The Cannabis Plant as a Complex System: Interrelationships between Cannabinoid Compositions, Morphological, Physiological and Phenological Traits
Abstract
1. Introduction
2. Results
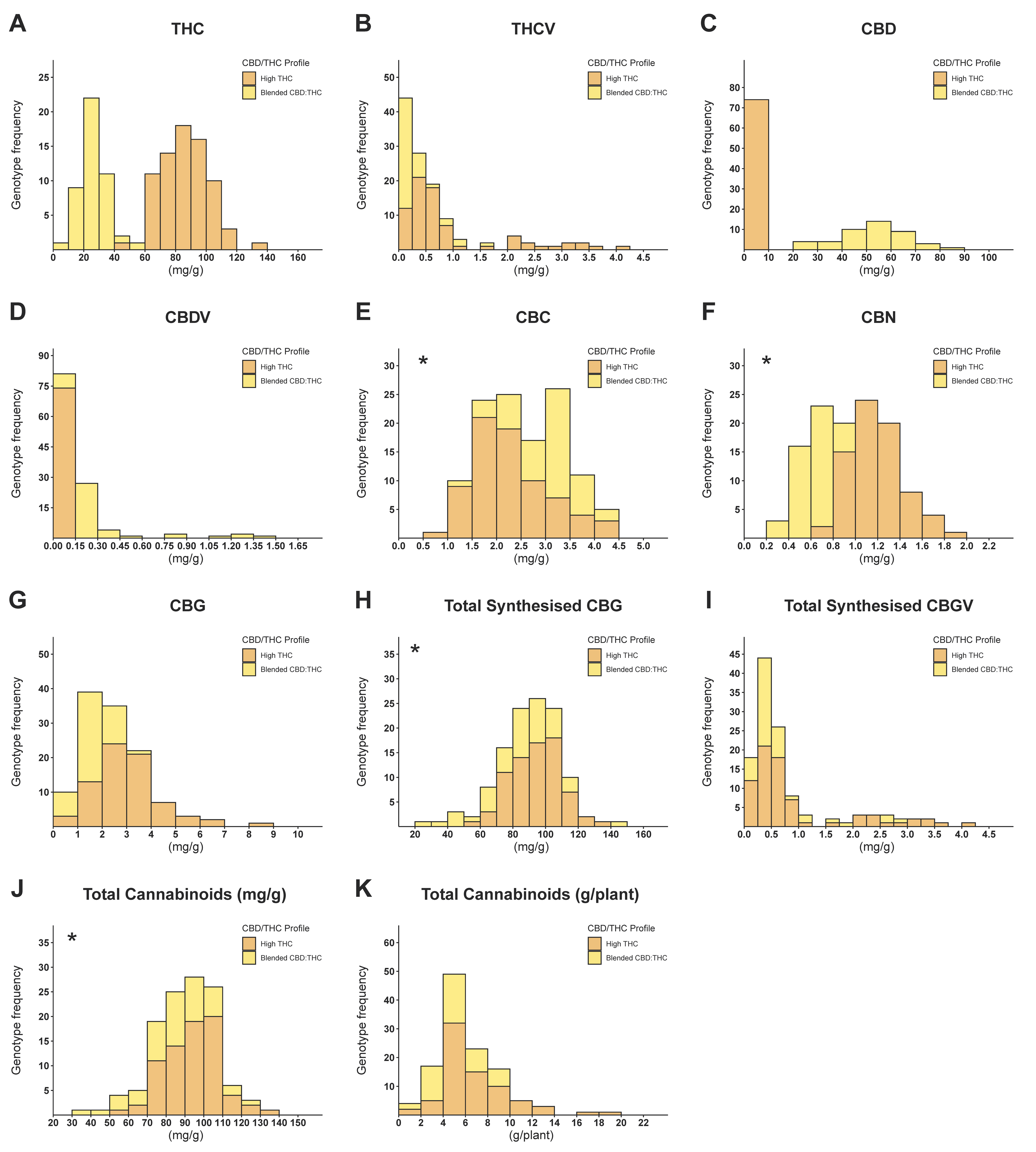
2.1. Phenotypic Diversity of Cannabinoid Compositions
2.2. Cannabinoids’ Broad-Sense Heritability
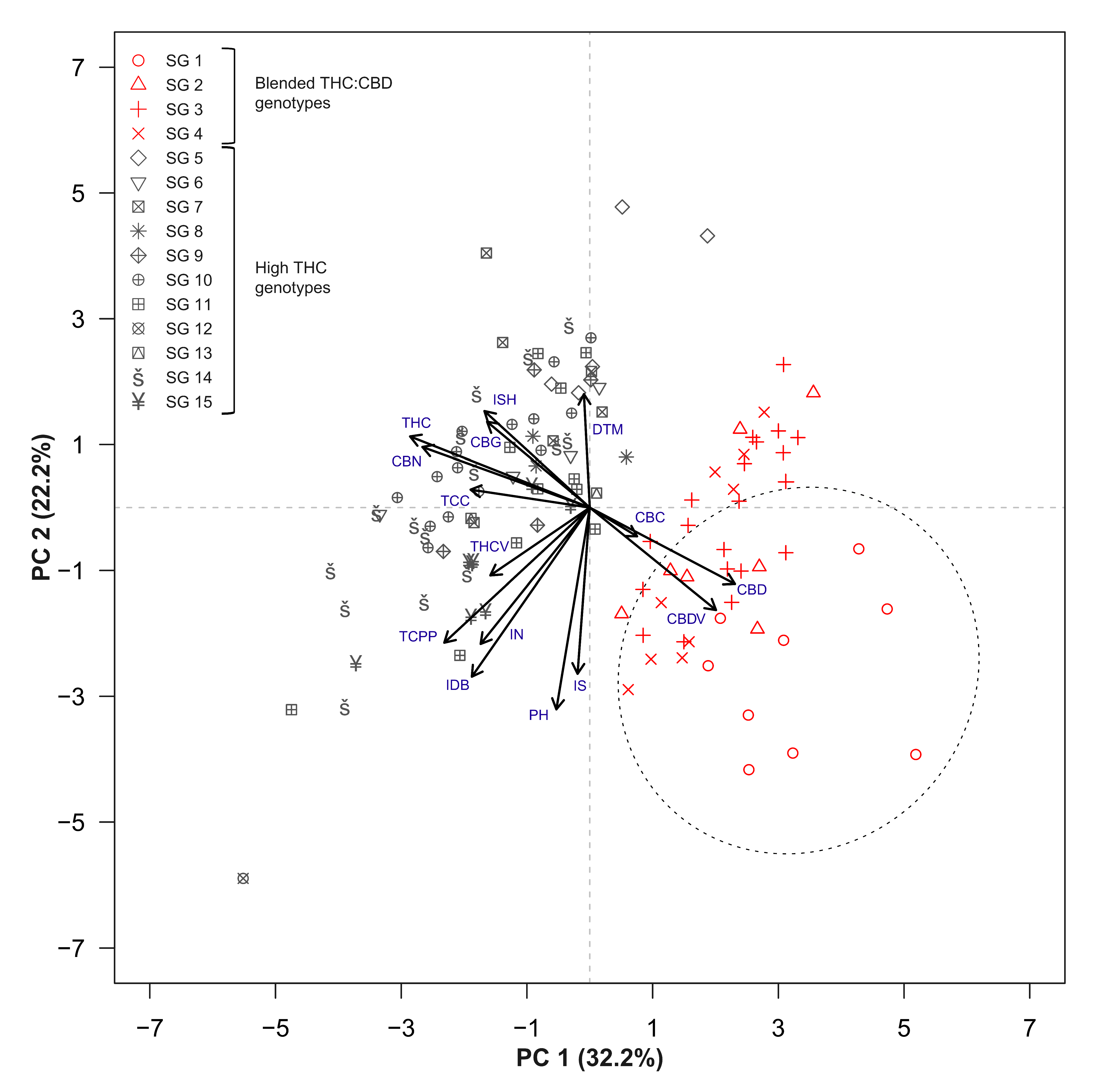
2.3. Trait Associations
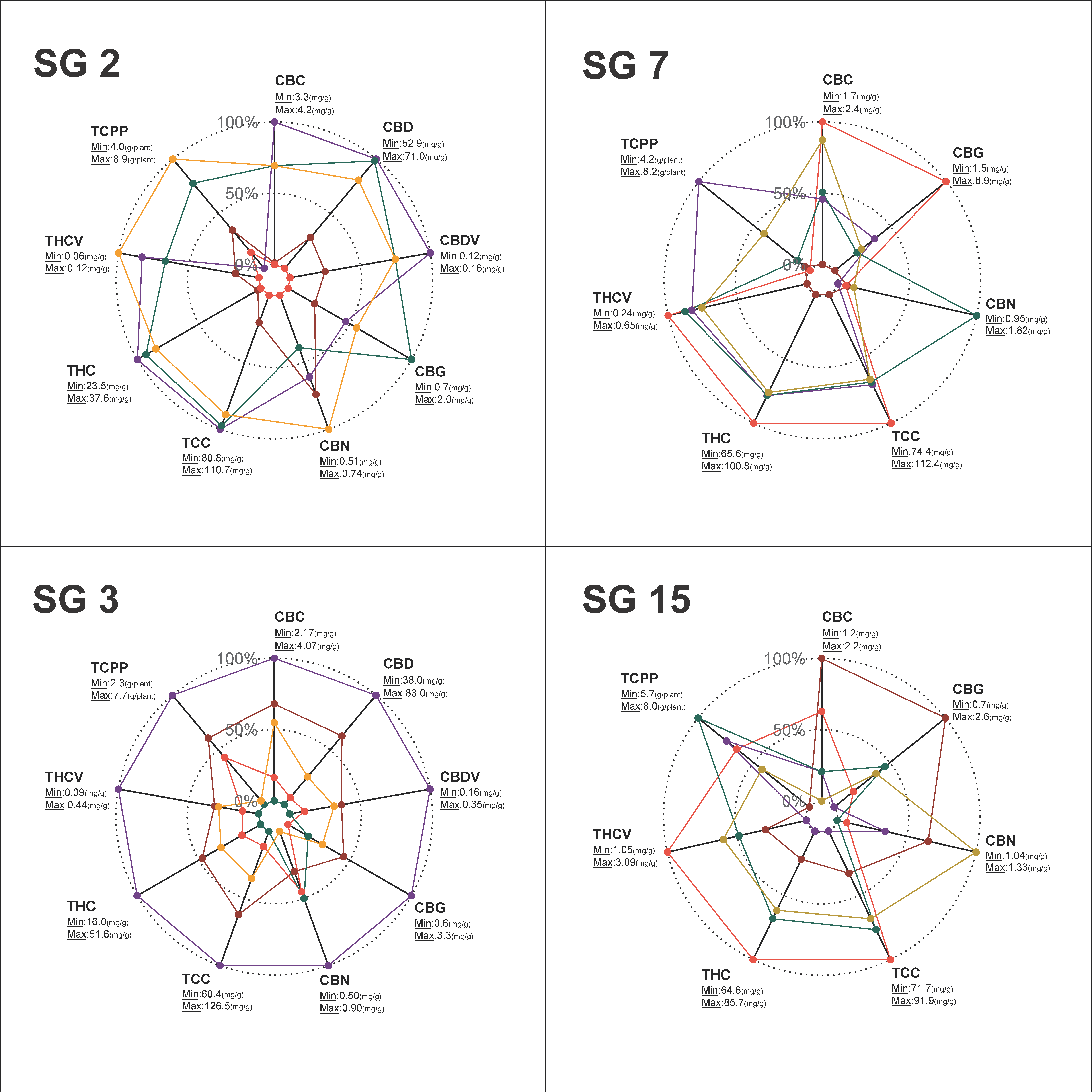
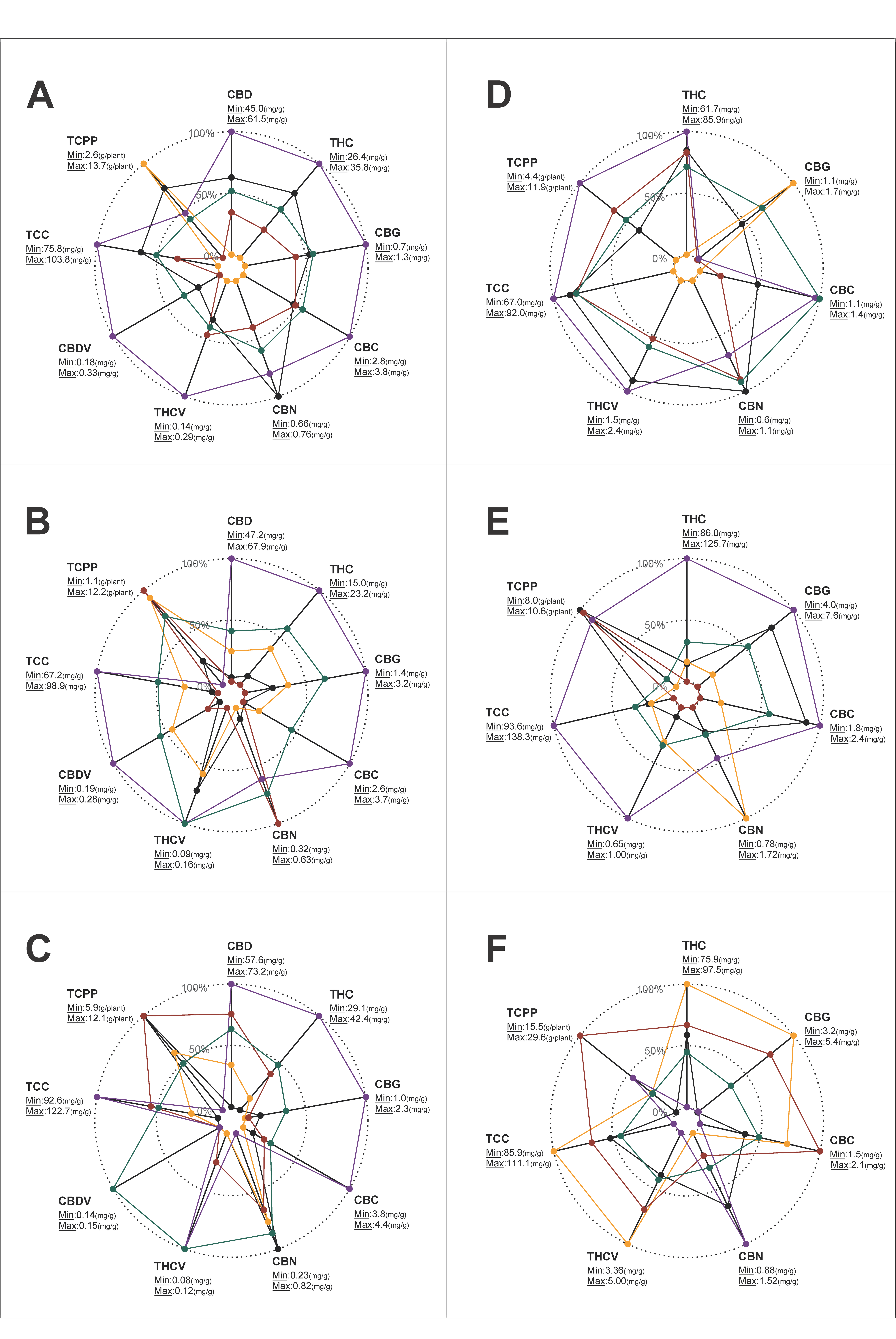
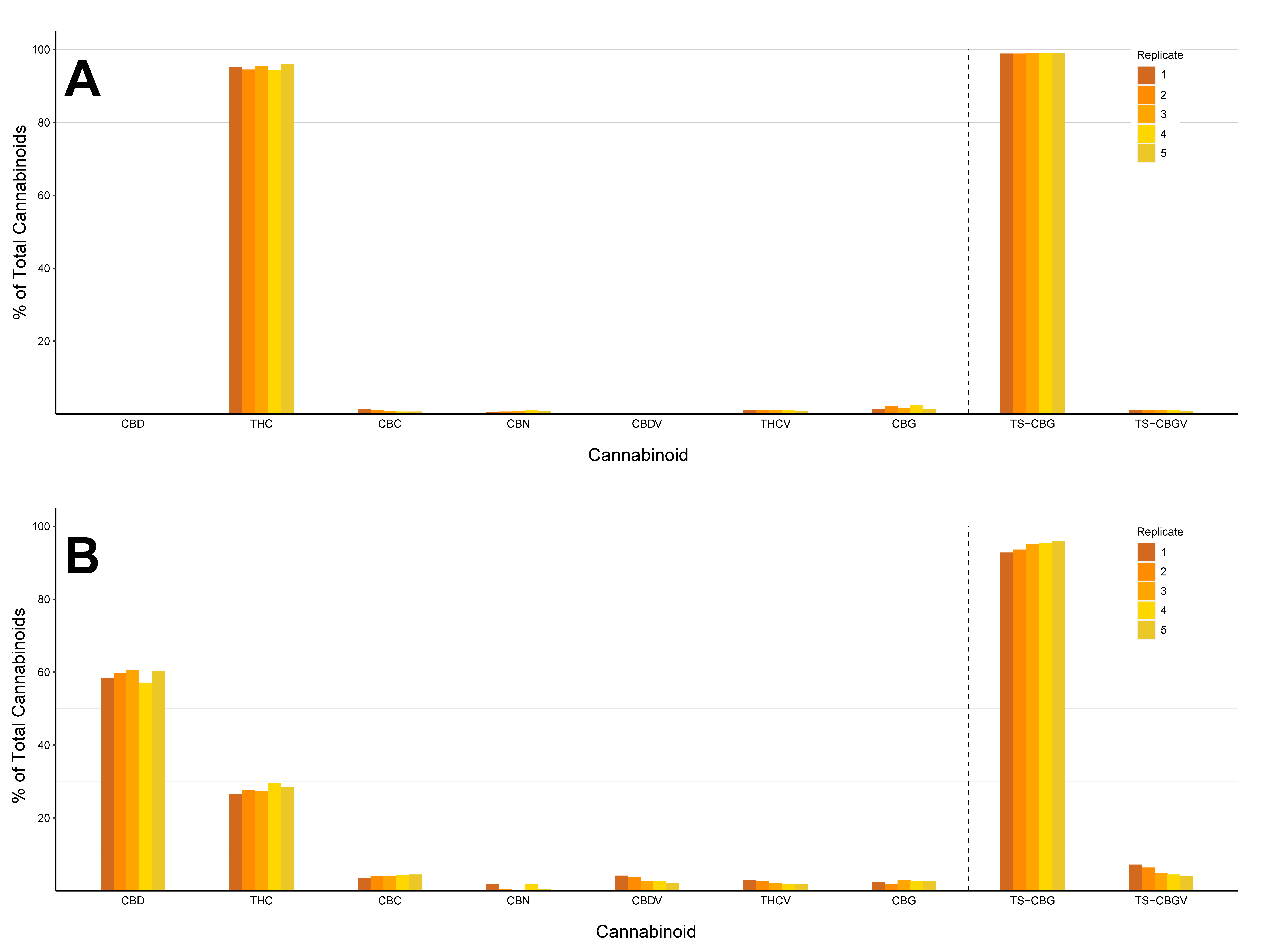
2.4. Variation in Cannabinoid Composition across Cultivars
2.5. Cannabinoid Variability within Genotypic Replicates
2.6. Prediction of Cannabinoid Production (TCPP) through Morphological, Physiological and Phenological Parameters
3. Discussion
3.1. Chemotypic Variation in the Examined Plant Population
3.2. Broad-Sense Heritability
3.3. Strain Classifications and Trait Associations
3.4. Cannabinoid Variation between and within Strain Groups
3.5. Prediction Equation
4. Materials and Methods
4.1. Plant Material, Experimental Design and Growth Conditions
4.2. Determination of Morphological and Phenological Parameters
4.3. Cannabinoid Quantification
4.4. Statistical Analysis and Spatial Adjustments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Karche, T.; Singh, M.R. The Application of Hemp (Cannabis sativa L.) for a Green Economy: A Review. Turk. J. Bot. 2019, 43, 710–723. [Google Scholar] [CrossRef]
- Ren, M.; Tang, Z.; Wu, X.; Spengler, R.; Jiang, H.; Yang, Y.; Boivin, N. The Origins of Cannabis Smoking: Chemical Residue Evidence from the First Millennium BCE in the Pamirs. Sci. Adv. 2019, 5, eaaw1391. [Google Scholar] [CrossRef]
- Koltai, H.; Albo, B.; Yaniv, Z. Traditional Uses of Cannabis in the Middle East and the Pathway to Cannabis-Based Healthcare in Israel. In Cannabis/Marijuana for Healthcare; Agrawal, C.D., Kumar, R., Dhanasekaran, M., Eds.; Springer Nature: Singapore, 2022; pp. 173–188. ISBN 9789811688218. [Google Scholar]
- Chandra, S.; Lata, H.; ElSohly, M.A.; Walker, L.A.; Potter, D. Cannabis Cultivation: Methodological Issues for Obtaining Medical-Grade Product. Epilepsy Behav. 2017, 70, 302–312. [Google Scholar] [CrossRef] [PubMed]
- O’Shaughnessy, W.B. On the Preparations of the Indian Hemp, or Gunjah. Prov. Med. J. 1843, 123, 363–369. [Google Scholar] [CrossRef]
- Ware, M.A.; Wang, T.; Shapiro, S.; Robinson, A.; Ducruet, T.; Huynh, T.; Gamsa, A.; Bennett, G.J.; Collet, J. Smoked Cannabis for Chronic Neuropathic Pain: A Randomized Controlled Trial. CMAJ 2010, 182, E694–E701. [Google Scholar] [CrossRef] [PubMed]
- Touw, M. The Religious and Medicinal Uses of Cannabis in China, India and Tibet. J. Psychoact. Drugs 1981, 13, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.; DeJean, D.; Clifford, T.; Coyle, D.; Potter, B.K.; Skidmore, B.; Alexander, C.; Repetski, A.E.; Shukla, V.; McCoy, B.; et al. Cannabis-Based Products for Pediatric Epilepsy: A Systematic Review. Epilepsia 2019, 60, 6–19. [Google Scholar] [CrossRef]
- Hussain, S.A.; Zhou, R.; Jacobson, C.; Weng, J.; Cheng, E.; Lay, J.; Hung, P.; Lerner, J.T.; Sankar, R. Perceived Efficacy of Cannabidiol-Enriched Cannabis Extracts for Treatment of Pediatric Epilepsy: A Potential Role for Infantile Spasms and Lennox-Gastaut Syndrome. Epilepsy Behav. 2015, 47, 138–141. [Google Scholar] [CrossRef]
- Pagano, C.; Navarra, G.; Coppola, L.; Avilia, G.; Bifulco, M.; Laezza, C. Cannabinoids: Therapeutic Use in Clinical Practice. Int. J. Mol. Sci. 2022, 23, 3344. [Google Scholar] [CrossRef]
- Hesami, M.; Pepe, M.; Baiton, A.; Jones, A.M.P. Current Status and Future Prospects in Cannabinoid Production through in Vitro Culture and Synthetic Biology. Biotechnol. Adv. 2023, 62, 108074. [Google Scholar] [CrossRef]
- Happyana, N.; Agnolet, S.; Muntendam, R.; Van Dam, A.; Schneider, B.; Kayser, O. Analysis of Cannabinoids in Laser-Microdissected Trichomes of Medicinal Cannabis sativa Using LCMS and Cryogenic NMR. Phytochemistry 2013, 87, 51–59. [Google Scholar] [CrossRef]
- Elsohly, M.A.; Slade, D. Chemical Constituents of Marijuana: The Complex Mixture of Natural Cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Hurgobin, B.; Tamiru-Oli, M.; Welling, M.T.; Doblin, M.S.; Bacic, A.; Whelan, J.; Lewsey, M.G. Recent Advances in Cannabis sativa Genomics Research. New Phytol. 2021, 230, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Pepe, M.; Alizadeh, M.; Rakei, A.; Baiton, A.; Phineas Jones, A.M. Recent Advances in Cannabis Biotechnology. Ind. Crops Prod. 2020, 158, 1–20. [Google Scholar] [CrossRef]
- Hussain, T.; Jeena, G.; Pitakbut, T.; Vasilev, N.; Kayser, O. Cannabis sativa Research Trends, Challenges, and New-Age Perspectives. iScience 2021, 24, 103391. [Google Scholar] [CrossRef]
- Jin, D.; Dai, K.; Xie, Z.; Chen, J. Secondary Metabolites Profiled in Cannabis Inflorescences, Leaves, Stem Barks, and Roots for Medicinal Purposes. Sci. Rep. 2020, 10, 3309. [Google Scholar] [CrossRef]
- Raman, V.; Lata, H.; Chandra, S.; Khan, I.A.; ElSohly, M.A. Morpho-Anatomy of Marijuana (Cannabis sativa L.). In Cannabis sativa L.—Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 123–136. ISBN 9783319545646. [Google Scholar]
- Huchelmann, A.; Boutry, M.; Hachez, C. Plant Glandular Trichomes: Natural Cell Factories of High Biotechnological Interest. Plant Physiol. 2017, 175, 6–22. [Google Scholar] [CrossRef]
- Gorelick, J.; Bernstein, N. Chemical and Physical Elicitation for Enhanced Cannabinoid Production in Cannabis. In Cannabis sativa L.-Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 439–456. ISBN 9783319545646. [Google Scholar]
- Bernstein, N.; Gorelick, J.; Koch, S. Interplay between Chemistry and Morphology in Medical Cannabis (Cannabis sativa L.). Ind. Crops Prod. 2019, 129, 185–194. [Google Scholar] [CrossRef]
- Small, E. Classification of Cannabis sativa L.: In Relation to Agricultural, Biotechnological, Medical and Recreational Utilization. In Cannabis sativa L.—Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–62. ISBN 9783319545646. [Google Scholar]
- Flores-sanchez, I.J.; Verpoorte, R. Secondary Metabolism in Cannabis. Phytochem. Rev. 2008, 7, 615–639. [Google Scholar] [CrossRef]
- Pate, D.W. Chemical Ecology of Cannabis. J. Int. Hemp Assoc. 1994, 2, 32–37. [Google Scholar] [CrossRef]
- Pate, D.W. Possible Role of Ultraviolet Radiation in Evolution of Cannabis Chemotypes. Econ. Bot. 1983, 37, 396–405. [Google Scholar] [CrossRef]
- Lydon, J.; Teramura, A.H.; Coffman, C.B. UV-B Radiation Effects on Photosynthesis, Growth and Cannabinoid Production of Two Cannabis sativa Chemotypes. Photochem. Photobiol. 1987, 46, 201–206. [Google Scholar] [CrossRef] [PubMed]
- ElSohly, M.A.; Radwan, M.M.; Gul, W.; Chandra, S.; Galal, A. Phytochemistry of Cannabis sativa L. In Phytocannabinoids. Progress in the Chemistry of Organic Natural Products; Kinghorn, A.D., Falk, H., Gibbons, S., Kobayashi, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 103, pp. 1–36. ISBN 9783319455419. [Google Scholar]
- Clarke, R.; Merlin, M. Hypotheses Concerning the Early Evolution of Cannabis. In Cannabis—Evolution and Ethnobotany; University of California Press: Berkeley, California, 2013; pp. 333–364. ISBN 9780520270480. [Google Scholar]
- de Meijer, E. The Chemical Phenotypes (Chemotypes) of Cannabis. In Handbook of Cannabis; Pertwee, R.G., Ed.; Oxford University Press: Oxford, UK, 2014; pp. 89–110. ISBN 9780199251841. [Google Scholar]
- de Meijer, E.; Bagatta, M.; Carboni, A.; Crucitti, P.; Moliterni, C.; Ranalli, P.; Mandolino, G. The Inheritance of Chemical Phenotype in Cannabis sativa L. Genetics 2003, 163, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Dowling, C.A.; Shi, J.; Ryan, L.; Hunt, D.J.L.; O’Reilly, E.; Perry, A.S.; Kinnane, O.; McCabe, P.F.; Melzer, R. The Cream of the Crop: Biology, Breeding, and Applications of Cannabis sativa. Annu. Plant Rev. Online 2021, 4, 471–528. [Google Scholar] [CrossRef]
- Nahtigal, I.; Blake, A.; Hand, A.; Florentinus-Mefailoski, A.; Haleh, H.; Friedberg, J. The Pharmacological Properties of Cannabis. J. Pain Manag. 2016, 9, 481–491. [Google Scholar]
- Chagas, M.H.N.; Eckeli, A.L.; Zuardi, A.W.; Pena-Pereira, M.A.; Sobreira-Neto, M.A.; Sobreira, E.T.; Camilo, M.R.; Bergamaschi, M.M.; Schenck, C.H.; Hallak, J.E.C.; et al. Cannabidiol Can Improve Complex Sleep-Related Behaviours Associated with Rapid Eye Movement Sleep Behaviour Disorder in Parkinson’s Disease Patients: A Case Series. J. Clin. Pharm. Ther. 2014, 39, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Machado Rocha, F.C.; Stéfano, S.C.; De Cássia Haiek, R.; Rosa Oliveira, L.M.Q.; Da Silveira, D.X. Therapeutic Use of Cannabis sativa on Chemotherapy-Induced Nausea and Vomiting among Cancer Patients: Systematic Review and Meta-Analysis. Eur. J. Cancer Care 2008, 17, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Mechoulam, R.; Ben-Shabat, S. From Gan-Zi-Gun-Nu to Anandamide and 2-Arachidonoylglycerol: The Ongoing Story of Cannabis. Nat. Prod. Rep. 1999, 16, 131–143. [Google Scholar] [CrossRef]
- Koltai, H.; Namdar, D. Cannabis Phytomolecule “Entourage”: From Domestication to Medical Use. Trends Plant Sci. 2020, 25, 976–984. [Google Scholar] [CrossRef]
- Russo, E.B. The Case for the Entourage Effect and Conventional Breeding of Clinical Cannabis: No “Strain”, No Gain. Front. Plant Sci. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Russo, E.B. Taming THC: Potential Cannabis Synergy and Phytocannabinoid-Terpenoid Entourage Effects. Br. J. Pharmacol. 2011, 163, 1344–1364. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.A.; Russo, E.B.; Smith, K.M. Pharmacological Foundations of Cannabis Chemovars. Planta Med. 2018, 84, 225–233. [Google Scholar] [CrossRef] [PubMed]
- de Meijer, E.P.M.; Hammond, K.M.; Sutton, A. The Inheritance of Chemical Phenotype in Cannabis sativa L. (IV): Cannabinoid-Free Plants. Euphytica 2009, 168, 95–112. [Google Scholar] [CrossRef]
- McPartland, J.M.; Russo, E.B. Cannabis and Cannabis Extracts: Greater than the Sum of Their Parts? J. Cannabis Ther. 2001, 1, 103–132. [Google Scholar] [CrossRef]
- Namdar, D.; Anis, O.; Poulin, P.; Koltai, H. Chronological Review and Rational and Future Prospects of Cannabis-Based Drug Development. Molecules 2020, 25, 4821. [Google Scholar] [CrossRef] [PubMed]
- Vergara, D.; Huscher, E.L.; Keepers, K.G.; Givens, R.M.; Cizek, C.G.; Torres, A.; Gaudino, R.; Kane, N.C. Gene Copy Number Is Associated with Phytochemistry in Cannabis sativa. AoB Plants 2019, 11, plz074. [Google Scholar] [CrossRef]
- Onofri, C.; De Meijer, E.P.M.; Mandolino, G. Sequence Heterogeneity of Cannabidiolic- and Tetrahydrocannabinolic Acid-Synthase in Cannabis sativa L. and Its Relationship with Chemical Phenotype. Phytochemistry 2015, 116, 57–68. [Google Scholar] [CrossRef]
- Small, E. Evolution and Classification of Cannabis sativa (Marijuana, Hemp) in Relation to Human Utilization. Bot. Rev. 2015, 81, 189–294. [Google Scholar] [CrossRef]
- McPartland, J.M. Cannabis sativa and Cannabis indica versus “Sativa” and “Indica.” In Cannabis sativa L.—Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 101–121. ISBN 9783319545646. [Google Scholar]
- Hillig, K. Letter to the Editor. J. Ind. Hemp 2002, 7, 5–6. [Google Scholar] [CrossRef]
- Grassi, G.; McPartland, J.M. Chemical and Morphological Phenotypes in Breeding of Cannabis sativa L. In Cannabis sativa L.-Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 137–160. ISBN 9783319545646. [Google Scholar]
- Chandra, S.; Lata, H.; Khan, I.A.; Mahmoud, E. Cannabis sativa L.: Botany and Horticulture. In Cannabis sativa L.—Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 79–100. ISBN 9783319545639. [Google Scholar]
- Parker, K.A.; Di Mattia, A.; Shaik, F.; Cerón Ortega, J.C.; Whittle, R. Risk Management within the Cannabis Industry: Building a Framework for the Cannabis Industry. Financ. Mark. Inst. Instrum. 2019, 28, 3–55. [Google Scholar] [CrossRef]
- Jin, D.; Jin, S.; Chen, J. Cannabis Indoor Growing Conditions, Management Practices, and Post-Harvest Treatment: A Review. Am. J. Plant Sci. 2019, 10, 925–946. [Google Scholar] [CrossRef]
- Lata, H.; Chandra, S.; Uchendu, E.E.; Khan, A.I.; ElSohly, M.A. Cultivating Research Grade Cannabis for the Development of Phytopharmaceuticals. In Medicinal Plants From Farm to Pharmacy; Joshee, N., Dhekney, S.A., Parajuli, P., Eds.; Springer: Cham, Switzerland, 2019; pp. 169–186. ISBN 9783030312688. [Google Scholar]
- Chandra, S.; Lata, H.; ElSohly, M.A. Propagation of Cannabis for Clinical Research: An Approach towards a Modern Herbal Medicinal Products Development. Front. Plant Sci. 2020, 11, 958. [Google Scholar] [CrossRef]
- Saloner, A.; Bernstein, N. Nitrogen Supply Affects Cannabinoid and Terpenoid Profile in Medical Cannabis (Cannabis sativa L.). Ind. Crops Prod. 2021, 167, 113516. [Google Scholar] [CrossRef]
- Shiponi, S.; Bernstein, N. The Highs and Lows of P Supply in Medical Cannabis: Effects on Cannabinoids, the Ionome, and Morpho-Physiology. Front. Plant Sci. 2021, 12, 657323. [Google Scholar] [CrossRef] [PubMed]
- Magagnini, G.; Grassi, G.; Kotiranta, S. The Effect of Light Spectrum on the Morphology and Cannabinoid Content of Cannabis sativa L. Med. Cannabis Cannabinoids 2018, 1, 19–27. [Google Scholar] [CrossRef]
- Caplan, D.; Dixon, M.; Zheng, Y. Increasing Inflorescence Dry Weight and Cannabinoid Content in Medical Cannabis Using Controlled Drought Stress. HortScience 2019, 54, 964–969. [Google Scholar] [CrossRef]
- Danziger, N.; Bernstein, N. Plant Architecture Manipulation Increases Cannabinoid Standardization in ‘Drug-Type’ Medical Cannabis. Ind. Crops Prod. 2021, 167, 113528. [Google Scholar] [CrossRef]
- Naim-Feil, E.; Breen, E.J.; Pembleton, L.W.; Spooner, L.E.; Spangenberg, G.C.; Cogan, N.O.I. Empirical Evaluation of Inflorescences’ Morphological Attributes for Yield Optimization of Medicinal Cannabis Cultivars. Front. Plant Sci. 2022, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Naim-Feil, E.; Pembleton, L.W.; Spooner, L.E.; Malthouse, A.L.; Miner, A.; Quinn, M.; Polotnianka, R.M.; Baillie, R.C.; Spangenberg, G.C.; Cogan, N.O.I. The Characterization of Key Physiological Traits of Medicinal Cannabis (Cannabis sativa L.) as a Tool for Precision Breeding. BMC Plant Biol. 2021, 21, 294. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.; Merlin, M. Recent History of Cannabis Breeding. In Cannabis—Evolution and Ethanobotany; University of California Press: Berkeley, CA, USA, 2013; pp. 295–310. ISBN 9780520270480. [Google Scholar]
- Welling, M.T.; Shapter, T.; Rose, T.J.; Liu, L.; Stanger, R.; King, G.J. A Belated Green Revolution for Cannabis: Virtual Genetic Resources to Fast-Track Cultivar Development. Front. Plant Sci. 2016, 7, 1113. [Google Scholar] [CrossRef]
- McPartland, J.M.; Guy, G.W. Models of Cannabis Taxonomy, Cultural Bias, and Conflicts between Scientific and Vernacular Names. Bot. Rev. 2017, 83, 327–381. [Google Scholar] [CrossRef]
- Chouvy, P.-A. Cannabis Cultivation in the World: Heritages, Trends and Challenges. EchoGéo 2019, 48, 1–20. [Google Scholar] [CrossRef]
- Clarke, R.; Merlin, M. The Cultural Diffusion of Cannabis. In Cannabis—Evolution and Ethanobotany; University of California Press: Berkeley, CA, USA, 2013; pp. 59–134. ISBN 9780520270480. [Google Scholar]
- van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The Draft Genome and Transcriptome of Cannabis sativa. Genome Biol. 2011, 12, R102. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Reiter, M.A.; d’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W.; et al. Complete Biosynthesis of Cannabinoids and Their Unnatural Analogues in Yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Weiblen, G.D.; Wenger, J.P.; Craft, K.J.; ElSohly, M.A.; Mehmedic, Z.; Treiber, E.L.; Marks, M.D. Gene Duplication and Divergence Affecting Drug Content in Cannabis sativa. New Phytol. 2015, 208, 1241–1250. [Google Scholar] [CrossRef]
- Jones, C.G.; Firn, R.D. On the Evolution of Plant Secondary Chemical Diversity. Philos. Trans.-R. Soc. London B 1991, 333, 273–280. [Google Scholar] [CrossRef]
- Ross, S.A.; Elsohly, M.A. CBN and Delta 9-THC Concentration Ratio as an Indicator of the Age of Stored Marijuana Samples. UNODC-Bull. Narc. 1997, 49, 139–147. [Google Scholar]
- Trofin, I.G.; Dabija, G.; Vaireanu, D.I.; Filipescu, L. Long-Term Storage and Cannabis Oil Stability. Rev. Chim. 2012, 63, 293–297. [Google Scholar] [CrossRef]
- Smith, C.J.; Vergara, D.; Keegan, B.; Jikomes, N. The Phytochemical Diversity of Commercial Cannabis in the United States. PLoS ONE 2022, 17, e0267498. [Google Scholar] [CrossRef]
- Hazekamp, A.; Fischedick, J.T. Cannabis-from Cultivar to Chemovar. Drug Test. Anal. 2012, 4, 660–667. [Google Scholar] [CrossRef]
- Hazekamp, A.; Tejkalová, K.; Papadimitriou, S. Cannabis: From Cultivar to Chemovar II— a Metabolomics Approach to Cannabis Classification. Cannabis Cannabinoid Res. 2016, 1, 202–215. [Google Scholar] [CrossRef]
- Chen, Y.; Lübberstedt, T. Molecular Basis of Trait Correlations. Trends Plant Sci. 2010, 15, 454–461. [Google Scholar] [CrossRef]
- Kearsey, M.J.; Pooni, H.S. Applications. In Genetical Analysis of Quantitative Traits; Chapman & Hall: London, UK, 1998; pp. 302–334. ISBN 0412609800. [Google Scholar]
- Woods, P.; Campbell, B.J.; Nicodemus, T.J.; Cahoon, E.B.; Mullen, J.L.; McKay, J.K. Quantitative Trait Loci Controlling Agronomic and Biochemical Traits in Cannabis sativa. Genetics 2021, 219, iyab099. [Google Scholar] [CrossRef] [PubMed]
- De Meijer, E.P.M.; Hammond, K.M.; Micheler, M. The Inheritance of Chemical Phenotype in Cannabis sativa L. (III): Variation in Cannabichromene Proportion. Euphytica 2009, 165, 293–311. [Google Scholar] [CrossRef]
- Namdar, D.; Charuvi, D.; Ajjampura, V.; Mazuz, M.; Ion, A.; Kamara, I.; Koltai, H. LED Lighting Affects the Composition and Biological Activity of Cannabis sativa Secondary Metabolites. Ind. Crop. Prod. 2019, 132, 177–185. [Google Scholar] [CrossRef]
- Erkelens, J.L.; Hazekamp, A. That Which We Call Indica, by Any Other Name Would Smell as Sweet. An Essay on the History of the Term Indica and the Taxonomical Conflict between the Monotypic and Polytypic Views of Cannabis. Cannabinoids 2014, 9, 9–15. [Google Scholar]
- Massuela, D.C.; Hartung, J.; Munz, S.; Erpenbach, F.; Graeff-Hönninger, S. Impact of Harvest Time and Pruning Technique on Total CBD Concentration and Yield of Medicinal Cannabis. Plants 2022, 11, 140. [Google Scholar] [CrossRef]
- Danziger, N.; Bernstein, N. Light Matters: Effect of Light Spectra on Cannabinoid Profile and Plant Development of Medical Cannabis (Cannabis sativa L.). Ind. Crops Prod. 2021, 164, 113351. [Google Scholar] [CrossRef]
- Clarke, R.; Merlin, M. Cannabis Domestication, Breeding History, Present-Day Genetic Diversity, and Future Prospects. Crit. Rev. Plant Sci. 2016, 35, 293–327. [Google Scholar] [CrossRef]
- Tran, J.; Elkins, A.C.; Spangenberg, G.C.; Rochfort, S.J. High-Throughput Quantitation of Cannabinoids by Liquid Chromatography Triple-Quadrupole Mass Spectrometry. Molecules 2022, 27, 742. [Google Scholar] [CrossRef]
- Elkins, A.C.; Deseo, M.A.; Rochfort, S.; Ezernieks, V.; Spangenberg, G. Development of a Validated Method for the Qualitative and Quantitative Analysis of Cannabinoids in Plant Biomass and Medicinal Cannabis Resin Extracts Obtained by Super-Critical Fluid Extraction. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1109, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, A.; Gogel, B.; Cullis, B.; Thompson, R. ASReml User Guide Release 3.0; VSN International Ltd.: Hemel Hempstead, UK, 2009. [Google Scholar]

| Category | Trait | H2 |
|---|---|---|
| Cannabinoids | THC | 0.89 |
| THCV | 0.95 | |
| CBD | 0.96 | |
| CBDV | 0.89 | |
| CBC | 0.79 | |
| CBG | 0.72 | |
| CBN | 0.51 | |
| TCC | 0.62 | |
| Phenology | DTM | 0.49 |
| Plant’s morphology | PH | 0.52 |
| IDB | 0.33 | |
| Int.L | 0.35 | |
| Inflorescences | ISH | 0.38 |
| IN | 0.29 | |
| IS | 0.15 | |
| Inf.W | 0.19 | |
| Inf.L | 0.18 |
| Strain Group | CBC | CBD | CBDV | CBG | CBN | THC | THCV | TCC | TCPP | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | Blended THC:CBD | 1.3 | 1.3 | 1.3 | 3.0 | 1.5 | 1.6 | 2.0 | 1.4 | 2.2 |
| 3 | 1.9 | 2.2 | 2.2 | 5.7 | 1.8 | 3.2 | 4.9 | 2.1 | 3.3 | |
| 7 | High THC | 1.4 | 5.9 | 1.9 | 1.5 | 2.7 | 1.5 | 1.9 | ||
| 15 | 1.9 | 3.9 | 1.3 | 1.3 | 2.9 | 1.3 | 1.4 | |||
| Mean Ratio | 1.6 | 1.8 | 1.8 | 4.6 | 1.6 | 1.9 | 3.1 | 1.6 | 2.2 | |
| Genotype | CBD | THC | CBG | CBC | THCV | CBDV | |
|---|---|---|---|---|---|---|---|
| ID:446060 (High-THC) | Proportions’ Mean | --- | 95.07% | 1.81% | 0.94% | 1.02% | --- |
| σ | --- | 0.62% | 0.52% | 0.24% | 0.07% | --- | |
| ID:446039 (Blended THC:CBD) | Proportions’ Mean | 59.16% | 27.90% | 2.51% | 4.08% | 2.32% | 3.09% |
| σ | 1.41% | 1.16% | 0.38% | 0.35% | 0.54% | 0.81% |
| TCPP | B | 95% CI for B | SE B | β | R2 | ΔR2 | |
|---|---|---|---|---|---|---|---|
| LL | UL | ||||||
| (A) | |||||||
| Model | 0.67 | 0.65 *** | |||||
| Constant | −21.64 *** | −31.96 | −11.32 | 5.17 | |||
| PH | 0.13 *** | 0.08 | 0.19 | 0.03 | 0.52 | ||
| Int.L | −1.11 * | −2.06 | −0.16 | 0.48 | −0.18 | ||
| DTM | 0.19 * | 0.05 | 0.34 | 0.07 | 0.22 | ||
| Inf.W10 | 2.87 *** | 1.74 | 4.00 | 0.57 | 0.49 | ||
| (B) | |||||||
| Model | 0.66 | 0.63 *** | |||||
| Constant | −13.68 *** | −19.91 | −7.45 | 3.06 | |||
| PH | 0.71 ** | 0.02 | 0.12 | 0.24 | 0.38 | ||
| Int.L | 1.13 | 0.14 | −0.38 | 0.74 | 0.2 | ||
| Inf.W10 | 1.77 ** | 0.55 | 2.98 | 0.6 | 0.4 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naim-Feil, E.; Elkins, A.C.; Malmberg, M.M.; Ram, D.; Tran, J.; Spangenberg, G.C.; Rochfort, S.J.; Cogan, N.O.I. The Cannabis Plant as a Complex System: Interrelationships between Cannabinoid Compositions, Morphological, Physiological and Phenological Traits. Plants 2023, 12, 493. https://doi.org/10.3390/plants12030493
Naim-Feil E, Elkins AC, Malmberg MM, Ram D, Tran J, Spangenberg GC, Rochfort SJ, Cogan NOI. The Cannabis Plant as a Complex System: Interrelationships between Cannabinoid Compositions, Morphological, Physiological and Phenological Traits. Plants. 2023; 12(3):493. https://doi.org/10.3390/plants12030493
Chicago/Turabian StyleNaim-Feil, Erez, Aaron C. Elkins, M. Michelle Malmberg, Doris Ram, Jonathan Tran, German C. Spangenberg, Simone J. Rochfort, and Noel O. I. Cogan. 2023. "The Cannabis Plant as a Complex System: Interrelationships between Cannabinoid Compositions, Morphological, Physiological and Phenological Traits" Plants 12, no. 3: 493. https://doi.org/10.3390/plants12030493
APA StyleNaim-Feil, E., Elkins, A. C., Malmberg, M. M., Ram, D., Tran, J., Spangenberg, G. C., Rochfort, S. J., & Cogan, N. O. I. (2023). The Cannabis Plant as a Complex System: Interrelationships between Cannabinoid Compositions, Morphological, Physiological and Phenological Traits. Plants, 12(3), 493. https://doi.org/10.3390/plants12030493







