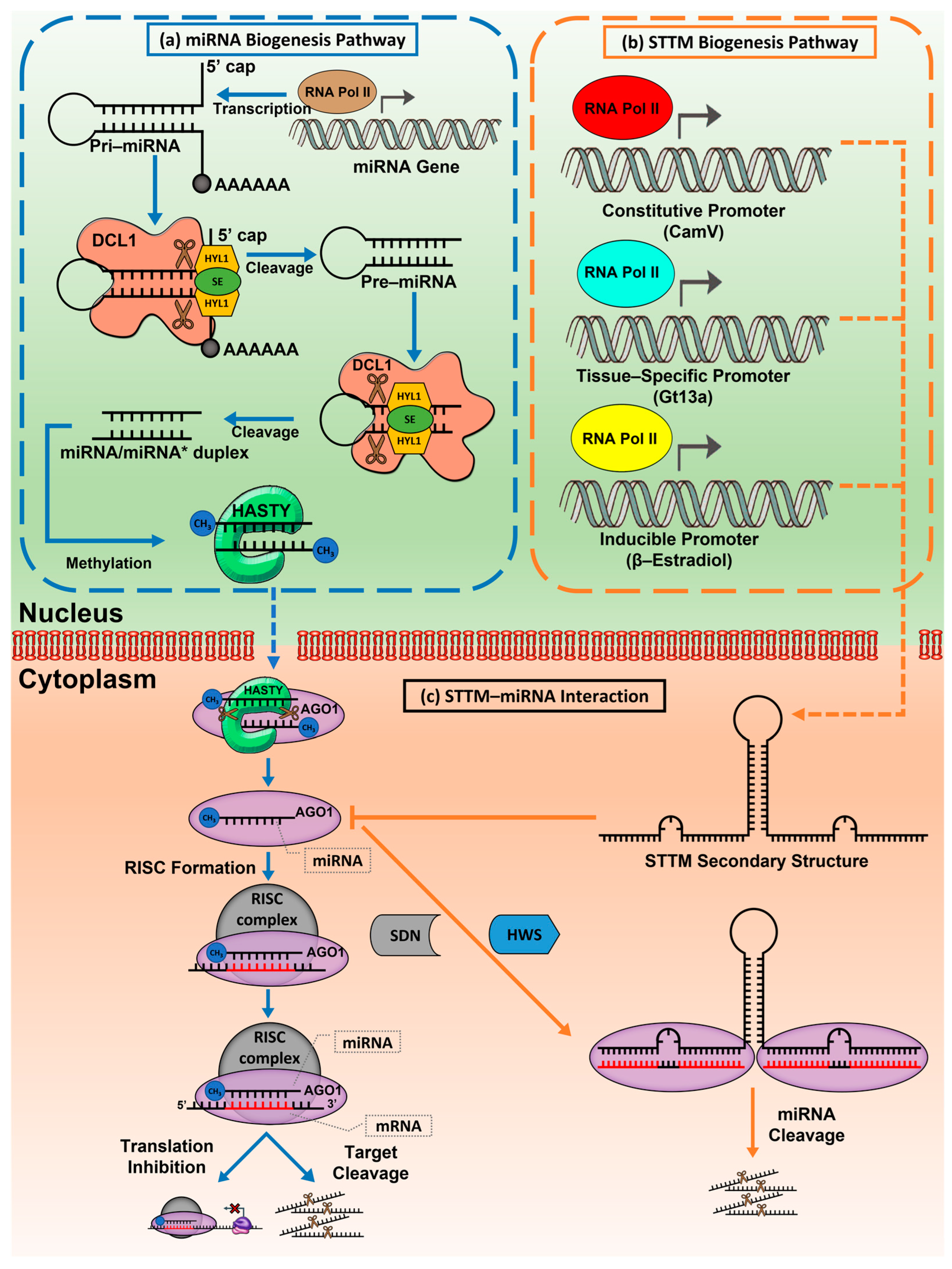
2. miRNA Biogenesis and Its Regulation in Plant Transcripts
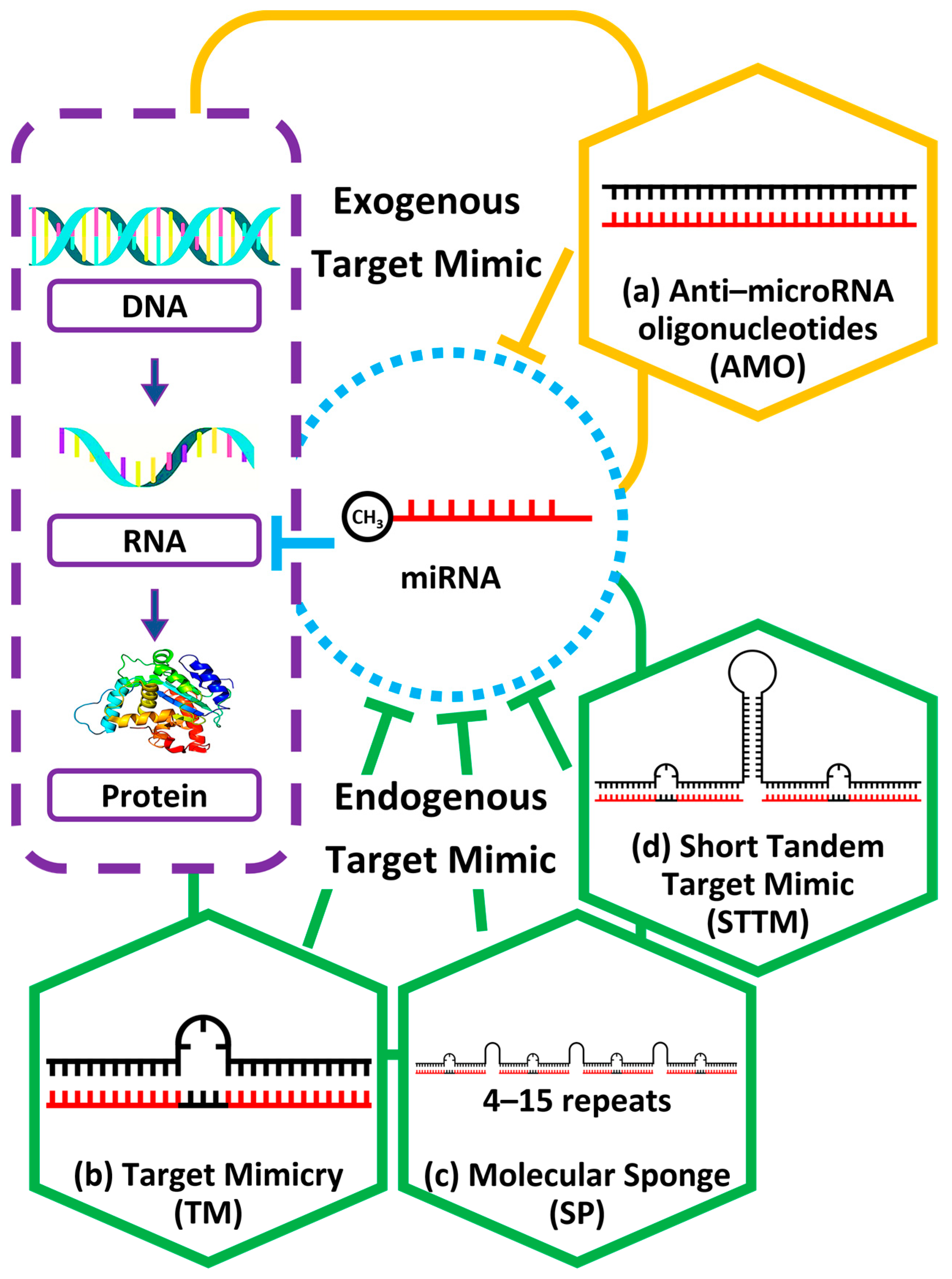
3. Inhibition Mechanism of Target Mimic That Confers Mature miRNA Repression in Plants
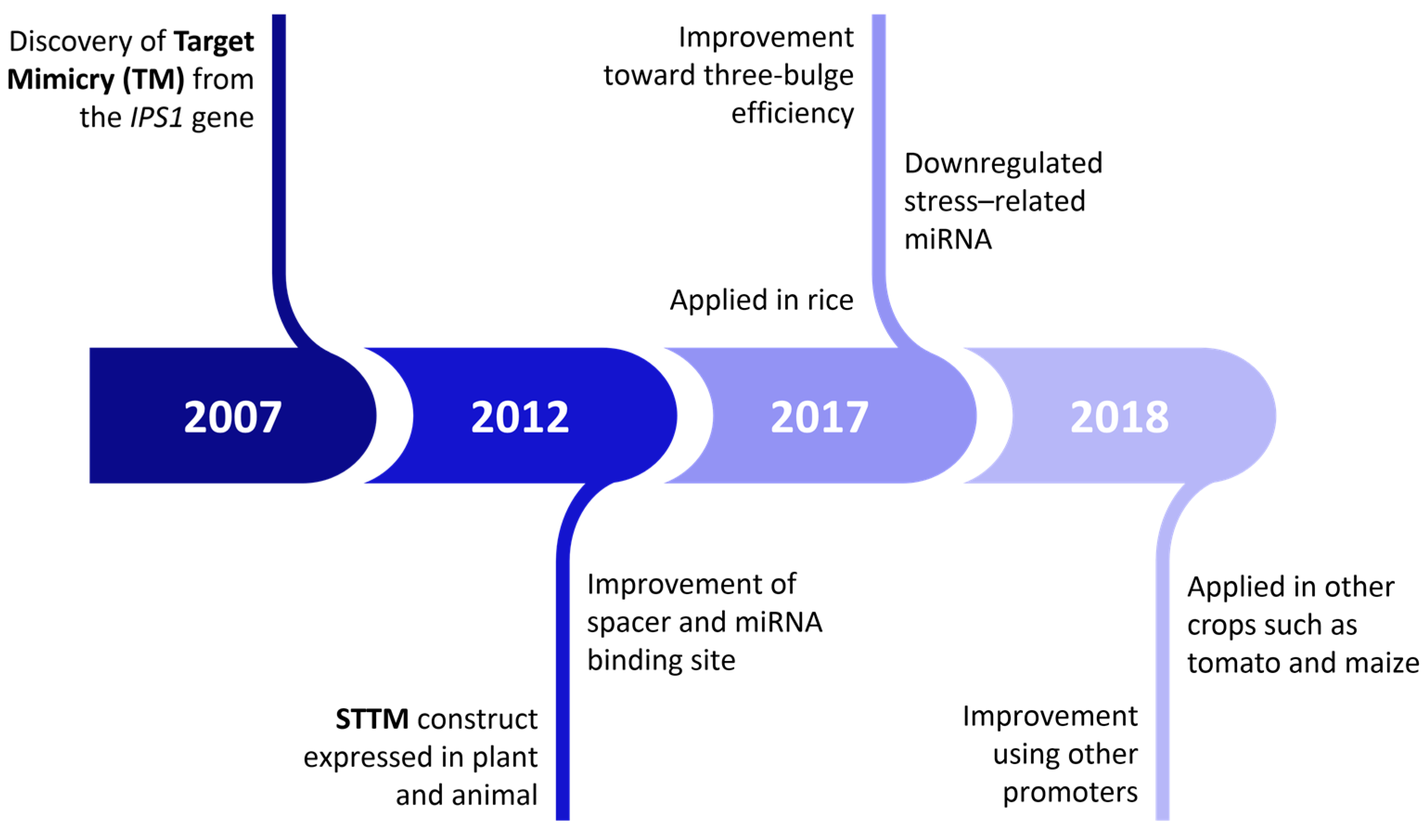
4. STTM Application in miRNA LoF Studies
5. Effectiveness of STTM in Regulating miRNA
6. Diverse Plant miRNA Functions Uncovered Using STTM
6.1. Plant Development and Architecture
6.2. Leaf Development
6.3. Root Development
6.4. Flower Development
6.5. Fruit Development
6.6. Secondary Metabolite
6.7. Biotic and Abiotic Stresses
6.8. Yield
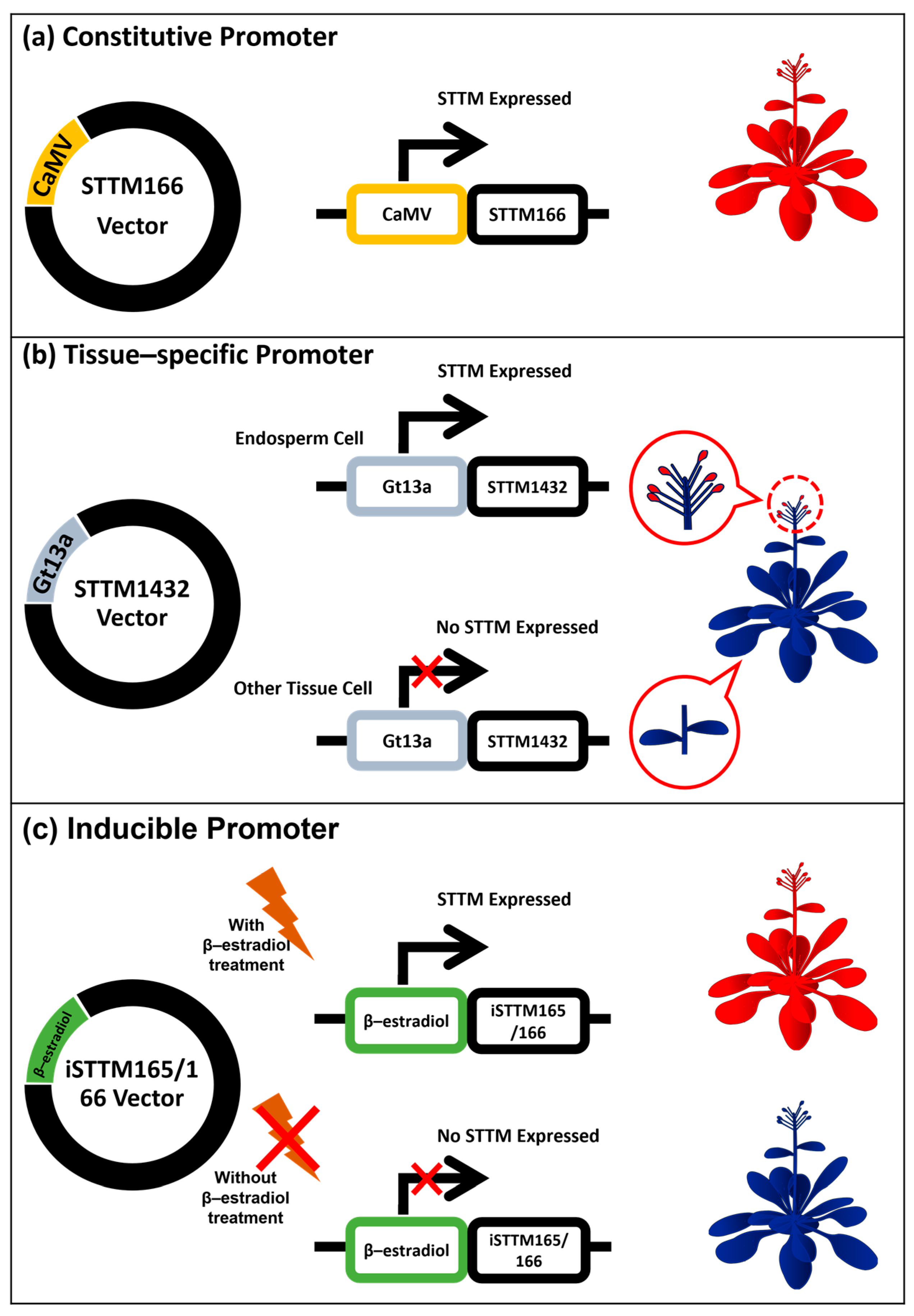
7. Application of STTM as a Constitutive and Spatial–Temporal miRNA Repressor
8. Future Direction of STTM as Alternative Ways to Explore miRNA in Different Spatial–Temporal Expressions
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, B.; Raina, A.; Khan, S. Impact of biotic and abiotic stresses on plants, and their responses. In Disease Resistance in Crop Plants; Wani, S.H., Ed.; Springer: Cham, Switzerland, 2019; pp. 1–19. [Google Scholar] [CrossRef]
- Deutsch, C.A.; Tewksbury, J.J.; Tigchelaar, M.; Battisti, D.S.; Merrill, S.C.; Huey, R.B.; Naylor, R.L. Increase in crop losses to insect pests in a warming climate. Science 2018, 361, 916–919. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Qin, N.; Wang, J.; Chen, X.; Yang, G.; Liang, H. Impacts of climate change on regional hydrological regimes of the Wujiang River watershed in the Karst Area, Southwest China. Geoenvironmental Disasters 2015, 2, 10. [Google Scholar] [CrossRef]
- Yadav, N.; Kumar, A.; Kumar, N.; Kumar, S.; Arya, S.S. Impacts on plant growth and development under stress. In Plant Stress Mitigators; Springer Nature Singapore: Singapore, 2022; pp. 61–100. [Google Scholar] [CrossRef]
- United Nation. Growing at a Slower Pace, World Population Is Expected to Reach 9.7 Billion in 2050 and Could Peak at Nearly 11 Billion around 2100; United Nation: New York, NY, USA, 2019. Available online: https://www.un.org/development/desa/en/news/population/world-population-prospects-2019.html (accessed on 20 July 2022).
- Kromdijk, J.; Long, S.P. One crop breeding cycle from starvation? How engineering crop photosynthesis for rising CO2 and temperature could be one important route to alleviation. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152578. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Wang, G.; Zhang, J.; Wang, G. Analysis of gene expression in early seed germination of rice: Landscape and genetic regulation. BMC Plant Biol. 2022, 22, 70. [Google Scholar] [CrossRef]
- Yang, J.; Xu, Y.; Wang, J.; Gao, S.; Huang, Y.; Hung, F.-Y.; Li, T.; Li, Q.; Yue, L.; Wu, K.; et al. The chromatin remodelling ATPase BRAHMA interacts with GATA-family transcription factor GNC to regulate flowering time in Arabidopsis. J. Exp. Bot. 2022, 73, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Fu, D. The roles of BLH transcription factors in plant development and environmental response. Int. J. Mol. Sci. 2022, 23, 3731. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Shi, H.; Zhang, Z.; Fan, T.; Dang, H.; Chen, P.; Xie, Y.; Bao, C.; Ma, F.; Guan, Q.; et al. MdMYB88/124 modulates apple tree microRNA biogenesis through post-transcription processing and/or transcription pathway. Acta Physiol. Plant. 2022, 44, 86. [Google Scholar] [CrossRef]
- Hu, Z.; Nie, Z.; Yan, C.; Huang, H.; Ma, X.; Wang, Y.; Ye, N.; Tuskan, G.A.; Yang, X.; Yin, H. Transcriptome and degradome profiling reveals a role of miR530 in the circadian regulation of gene expression in Kalanchoë marnieriana. Cells 2021, 10, 1526. [Google Scholar] [CrossRef]
- Xu, J.; Du, R.; Meng, X.; Zhao, W.; Kong, L.; Chen, J. Third-Generation sequencing indicated that LncRNA could regulate eIF2D to enhance protein translation under heat stress in Populus Simonii. Plant Mol. Biol. Rep. 2021, 39, 240–250. [Google Scholar] [CrossRef]
- Dong, Q.; Hu, B.; Zhang, C. microRNAs and their roles in plant development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef]
- Fukunaga, R. miRNAs/small noncoding RNAs. In Encyclopedia of Cell Biology; Elsevier: Baltimore, MD, USA, 2016; pp. 354–363. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Dubery, I.A. Lipopolysaccharide perception leads to dynamic alterations in the microtranscriptome of Arabidopsis thaliana cells and leaf tissues. BMC Plant Biol. 2015, 15, 79. [Google Scholar] [CrossRef]
- Ding, X.; Guo, J.; Zhang, Q.; Yu, L.; Zhao, T.; Yang, S. Heat-responsive miRNAs participate in the regulation of male fertility stability in soybean CMS-based F1 under high temperature stress. Int. J. Mol. Sci. 2021, 22, 2446. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Nong, W.; Zhao, S.; Lin, X.; Xie, Y.; Cheung, M.-Y.; Xiao, Z.; Wong, A.Y.P.; Chan, T.F.; Hui, J.H.L.; et al. Differential microRNA expression, microRNA arm switching, and microRNA: Long noncoding RNA interaction in response to salinity stress in soybean. BMC Genom. 2022, 23, 65. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, Y.; Pan, H.; Zhu, J.; Zhu, M.; Xu, T.; Li, Z.; Dong, T. Molecular characterization and target prediction of candidate miRNAs related to abiotic stress responses and/or storage root development in sweet potato. Genes 2022, 13, 110. [Google Scholar] [CrossRef]
- Zhang, Y.; Waseem, M.; Zeng, Z.; Xu, J.; Chen, C.; Liu, Y.; Zhai, J.; Xia, R. microRNA482/2118, a miRNA superfamily essential for both disease resistance and plant development. New Phytol. 2022, 233, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.-L.; Qu, L.-H. Application of microRNA gene resources in the improvement of agronomic traits in rice. Plant Biotechnol. J. 2015, 13, 329–336. [Google Scholar] [CrossRef]
- Peng, T.; Teotia, S.; Tang, G.; Zhao, Q. microRNAs meet with quantitative trait loci: Small powerful players in regulating quantitative yield traits in rice. WIREs RNA 2019, 10, e1556. [Google Scholar] [CrossRef]
- Yu, Y.; Ni, Z.; Wang, Y.; Wan, H.; Hu, Z.; Jiang, Q.; Sun, X.; Zhang, H. Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Sci. 2019, 285, 68–78. [Google Scholar] [CrossRef]
- Zubair, M.; Khan, M.Z.; Rauf, I.; Raza, A.; Shah, A.H.; Hassan, I.; Amin, I.; Mansoor, S. Artificial micro RNA (AmiRNA)-mediated resistance against whitefly (Bemisia tabaci) targeting three genes. Crop Prot. 2020, 137, 105308. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, F. Different roles of a novel shrimp microRNA in white spot syndrome virus (WSSV) and Vibrio alginolyticus infection. Dev. Comp. Immunol. 2018, 79, 21–30. [Google Scholar] [CrossRef]
- Tsygankova, V.A.; Andrusevich, Y.V.; Shysha, E.N.; Biliavska, L.O.; Galagan, T.O.; Galkin, A.P.; Yemets, A.I.; Iutynska, G.A.; Blume, Y.B. RNAi-mediated resistance against plant parasitic nematodes of wheat plants obtained in vitro using bioregulators of microbiological origin. Curr. Chem. Biol. 2019, 13, 73–89. [Google Scholar] [CrossRef]
- Bi, H.; Fei, Q.; Li, R.; Liu, B.; Xia, R.; Char, S.N.; Meyers, B.C.; Yang, B. Disruption of miRNA sequences by TALENs and CRISPR/Cas9 induces varied lengths of miRNA production. Plant Biotechnol. J. 2020, 18, 1526–1536. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
- Morea, E.G.O.; da Silva, E.M.; e Silva, G.F.F.; Valente, G.T.; Barrera Rojas, C.H.; Vincentz, M.; Nogueira, F.T.S. Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol. 2016, 16, 40. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Gangadhar, B.H.; Venkidasamy, B.; Samynathan, R.; Saranya, B.; Chung, I.-M.; Thiruvengadam, M. Overview of miRNA biogenesis and applications in plants. Biologia 2021, 76, 2309–2327. [Google Scholar] [CrossRef]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive La Différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef]
- Cui, J.; You, C.; Chen, X. The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 2017, 35, 61–67. [Google Scholar] [CrossRef]
- Wei, X.; Ke, H.; Wen, A.; Gao, B.; Shi, J.; Feng, Y. Structural basis of microRNA processing by Dicer-like 1. Nat. Plants 2021, 7, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dong, W.; Ren, W.; Zhao, Q.; Wu, F.; He, Y. Cytoplasmic HYL1 modulates miRNA-mediated translational repression. Plant Cell 2021, 33, 1980–1996. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, H.; Liu, K.; Yang, W.; Zhou, B.; Gan, L.; Li, S.; Zhang, C.; Yu, B. Serrate-associated protein 1, a splicing-related protein, promotes miRNA biogenesis in Arabidopsis. New Phytol. 2021, 232, 1959–1973. [Google Scholar] [CrossRef] [PubMed]
- Hossain, R.; Quispe, C.; Saikat, A.S.M.; Jain, D.; Habib, A.; Janmeda, P.; Islam, M.T.; Daştan, S.D.; Kumar, M.; Butnariu, M.; et al. Biosynthesis of secondary metabolites based on the regulation of microRNAs. BioMed Res. Int. 2022, 2022, 9349897. [Google Scholar] [CrossRef]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef]
- Liu, C.; Xin, Y.; Xu, L.; Cai, Z.; Xue, Y.; Liu, Y.; Xie, D.; Liu, Y.; Qi, Y. Arabidopsis Argonaute 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev. Cell 2018, 44, 348–361.e7. [Google Scholar] [CrossRef] [PubMed]
- Baranauskė, S.; Mickutė, M.; Plotnikova, A.; Finke, A.; Venclovas, Č.; Klimašauskas, S.; Vilkaitis, G. Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. 2015, 43, 2802–2812. [Google Scholar] [CrossRef]
- Cambiagno, D.A.; Giudicatti, A.J.; Arce, A.L.; Gagliardi, D.; Li, L.; Yuan, W.; Lundberg, D.S.; Weigel, D.; Manavella, P.A. HASTY modulates miRNA biogenesis by linking pri-miRNA transcription and processing. Mol. Plant 2021, 14, 426–439. [Google Scholar] [CrossRef]
- Dalmadi, Á.; Miloro, F.; Bálint, J.; Várallyay, É.; Havelda, Z. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021, 49, 12912–12928. [Google Scholar] [CrossRef]
- Fang, X.; Zhao, G.; Zhang, S.; Li, Y.; Gu, H.; Li, Y.; Zhao, Q.; Qi, Y. Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev. Cell 2019, 48, 371–382.e4. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. microRNAs and their regulatory roles in plant. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Bansal, C.; Balyan, S.; Mathur, S. Inferring the regulatory network of the miRNA-mediated response to individual and combined heat and drought stress in tomato. J. Plant Biochem. Biotechnol. 2021, 30, 862–877. [Google Scholar] [CrossRef]
- Şanlı, B.A.; Öztürk Gökçe, Z.N. Investigating effect of miR160 through overexpression in potato cultivars under single or combination of heat and drought stresses. Plant Biotechnol. Rep. 2021, 15, 335–348. [Google Scholar] [CrossRef]
- Liu, X.; Xia, B.; Purente, N.; Chen, B.; Zhou, Y.; He, M. Transgenic Chrysanthemum indicum overexpressing cin-miR396a exhibits altered plant development and reduced salt and drought tolerance. Plant Physiol. Biochem. 2021, 168, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Yadav, K.; Srivastava, A.K.; Suprasanna, P.; Ganapathi, T.R. Overexpression of native Musa-miR397 enhances plant biomass without compromising abiotic stress tolerance in banana. Sci. Rep. 2019, 9, 16434. [Google Scholar] [CrossRef]
- Basso, M.F.; Ferreira, P.C.G.; Kobayashi, A.K.; Harmon, F.G.; Nepomuceno, A.L.; Molinari, H.B.C.; Grossi-de-Sa, M.F. microRNAs and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechnol. J. 2019, 17, 1482–1500. [Google Scholar] [CrossRef]
- Vaistij, F.E.; Elias, L.; George, G.L.; Jones, L. Suppression of microRNA accumulation via RNA interference in Arabidopsis thaliana. Plant Mol. Biol. 2010, 73, 391–397. [Google Scholar] [CrossRef]
- Tang, G.; Yan, J.; Gu, Y.; Qiao, M.; Fan, R.; Mao, Y.; Tang, X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef]
- Zhao, Y.; Wen, H.; Teotia, S.; Du, Y.; Zhang, J.; Li, J.; Sun, H.; Tang, G.; Peng, T.; Zhao, Q. Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol. 2017, 17, 215. [Google Scholar] [CrossRef]
- Villar-Martin, L.M.; Rubio-Somoza, I. Mimicry technology: A versatile tool for small RNA suppression. In Plant microRNAs: Methods and Protocols; de Folter, S., Ed.; Springer New York: New York, NY, USA, 2019; pp. 239–245. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. A direct comparison of anti-microRNA oligonucleotide potency. Pharm. Res. 2010, 27, 1788–1799. [Google Scholar] [CrossRef]
- He, L.; Xie, M.; Huang, J.; Zhang, T.; Shi, S.; Tang, T. Efficient and specific inhibition of plant microRNA function by anti-microrna oligonucleotides (AMOs) in vitro and in vivo. Plant Cell Rep. 2016, 35, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Borah, P.; Das, A.; Milner, M.; Ali, A.; Bentley, A.; Pandey, R. Long non-coding RNAs as endogenous target mimics and exploration of their role in low nutrient stress tolerance in plants. Genes 2018, 9, 459. [Google Scholar] [CrossRef] [PubMed]
- Karakülah, G.; Yücebilgili Kurtoğlu, K.; Unver, T. PeTMbase: A database of plant endogenous target mimics (ETMs). PLoS ONE 2016, 11, e0167698. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, X.; Dobrovolskaya, O.B.; Orlov, Y.L.; Chen, M. Non-coding RNAs and their roles in stress response in plants. Genom. Proteom. Bioinform. 2017, 15, 301–312. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Weigel, D.; Franco-Zorilla, J.-M.; García, J.A.; Paz-Ares, J. CeRNAs: miRNA target mimic mimics. Cell 2011, 147, 1431–1432. [Google Scholar] [CrossRef]
- Gupta, P.K. microRNAs and target mimics for crop improvement. Curr. Sci. 2015, 108, 1624–1633. [Google Scholar]
- Ye, C.; Chen, L.; Liu, C.; Zhu, Q.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol. 2015, 208, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Kluiver, J.; Gibcus, J.H.; Hettinga, C.; Adema, A.; Richter, M.K.S.; Halsema, N.; Slezak-Prochazka, I.; Ding, Y.; Kroesen, B.-J.; van den Berg, A. Rapid generation of microRNA sponges for microRNA inhibition. PLoS ONE 2012, 7, e29275. [Google Scholar] [CrossRef]
- Reichel, M.; Li, Y.; Li, J.; Millar, A.A. Inhibiting plant microRNA activity: Molecular SPONGEs, Target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol. J. 2015, 13, 915–926. [Google Scholar] [CrossRef]
- Peng, T.; Qiao, M.; Liu, H.; Teotia, S.; Zhang, Z.; Zhao, Y.; Wang, B.; Zhao, D.; Shi, L.; Zhang, C.; et al. A resource for inactivation of microRNAs using short tandem target mimic technology in model and crop plants. Mol. Plant 2018, 11, 1400–1417. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Verma, S.; Mantri, S.; Berman, N.E.; Sandhir, R. Targeting microRNAs in prevention and treatment of neurodegenerative disorders. Drug Dev. Res. 2015, 76, 397–418. [Google Scholar] [CrossRef] [PubMed]
- Teotia, S.; Singh, D.; Tang, G. Technologies to address plant microRNA functions. In Plant Sciences; Miguel, C., Dalmay, T., Chaves, I., Eds.; Plant microRNAs, Concepts and Strategies; Springer Nature: Cham, Switzerland, 2020; pp. 25–43. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R.; Zhu, J.-K. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. Silencing of stress-regulated miRNAs in plants by short tandem target mimic (STTM) approach. In Plant Stress Tolerance: Methods and Protocols; Sunkar, R., Ed.; Springer New York: New York, NY, USA, 2017; pp. 337–348. [Google Scholar] [CrossRef]
- Yu, C.; Chen, Y.; Cao, Y.; Chen, H.; Wang, J.; Bi, Y.-M.; Tian, F.; Yang, F.; Rothstein, S.J.; Zhou, X.; et al. Overexpression of miR169o, an overlapping microRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice. Plant Cell Physiol. 2018, 59, 1234–1247. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Jiang, N.; Ren, G. The F-box protein HAWAIIAN SKIRT is required for mimicry target-induced microRNA degradation in Arabidopsis. J. Integr. Plant Biol. 2019, 61, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.L.M.; Christie, M.D.; Dogan, E.S.; Schwab, R.; Hagmann, J.; van de Weyer, A.-L.; Scacchi, E.; Weigel, D. A role for the F-Box protein HAWAIIAN SKIRT in plant microRNA function. Plant Physiol. 2018, 176, 730–741. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.; Arazi, T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.-K. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef]
- Zhang, M.; Zheng, H.; Jin, L.; Xing, L.; Zou, J.; Zhang, L.; Liu, C.; Chu, J.; Xu, M.; Wang, L. miR169o and ZmNF-YA13 act in concert to coordinate the expression of ZmYUC1 that determines seed size and weight in maize kernels. New Phytol. 2022, 235, 2270–2284. [Google Scholar] [CrossRef]
- Xing, L.; Zhu, M.; Luan, M.; Zhang, M.; Jin, L.; Liu, Y.; Zou, J.; Wang, L.; Xu, M. miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol. 2022, 188, 608–623. [Google Scholar] [CrossRef]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, N.; Zhang, J.; Jin, X.; Zhu, X.; Ma, R.; Li, S.; Lui, S.; Yue, Y.; Si, H. Knockdown of microRNA160a/b by STTM leads to root architecture changes via auxin signaling in Solanum tuberosum. Plant Physiol. Biochem. 2021, 166, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Kar, M.M.; Raichaudhuri, A. Overview of Arabidopsis as a genetics model system and its limitation, leading to the development of emerging plant model systems. In Model Organisms in Plant Genetics; Abdurakhmonov, I., Ed.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Zhan, J.; Chu, Y.; Wang, Y.; Diao, Y.; Zhao, Y.; Liu, L.; Wei, X.; Meng, Y.; Li, F.; Ge, X. The MiR164-GhCUC2-GhBRC1 module regulates plant architecture through abscisic acid in cotton. Plant Biotechnol. J. 2021, 19, 1839–1851. [Google Scholar] [CrossRef]
- Wu, F.; Tanksley, S.D. Chromosomal evolution in the plant family Solanaceae. BMC Genom. 2010, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Sunarti, S.; Kissoudis, C.; Van Der Hoek, Y.; Van Der Schoot, H.; Visser, R.G.F.; Van Der Linden, C.G.; Van De Wiel, C.; Bai, Y. Drought stress interacts with powdery mildew infection in tomato. Front. Plant Sci. 2022, 13, 845379. [Google Scholar] [CrossRef]
- Chen, L.; Meng, J.; He, X.L.; Zhang, M.; Luan, Y.S. Solanum lycopersicum microRNA1916 targets multiple target genes and negatively regulates the immune response in tomato. Plant. Cell Environ. 2019, 42, 1393–1407. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, K.; Gong, Z.; Zhang, Y.; Meng, Y.; Liu, Q. Molecular manipulation of miR398 increases rice grain yield under different conditions. Front. Plant Sci. 2022, 13, 1037604. [Google Scholar] [CrossRef]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q.; et al. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol. 2021, 230, 1017–1033. [Google Scholar] [CrossRef]
- Chi, Q.; Du, L.; Ma, W.; Niu, R.; Wu, B.; Guo, L.; Ma, M.; Liu, X.; Zhao, H. miR164-TaNAC14 module regulates root development and abiotic-stress tolerance of wheat seedlings. J. Integr. Agric. 2022, 1–20, in press. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q.; Hao, L.; Wang, S.; Wang, S.; Zhang, W.; Xu, C.; Yu, Y.; Li, T. A novel miRNA negatively regulates resistance to Glomerella Leaf Spot by suppressing expression of an NBS gene in apple. Hortic. Res. 2019, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, Y.; Zhang, Y.; Wu, C.; Wang, S.; Hao, L.; Wang, S.; Li, T. Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front. Plant Sci. 2017, 8, 526. [Google Scholar] [CrossRef] [PubMed]
- Nie, P.; Chen, C.; Yin, Q.; Jiang, C.; Guo, J.; Zhao, H.; Niu, D. Function of miR825 and miR825* as negative regulators in Bacillus cereus AR156-elicited systemic resistance to Botrytis cinerea in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 5032. [Google Scholar] [CrossRef]
- Jiang, C.; Fan, Z.; Li, Z.; Niu, D.; Li, Y.; Zheng, M.; Wang, Q.; Jin, H.; Guo, J. Bacillus cereus AR156 triggers induced systemic resistance against Pseudomonas syringae Pv. tomato DC3000 by suppressing miR472 and activating CNLs-mediated basal immunity in Arabidopsis. Mol. Plant Pathol. 2020, 21, 854–870. [Google Scholar] [CrossRef]
- Wang, X.; Yu, G.; Zhao, J.; Cui, N.; Yu, Y.; Fan, H. Functional identification of Corynespora cassiicola-responsive miRNAs and their targets in cucumber. Front. Plant Sci. 2019, 10, 668. [Google Scholar] [CrossRef]
- Li, C.; Liu, X.; Abouelnasr, H.; Arisha, M.H.; Kou, M.; Tang, W.; Yan, H.; Wang, X.; Wang, X.; Zhang, Y.; et al. Inhibition of miR397 by STTM technology to increase sweetpotato resistance to SPVD. J. Integr. Agric. 2022, 21, 2865–2875. [Google Scholar] [CrossRef]
- Zhou, L.; Deng, S.; Xuan, H.; Fan, X.; Sun, R.; Zhao, J.; Wang, H.; Guo, N.; Xing, H. A novel TIR-NBS-LRR gene regulates immune response to Phytophthora root rot in soybean. Crop J. 2022, 10, 1644–1653. [Google Scholar] [CrossRef]
- Bao, D.; Ganbaatar, O.; Cui, X.; Yu, R.; Bao, W.; Falk, B.W.; Wuriyanghan, H. Down-regulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful soybean mosaic virus infection in soybean. Mol. Plant Pathol. 2018, 19, 948–960. [Google Scholar] [CrossRef]
- Canto-Pastor, A.; Santos, B.A.M.C.; Valli, A.A.; Summers, W.; Schornack, S.; Baulcombe, D.C. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. USA 2019, 116, 2755–2760. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Meng, J.; Cui, J.; Sun, G.; Luan, Y. Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic. Res. 2018, 5, 9. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, Y.; Cui, J.; Meng, J.; Chen, Y.; Zhang, C.; Yang, J.; Luan, Y. The LncRNA39896–miR166b–HDZs module affects tomato resistance to Phytophthora infestans. J. Integr. Plant Biol. 2022, 64, 1979–1993. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Fang, Y.-N.; Wu, X.-M.; Qing, M.; Li, C.-C.; Xie, K.-D.; Deng, X.-X.; Guo, W.-W. The miR399-CsUBC24 module regulates reproductive development and male fertility in citrus. Plant Physiol. 2020, 183, 1681–1695. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular Mechanism of MicroRNA396 Mediating Pistil Development in Arabidopsis. Plant Physiol. 2014, 164, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Myat, A.A.; Liang, C.; Meng, Z.; Guo, S.; Wei, Y.; Sun, G.; Wang, Y.; Zhang, R. Insights into microRNA-mediated regulation of flowering time in cotton through small RNA sequencing. Front. Plant Sci. 2022, 13, 761244. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Liu, H.; Zhang, W.; Chai, M.; Tang, G.; Zhang, Z. microRNA1917-CTR1-LIKE PROTEIN KINASE 4 impacts fruit development via tuning ethylene synthesis and response. Plant Sci. 2020, 291, 110334. [Google Scholar] [CrossRef]
- Zhao, K.; Song, H.; Wang, Z.; Xing, Z.; Tian, J.; Wang, Q.; Meng, L.; Xu, X. Knockdown of sly-miR164a by short tandem target mimic (STTM) enhanced postharvest chilling tolerance of tomato fruit under low temperature storage. Postharvest Biol. Technol. 2022, 187, 111872. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, R.; Duan, W.; Meng, L.; Song, H.; Wang, Q.; Li, J.; Xu, X. Chilling injury of tomato fruit was alleviated under low-temperature storage by silencing sly-miR171e with short tandem target mimic technology. Front. Nutr. 2022, 9, 906227. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Wu, Y.; Li, D.; Allan, A.C.; Yin, X. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020, 225, 1618–1634. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, F.; Deng, Y.; Zhong, F.; Tian, P.; Lin, D.; Deng, J.; Zhang, Y.; Huang, T. Sly-miR159 regulates fruit morphology by modulating GA biosynthesis in tomato. Plant Biotechnol. J. 2022, 20, 833–845. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, M.; Li, W.; Ma, J.; Liu, Y.; Jiang, H.; Wang, L.; Xu, M. Molecular regulation mechanism of leaf development mediated by Ath-miR169d in Arabidopsis thaliana. Sci. Agric. Sin. 2017, 50, 3063–3070. [Google Scholar]
- Ma, C.; Chen, Q.; Wang, S.; Lers, A. Downregulation of GeBP-like α factor by miR827 suggests their involvement in senescence and phosphate homeostasis. BMC Biol. 2021, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xu, H.; Han, R.; Wang, S.; Liu, G.; Chen, S.; Chen, J.; Bian, X.; Jiang, J. Overexpression of BpCUC2 influences leaf shape and internode development in Betula pendula. Int. J. Mol. Sci. 2019, 20, 4722. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Lin, L.; Xue, H. Rice miR394 suppresses leaf inclination through targeting an F-box Gene, LEAF INCLINATION 4. J. Integr. Plant Biol. 2019, 61, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Wei, X.-Y.; Liu, L.-Z.; Zhou, L.-X.; Tian, Y.-P.; Geng, C.; Li, X.-D. A tobacco ringspot virus-based vector system for gene and microRNA function studies in cucurbits. Plant Physiol. 2021, 186, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fang, R.; Deng, R.; Li, J. The OsmiRNA166b-OsHox32 pair regulates mechanical strength of rice plants by modulating cell wall biosynthesis. Plant Biotechnol. J. 2021, 19, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, X.; Wang, M.; Xie, L.; Wu, Z.; Yu, J.; Wang, Y.; Zhang, Z.; Jia, Y.; Liu, Q. The miR528-D3 module regulates plant height in rice by modulating the gibberellin and abscisic acid metabolisms. Rice 2022, 15, 27. [Google Scholar] [CrossRef]
- Jian, C.; Hao, P.; Hao, C.; Liu, S.; Mao, H.; Song, Q.; Zhou, Y.; Yin, S.; Hou, J.; Zhang, W.; et al. The miR319/TaGAMYB3 module regulates plant architecture and improves grain yield in common wheat (Triticum aestivum). New Phytol. 2022, 235, 1515–1530. [Google Scholar] [CrossRef]
- Wang, X.; Yao, S.; Htet, W.P.P.M.; Yue, Y.; Zhang, Z.; Sun, K.; Chen, S.; Luo, K.; Fan, D. microRNA828 negatively regulates lignin biosynthesis in stem of Populus tomentosa through MYB targets. Tree Physiol. 2022, 42, 1646–1661. [Google Scholar] [CrossRef]
- Jia, X.; Ding, N.; Fan, W.; Yan, J.; Gu, Y.; Tang, X.; Li, R.; Tang, G. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015, 233, 11–21. [Google Scholar] [CrossRef]
- Kravchik, M.; Stav, R.; Belausov, E.; Arazi, T. Functional characterization of microRNA171 family in tomato. Plants 2019, 8, 10. [Google Scholar] [CrossRef]
- Chu, L.; He, X.; Shu, W.; Wang, L.; Tang, F. Knockdown of miR393 promotes the growth and biomass production in poplar. Front. Plant Sci. 2021, 12, 714907. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ma, J.; Zhang, Y.; Yang, S.; Feng, X.; Yan, J. The miR166 mediated regulatory module controls plant height by regulating gibberellic acid biosynthesis and catabolism in soybean. J. Integr. Plant Biol. 2022, 64, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. miR396a-mediated basic helix–loop–helix transcription factor BHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y. LACCASE2 negatively regulates lignin deposition of Arabidopsis roots. Plant Physiol. 2020, 182, 1190–1191. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Xu, C.; Fu, X.; Shen, Y.; Guo, L.; Leng, M.; Luo, K. The microRNA390/TRANS-ACTING SHORT INTERFERING RNA3 module mediates lateral root growth under salt stress via the auxin pathway. Plant Physiol. 2018, 177, 775–791. [Google Scholar] [CrossRef]
- Jia, X.; Shen, J.; Liu, H.; Li, F.; Ding, N.; Gao, C.; Pattanaik, S.; Patra, B.; Li, R.; Yuan, L. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 2015, 242, 283–293. [Google Scholar] [CrossRef]
- Li, F.; Wang, W.; Zhao, N.; Xiao, B.; Cao, P.; Wu, X.; Ye, C.; Shen, E.; Qiu, J.; Zhu, Q.-H.; et al. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco. Plant Physiol. 2015, 169, 1062–1071. [Google Scholar] [CrossRef]
- Zaman, F.; Zhang, M.; Liu, Y.; Wang, Z.; Xu, L.; Guo, D.; Luo, Z.; Zhang, Q. DkmiR397 regulates proanthocyanidin biosynthesis via negative modulating DkLAC2 in chinese persimmon. Int. J. Mol. Sci. 2022, 23, 3200. [Google Scholar] [CrossRef]
- Shi, Q.-F.; Long, J.-M.; Yin, Z.-P.; Jiang, N.; Feng, M.-Q.; Zheng, B.; Guo, W.-W.; Wu, X.-M. miR171 modulates induction of somatic embryogenesis in citrus callus. Plant Cell Rep. 2022, 41, 1403–1415. [Google Scholar] [CrossRef]
- Wang, R.; Yang, X.; Guo, S.; Wang, Z.; Zhang, Z.; Fang, Z. miR319-targeted OsTCP21 and OsGAmyb regulate tillering and grain yield in rice. J. Integr. Plant Biol. 2021, 63, 1260–1272. [Google Scholar] [CrossRef]
- Zhao, Y.; Peng, T.; Sun, H.; Teotia, S.; Wen, H.; Du, Y.; Zhang, J.; Li, J.; Tang, G.; Xue, H.; et al. miR1432-OsACOT (Acyl-CoA Thioesterase) module determines grain yield via enhancing grain filling rate in rice. Plant Biotechnol. J. 2019, 17, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, N.K.; Ziska, L.H. Rice: Importance for global nutrition. J. Nutr. Sci. Vitaminol. 2019, 65, S2–S3. [Google Scholar] [CrossRef]
- Tiwari, R.; Rajam, M.V. RNA and miRNA-interference to enhance abiotic stress tolerance in plants. J. Plant Biochem. Biotechnol. 2022, 31, 689–704. [Google Scholar] [CrossRef]
- Mahanty, B.; Bharat, S.S.; Mohanty, J.N.; Mishra, R.; Joshi, R.K. microRNA-mediated insect resistance in field crops. In Molecular Advances in Insect Resistance of Field Crops: Modern and Applied Approaches; Tanda, A.S., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 369–392. [Google Scholar] [CrossRef]
- Pandita, D. Role of miRNA technology and miRNAs in abiotic and biotic stress resilience. In Plant Perspectives to Global Climate Changes; Aftab, T., Roychoudhury, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 303–330. [Google Scholar] [CrossRef]
- Li, Y.; Tong, Y.; He, X.; Zhu, Y.; Li, T.; Lin, X.; Mao, W.; Gishkori, Z.G.N.; Zhao, Z.; Zhang, J.; et al. The rice miR171b–SCL6-IIs module controls blast resistance, grain yield, and flowering. Crop J. 2022, 10, 117–127. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, J.; Wang, K.; Li, D.; Yang, R.; Luo, H.; Zhang, W. miR396-GRF module associates with switchgrass biomass yield and feedstock quality. Plant Biotechnol. J. 2021, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Shu, Y.; Peng, S.; Li, Y. Leaf photosynthesis is positively correlated with xylem and phloem areas in leaf veins in rice (Oryza sativa) plants. Ann. Bot. 2022, 129, 619–631. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, S.; Tu, Y.; Mei, H.; Yang, Y. A novel microRNA, slymiR208, promotes leaf senescence via regulating cytokinin biosynthesis in tomato. Physiol. Plant. 2020, 169, 143–155. [Google Scholar] [CrossRef]
- Malamy, J.E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005, 28, 67–77. [Google Scholar] [CrossRef]
- Barrera-Rojas, C.H.; Rocha, G.H.B.; Polverari, L.; Pinheiro Brito, D.A.; Batista, D.S.; Notini, M.M.; da Cruz, A.C.F.; Morea, E.G.O.; Sabatini, S.; Otoni, W.C.; et al. miR156-targeted SPL10 controls Arabidopsis root meristem activity and root-derived de novo shoot regeneration via cytokinin responses. J. Exp. Bot. 2020, 71, 934–950. [Google Scholar] [CrossRef]
- Cai, H.; Yang, C.; Liu, S.; Qi, H.; Wu, L.; Xu, L.-A.; Xu, M. miRNA-target pairs regulate adventitious rooting in populus: A functional role for miR167a and its target auxin response factor 8. Tree Physiol. 2019, 39, 1922–1936. [Google Scholar] [CrossRef]
- Rao, S.; Li, Y.; Chen, J. Combined analysis of microRNAs and target genes revealed miR156-SPLs and miR172-AP2 are involved in a delayed flowering phenomenon after chromosome doubling in black goji (Lycium ruthencium). Front. Genet. 2021, 12, 706930. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.M.; Silva, G.F.F.e; Bidoia, D.B.; Silva Azevedo, M.; Jesus, F.A.; Pino, L.E.; Peres, L.E.P.; Carrera, E.; López-Díaz, I.; Nogueira, F.T.S. microRNA159-targeted SlGAMYB transcription factors are required for fruit set in tomato. Plant J. 2017, 92, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Kong, J.; Lai, T.; Manning, K.; Wu, C.; Wang, Y.; Qin, C.; Li, B.; Yu, Z.; Zhang, X.; et al. Tuning LeSPL-CNR expression by slymiR157 affects tomato fruit ripening. Sci. Rep. 2015, 5, 7852. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liu, C.; Wang, Y.; Dong, Q.; Gai, Y.; Ji, X. microRNA profiling during mulberry (Morus atropurpurea Roxb) fruit development and regulatory pathway of miR477 for anthocyanin accumulation. Front. Plant Sci. 2021, 12, 687364. [Google Scholar] [CrossRef]
- Kong, X.; Zeng, J.; Yun, Z.; Hu, C.; Yang, B.; Qu, H.; Jiang, Y.; Zhu, H. Characterization of miRNA-mediated auxin signaling during banana (Musa Spp.) fruit ripening. Postharvest Biol. Technol. 2022, 193, 112045. [Google Scholar] [CrossRef]
- Ding, T.; Tomes, S.; Gleave, A.P.; Zhang, H.; Dare, A.P.; Plunkett, B.; Espley, R.V.; Luo, Z.; Zhang, R.; Allan, A.C.; et al. microRNA172 targets APETALA2 to regulate flavonoid biosynthesis in apple (Malus domestica). Hortic. Res. 2022, 9, uhab007. [Google Scholar] [CrossRef]
- Du, Q.; Lv, W.; Guo, Y.; Yang, J.; Wang, S.; Li, W.-X. MIR164b represses iron uptake by regulating the NAC domain transcription factor5-nuclear factor y, subunit A8 module in Arabidopsis. Plant Physiol. 2022, 189, 1095–1109. [Google Scholar] [CrossRef]
- Liu, W.; Cui, J.; Luan, Y. Overexpression of LncRNA08489 enhances tomato immunity against Phytophthora infestans by decoying miR482e-3p. Biochem. Biophys. Res. Commun. 2022, 587, 36–41. [Google Scholar] [CrossRef]
- Dey, S.; Sarkar, A.; Chowdhury, S.; Singh, R.; Mukherjee, A.; Ghosh, Z.; Kundu, P. Heightened miR6024-NLR interactions facilitate necrotrophic pathogenesis in tomato. Plant Mol. Biol. 2022, 109, 717–739. [Google Scholar] [CrossRef]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef]
- Peng, T.; Lv, Q.; Zhang, J.; Li, J.; Du, Y.; Zhao, Q. Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa). J. Exp. Bot. 2011, 62, 4943–4954. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Sun, H.; Qiao, M.; Zhao, Y.; Du, Y.; Zhang, J.; Li, J.; Tang, G.; Zhao, Q. Differentially expressed microRNA cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice. BMC Plant Biol. 2014, 14, 196. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Ahanger, M.A.; Zhou, J.; Ahmed, N.; Wei, C.; Yuan, S.; Ashraf, M.; Zhang, L. Beneficial role of acetylcholine in chlorophyll metabolism and photosynthetic gas exchange in Nicotiana benthamiana seedlings under salinity stress. Plant Biol. 2020, 22, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Hussain, M.; Irfan, M.; Shi, B. Current understanding of the regulatory roles of miRNAs for enhancing photosynthesis in plants under environmental stresses. In Photosynthesis, Productivity and Environmental Stress; Wiley: Hoboken, NJ, USA, 2019; pp. 163–195. [Google Scholar] [CrossRef]
- Sekhar, S.; Das, S.; Panda, D.; Mohanty, S.; Mishra, B.; Kumar, A.; Navadagi, D.B.; Sah, R.P.; Pradhan, S.K.; Samantaray, S.; et al. Identification of microRNAs that provide a low light stress tolerance-mediated signaling pathway during vegetative growth in rice. Plants 2022, 11, 2558. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef]
- Della Rosa, M.; Spivakov, M. Silencers in the spotlight. Nat. Genet. 2020, 52, 244–245. [Google Scholar] [CrossRef]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef]
- Rombauts, S.; Florquin, K.; Lescot, M.; Marchal, K.; Rouzé, P.; Van de Peer, Y. Computational approaches to identify promoters and Cis-regulatory elements in plant genomes. Plant Physiol. 2003, 132, 1162–1176. [Google Scholar] [CrossRef]
- Jiang, P.; Zhang, K.; Ding, Z.; He, Q.; Li, W.; Zhu, S.; Cheng, W.; Zhang, K.; Li, K. Characterization of a strong and constitutive promoter from the Arabidopsis serine carboxypeptidase-like gene AtSCPL30 as a potential tool for crop transgenic breeding. BMC Biotechnol. 2018, 18, 59. [Google Scholar] [CrossRef]
- Dutt, M.; Dhekney, S.A.; Soriano, L.; Kandel, R.; Grosser, J.W. Temporal and spatial control of gene expression in horticultural crops. Hortic. Res. 2014, 1, 14047. [Google Scholar] [CrossRef]
- Forlani, S.; Mizzotti, C.; Masiero, S. The NAC side of the fruit: Tuning of fruit development and maturation. BMC Plant Biol. 2021, 21, 238. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Y.P.; Zhou, X.H.; Yang, X.M.; He, X.R.; Feng, Q.; Zhu, Y.; Li, G.B.; Wang, H.; Zhao, J.-H.; et al. Rice miR1432 fine-tunes the balance of yield and blast disease resistance via different modules. Rice 2021, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zhang, L.; Wang, J.; Liu, Z.; Zhang, Y.; Xue, C.; Liu, M.; Zhao, J. ZjSEP3 modulates flowering time by regulating the LHY promoter. BMC Plant Biol. 2021, 21, 527. [Google Scholar] [CrossRef]
- Huang, S.; Zheng, C.; Zhao, Y.; Li, Q.; Liu, J.; Deng, R.; Lei, T.; Wang, S.; Wang, X. RNA interference knockdown of the brassinosteroid receptor BRI1 in potato (Solanum tuberosum L.) reveals novel functions for brassinosteroid signaling in controlling tuberization. Sci. Hortic. 2021, 290, 110516. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, Y.; Huang, S.; Liu, Z.; Feng, H. Identification of an anther-specific promoter from a male sterile AB line in chinese cabbage (Brassica rapa L. Ssp. pekinensis). 3 Biotech 2022, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Tue, N.H.; Tuong, T.G.C.; Trang, P.T.H.; Chung, N.D.; Hoa, P.T.; Tien, N.Q.D.; Loc, N.H. Cloning the root-specific Asy promoter and genes encoding chitinase 42 KDa of Trichoderma asperellum into the plant expression vector. J. Appl. Biol. Biotechnol. 2022, 10, 7–11. [Google Scholar] [CrossRef]
- Xun, H.; Zhang, X.; Yu, J.; Pang, J.; Wang, S.; Liu, B.; Dong, Y.; Jiang, L.; Guo, D. Analysis of expression characteristics of soybean leaf and root tissue-specific promoters in Arabidopsis and soybean. Transgen. Res. 2021, 30, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.-T.; Skinner, J.S.; Park, E.-J.; Jeknić, Z.; Hayes, P.M.; Thomashow, M.F.; Chen, T.H.H. Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol. J. 2007, 5, 591–604. [Google Scholar] [CrossRef]
- Mishra, D.K.; Srivastava, R.; Pandey, B.K.; Verma, P.C.; Sawant, S.V. Identification and validation of the wound and insect bite early inducible promoter from Arabidopsis thaliana. 3 Biotech 2022, 12, 74. [Google Scholar] [CrossRef]
- Xu, C.; Tao, Y.; Fu, X.; Guo, L.; Xing, H.; Li, C.; Yang, Z.; Su, H.; Wang, X.; Hu, J.; et al. The microRNA476a-RFL module regulates adventitious root formation through a mitochondria-dependent pathway in populus. New Phytol. 2021, 230, 2011–2028. [Google Scholar] [CrossRef]
- Yang, Y.; Lee, J.H.; Poindexter, M.R.; Shao, Y.; Liu, W.; Lenaghan, S.C.; Ahkami, A.H.; Blumwald, E.; Stewart, C.N., Jr. Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements. Plant Biotechnol. J. 2021, 19, 1354–1369. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Huang, Y.; Yang, J.; Yao, S.; Zhao, K.; Wang, D.; Qin, Q.; Bian, Z.; Li, Y.; Lan, Y.; et al. Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 2020, 28, 89–103.e8. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Reyes-Pérez, P.; Angulo-Bejarano, P.I.; Srivastava, A.; Ramalingam, S.; Sharma, A. Characterization of microRNAs from neem (Azadirachta indica) and their tissue-specific expression study in leaves and stem. 3 Biotech 2021, 11, 277. [Google Scholar] [CrossRef]
- Premachandran, Y. Triggered in Distress: A miRNA-controlled switch for drought-induced ABA biosynthesis in rice. Plant Physiol. 2022, 189, 447–449. [Google Scholar] [CrossRef] [PubMed]
- Pawełkowicz, M.E.; Skarzyńska, A.; Koter, M.D.; Turek, S.; Pląder, W. miRNA profiling and its role in multi-omics regulatory networks connected with somaclonal variation in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2022, 23, 4317. [Google Scholar] [CrossRef]
- Gutiérrez-García, C.; Ahmed, S.S.S.J.; Ramalingam, S.; Selvaraj, D.; Srivastava, A.; Paul, S.; Sharma, A. Identification of microRNAs from medicinal plant Murraya koenigii by high-throughput sequencing and their functional implications in secondary metabolite biosynthesis. Plants 2021, 11, 46. [Google Scholar] [CrossRef]
- Öztürk Gökçe, Z.N.; Aksoy, E.; Bakhsh, A.; Demirel, U.; Çalışkan, S.; Çalışkan, M.E. Combined drought and heat stresses trigger different sets of miRNAs in contrasting potato cultivars. Funct. Integr. Genom. 2021, 21, 489–502. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Yuan, L.; Liu, Y.; Shen, T.; Zhang, Y. Comparative small RNA profiling and functional exploration on wheat with high- and low-cadmium accumulation. Front. Genet. 2021, 12, 635599. [Google Scholar] [CrossRef]

| Component | No/Weak Phenotypic Alteration | Strong Phenotypic Alteration |
|---|---|---|
| Spacer construct | Absence | Present |
| STTM expression | Low expression | High expression |
| Spacer lengths | Lengths of 8 and 31 nucleotides | Lengths of 48, 88 and 96 nucleotides |
| Stem region structure | Stem region disrupted via mutation, even though a long spacer was used | Stable stem region with no mutation |
| Number of miRNA binding sites | One binding site | Two binding sites |
| Mutation of miRNA binding sites | Mutation occurs | No mutation occurs |
| Tri-nucleotide bulge | Complementary to 10th to 11th | Not complementary to 10th to 11th |
| Plant | STTM Target(s) | Target Gene(s) | Trait | Observed Effects | References |
|---|---|---|---|---|---|
| Rice | miR398 | CSD1–2, CCS | Abiotic resistance | Tolerance to salinity but growth inhibited under normal conditions | [86] |
| Maize | ZmmiR169q | ZmNF-YA8 | Abiotic resistance | Enhanced plant salt resistance | [78] |
| Rice | miR172 | IDS1 | Abiotic resistance | Reactive oxygen species (ROS) regulation | [87] |
| Wheat | miR164 | TaNAC14 | Abiotic resistance | Impacted root development and growth and stress (drought and salinity) | [88] |
| Apple | MdmiRln20 | Md-TN1-GLS | Biotic resistance | Reduced Glomerella leaf spot (GLS) incidence | [89] |
| Apple | MdmiR156ab MdmiR395 | MdWRKYN1MdWRKY26 | Biotic resistance | WRKY-regulated pathogenesis-related (PR) protein-encoding genes boost plant biotic resistance | [90] |
| Arabidopsis | miR825/825 | AT5G40910 AT5G38850AT3G04220AT5G44940 | Biotic resistance | Increased resistance to Botrytis cinerea B1301 strain | [91] |
| Arabidopsis | miR472 | NBS-LRR | Biotic resistance | Increased resistance to Pseudomonas syringae Pv. tomato (Pst) DC3000 | [92] |
| Cucumber | miR164d miR396b NovelmiR1NovelmiR7 | NAC, APE, 4CL, PAL | Biotic resistance | Increased resistance to Corynespora cassiicola | [93] |
| Potato | miR397 | IbLACs | Biotic resistance | Upregulates lignin content that provides physical defence and reduces the accumulation of sweet potato virus disease (SPVD) | [94] |
| Soybean | miR1510 | GmTNL16 | Biotic resistance | Plant hormone signaling and secondary metabolism interact with Phytophthora sojae infection | [95] |
| Soybean | miR1507a, miR1507c, miR482a, miR168a, miR1515a | Five NBS-LRR family genes | Biotic resistance | Increased resistance towards soybean mosaic virus (SMV) | [96] |
| Tomato | miR482, miR2118b | NLR | Biotic resistance | Enhanced resistance toward oomycete and bacterial pathogens | [97] |
| Tomato | miR482b | NBS-LRR | Biotic resistance | Enhancement of tomato resistance to Phytophthora infestans | [98] |
| Tomato | sly-miR1916 | STR-2, UGTs, MYB12 | Biotic resistance | Accumulation of α-tomatine and anthocyanins confers biotic stress tolerance | [85] |
| Tomato | miR166b | SlHDZ34/45 | Biotic resistance | Reduced pathogen accumulation in P. infestans-infected plants | [99] |
| Tomato | miR482/2118 | NLR | Biotic resistance | Improved resistance to oomycete and bacterial pathogen infection | [97] |
| Citrus | CsmiR399a.1 | CsUBC24 | Flower development | Abnormal floral development, suppression of anther dehiscence, and diminished pollen productivity | [100] |
| Arabidopsis | miR396 | GRF1, GRF2, GRF3, GRF4, GRF7, GRF8, GRF9 | Flower development | Rescues abnormal pistils and siliques | [101] |
| Cotton | Gh-miR156 | CHLI | Flower development | Early flowering | [102] |
| Tomato | miR1917 | SlCTR4 | Fruit development | Increased biomass, longer terminal leaflet, bigger floral organ and better fruit and seed size | [103] |
| Tomato | Sly-miR164a | NAC1, NAM3 | Fruit development | The amount of hydrogen peroxide (H2O2) in the fruit decreased, and its firmness increased in post-harvest chilling | [104] |
| Tomato | Sly-miR171e | GRAS24, CBF1, GA2ox1, COR, GA3, GA20ox1, GA3ox1 | Fruit development | Reduced chilling injury (CI) index, lower hydrogen peroxide (H2O2) level, and greater fruit firmness after harvest | [105] |
| Kiwi | miR164 | AdNAC6, AdNAC7 | Fruit development | Faster fruit ripening | [106] |
| Tomato | sly-miR159 | SlGA3ox1, SlGA3ox2 | Fruit morphology | Larger fruit | [107] |
| Arabidopsis | miR169d | YUC2, PIN1, ARF1, ARF2 | Leaf development | More and larger leaves | [108] |
| Arabidopsis | miR827 | GPLα | Leaf development | Reduced PTP1 gene expression, decreased leaf phosphate | [109] |
| Silver birch | BpmiR164 | BpCUC2 | Leaf development | Reduced internodes and irregular leaf forms | [110] |
| Rice | miR394 | LC4 | Leaf development | Increased leaf angles | [111] |
| Arabidopsis | miR164 | GhCUC2 | Plant architecture | Decreased length and number of lateral branches | [82] |
| Cotton | miR164 | GhCUC2 | Plant architecture | Decreased length and number of lateral branches | [82] |
| Cucurbit | miR159 | GAMYB | Plant architecture | Dwarf with smaller leaves | [112] |
| Potato | miR160a/b | StARF10, StARF16 | Plant architecture | Shorter roots, more lateral roots, and less fresh root weight | [80] |
| Rice | OsmiR166 | OsHox32 | Plant architecture | Increased thickness of cell wall and culm strength | [113] |
| Rice | miR166 | OsHB4 | Plant architecture | More drought tolerance with rolled-leaf phenotype and reduced xylem vessel diameter | [76] |
| Rice | miR159 | OsGAMYB, OsGAMYBL1 | Plant architecture | Reduced height and stem diameter, flag leaf length, main panicle, spikelet hulls, and grain size | [53] |
| Rice | miR528 | DWARF3 (D3) | Plant architecture | Higher plant height due to lower abscisic acid and higher gibberellin | [114] |
| Wheat | miR319 | TaPCF8, TaGAMYB3 | Plant architecture | Increased plant height, reduced tiller number, spikes and flag leaves, thicker culms, and higher grain output | [115] |
| Poplar | miR828 | MYB171, MYB011 | Plant architecture | Increased expression of lignin biosynthesis genes and lignin accumulation | [116] |
| Arabidopsis | miR165/166 | HD-ZIP III | Plant development | Increased auxin content and decreased auxin sensitivity | [117] |
| Tomato | sly-miR171a-b | SlHAM, SlHAM2 | Plant development | Delayed anther development, increased shoot branching and compound leaf morphogenesis | [118] |
| Poplar | miR393 | FBL family members | Plant development | Taller, thicker, more internodes, broader phloem, xylem, and cambium cell layers, higher lignin content | [119] |
| Soybean | GmSTTM166 | ATHB14-LIKE | Plant development | Stunted growth | [120] |
| Arabidopsis | AtMIR396a | bHLH74 | Root development | Longer roots | [121] |
| Arabidopsis | miR397b | LAC2OX | Root development | Elevated LAC2 transcript, decreased lignification in root xylem, and lengthened primary roots | [122] |
| Rice | miR390 | ARFs | Root development | Decreased salt tolerance with inhibited lateral root growth | [123] |
| Tomato | miR858 | SlMYB7-like | Secondary metabolite | Increased anthocyanin biosynthesis gene expression under normal conditions | [124] |
| Tobacco | nta-miRX27 | QPT2 | Secondary metabolite | Increased nicotine biosynthesis | [125] |
| Persimmon | DkmiR397 | DkLAC2 | Secondary metabolite | Increased accumulation of proanthocyanins | [126] |
| Citrus | miR171 | CsSCL | Somatic embryogenesis (SE) | Weakened somatic embryogenesis (SE) competence | [127] |
| Rice | miR319 | OsTCP21, OsGAmyb | Yield | Improved tiller number and grain yield | [128] |
| Rice | miR398 | Os07g46990 | Yield | Smaller panicles with fewer grains and late flowering | [69] |
| Rice | miR1432 | OsACOT | Yield | Increased grain weight by increasing grain filling rate | [129] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Othman, S.M.I.S.; Mustaffa, A.F.; Che-Othman, M.H.; Samad, A.F.A.; Goh, H.-H.; Zainal, Z.; Ismail, I. Overview of Repressive miRNA Regulation by Short Tandem Target Mimic (STTM): Applications and Impact on Plant Biology. Plants 2023, 12, 669. https://doi.org/10.3390/plants12030669
Othman SMIS, Mustaffa AF, Che-Othman MH, Samad AFA, Goh H-H, Zainal Z, Ismail I. Overview of Repressive miRNA Regulation by Short Tandem Target Mimic (STTM): Applications and Impact on Plant Biology. Plants. 2023; 12(3):669. https://doi.org/10.3390/plants12030669
Chicago/Turabian StyleOthman, Syed Muhammad Iqbal Syed, Arif Faisal Mustaffa, M. Hafiz Che-Othman, Abdul Fatah A. Samad, Hoe-Han Goh, Zamri Zainal, and Ismanizan Ismail. 2023. "Overview of Repressive miRNA Regulation by Short Tandem Target Mimic (STTM): Applications and Impact on Plant Biology" Plants 12, no. 3: 669. https://doi.org/10.3390/plants12030669
APA StyleOthman, S. M. I. S., Mustaffa, A. F., Che-Othman, M. H., Samad, A. F. A., Goh, H.-H., Zainal, Z., & Ismail, I. (2023). Overview of Repressive miRNA Regulation by Short Tandem Target Mimic (STTM): Applications and Impact on Plant Biology. Plants, 12(3), 669. https://doi.org/10.3390/plants12030669







