Genome-Wide Association Analysis of Freezing Tolerance and Winter Hardiness in Winter Wheat of Nordic Origin
Abstract
1. Introduction
2. Results
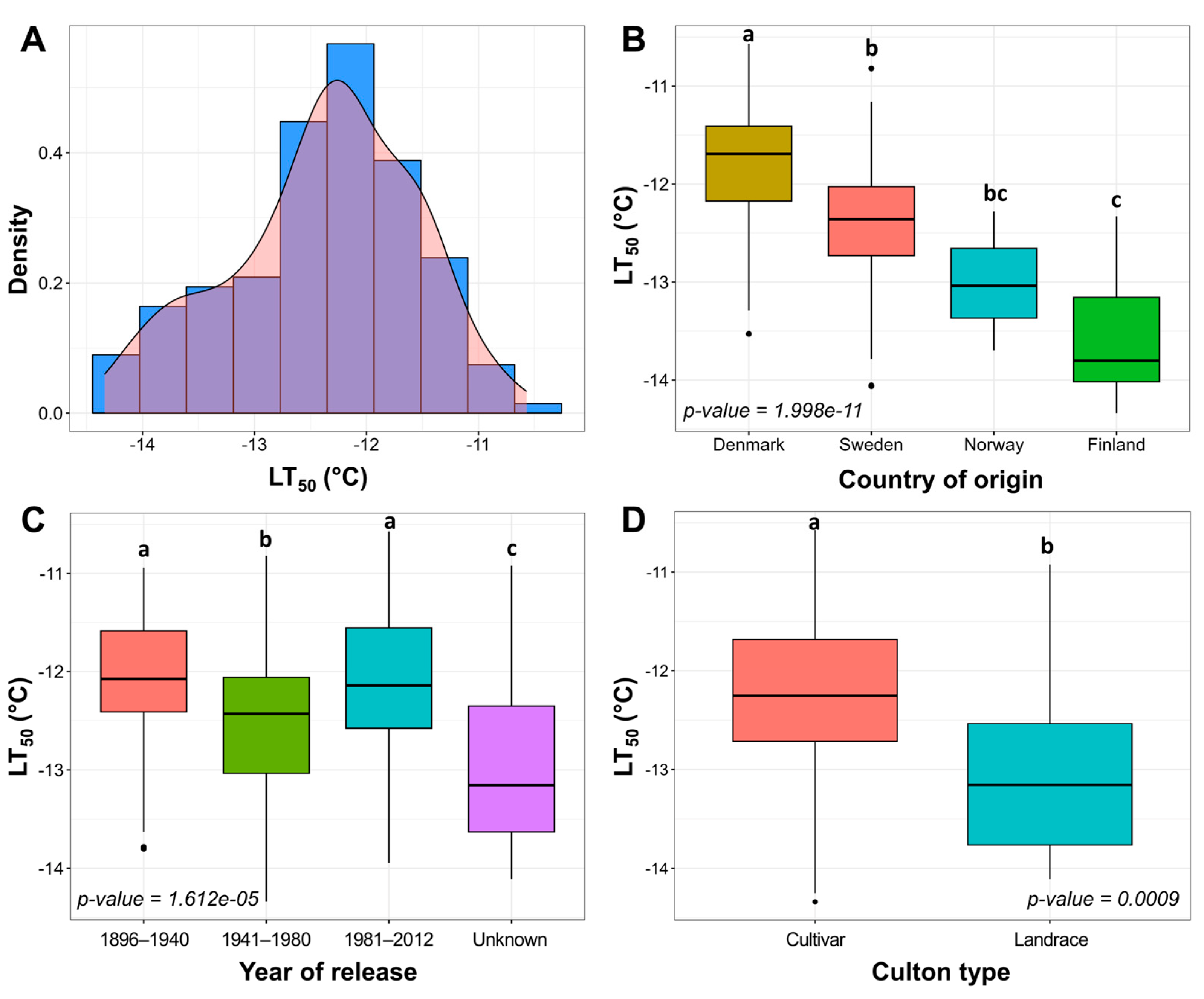
2.1. Freezing Tolerance of Winter Wheat under Controlled Conditions
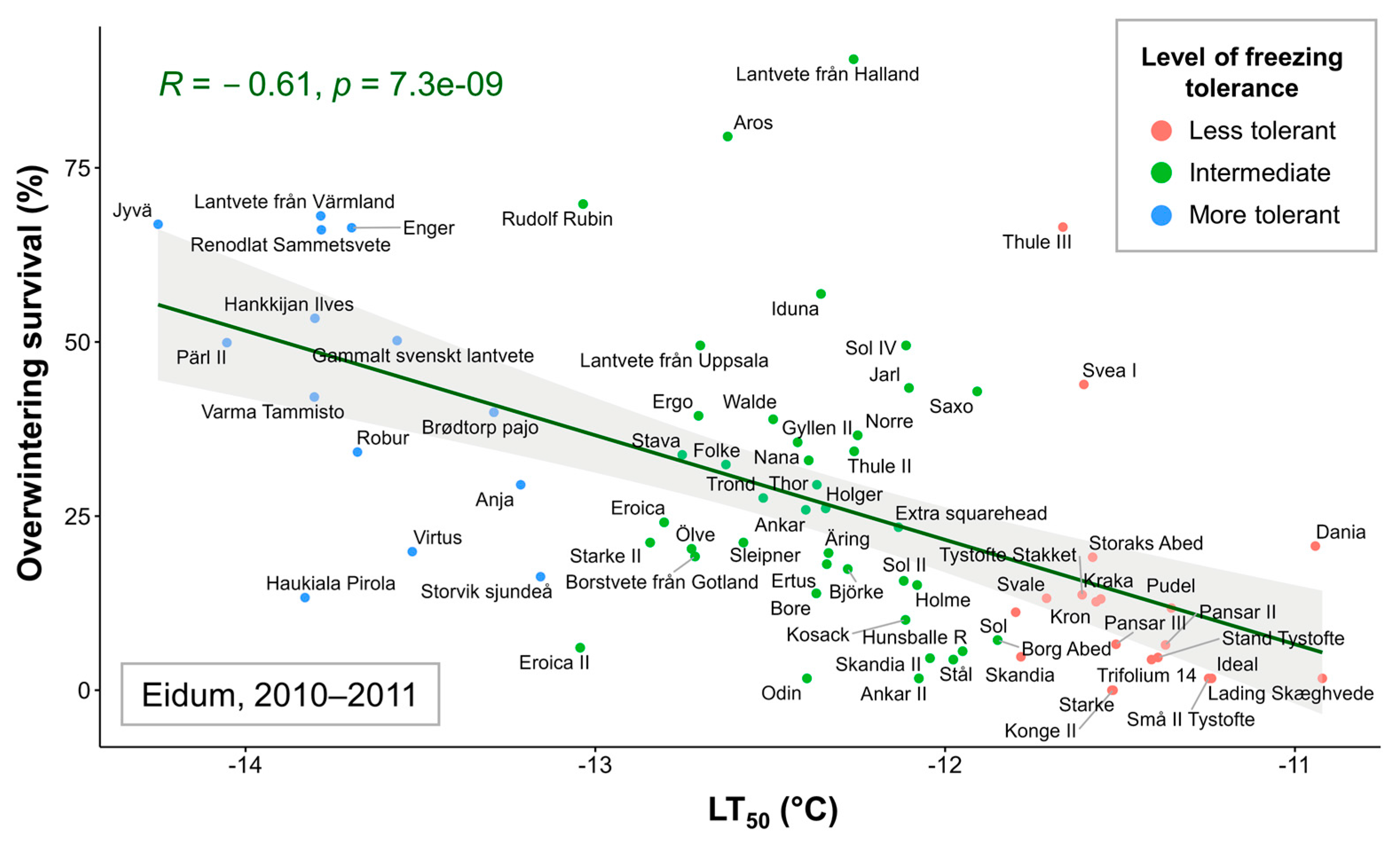
2.2. Overwintering of Winter Wheat in the Field
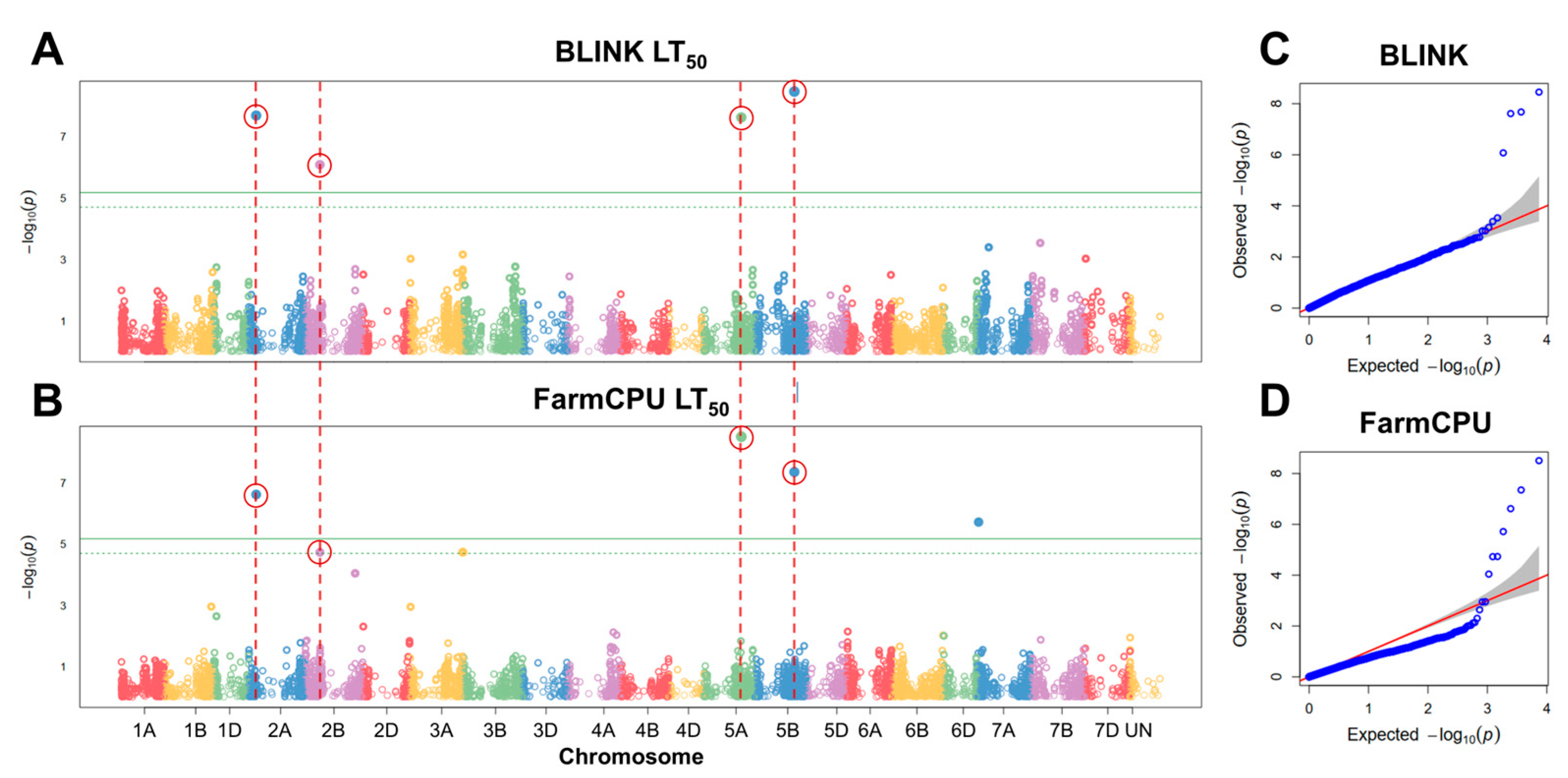
2.3. GWAS of Freezing Tolerance under Controlled Conditions
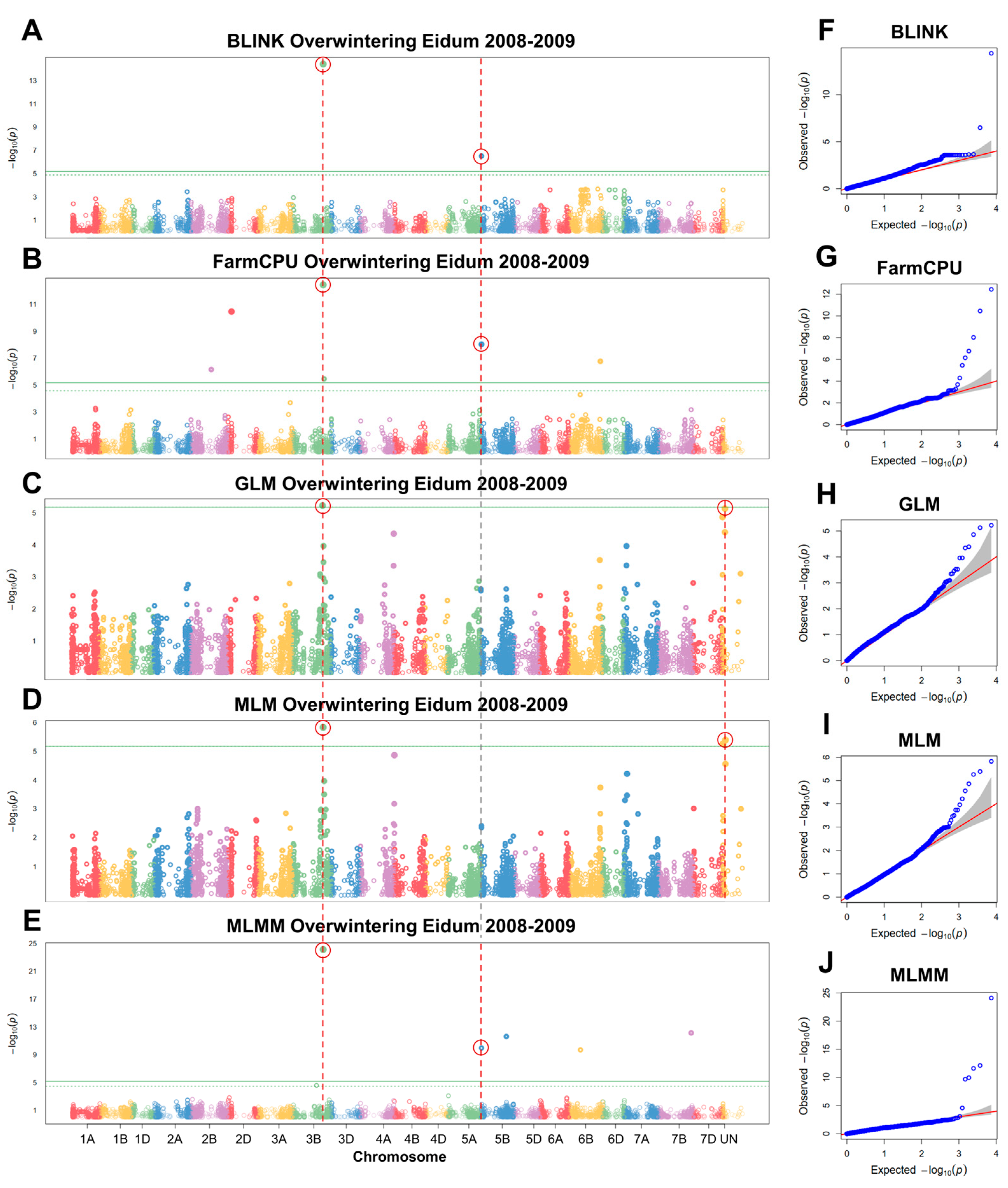
2.4. GWAS of Overwintering in the Field
3. Discussion
3.1. The Geographical and Temporal Trends of Freezing Tolerance in Winter Wheat
3.2. SNPs Associated with Freezing Tolerance under Controlled Conditions
3.3. SNPs Associated with Overwintering in the Field
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Freezing Tolerance Tests under Controlled Conditions
4.3. Overwintering Field Experiments
4.4. Genome-Wide Association Studies
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Discussion—Supplementary Information
References
- Entz, M.H.; Fowler, D.B. Agronomic Performance of Winter versus Spring Wheat. Agron. J. 1991, 83, 527–532. [Google Scholar] [CrossRef]
- Minoli, S.; Jägermeyr, J.; Asseng, S.; Urfels, A.; Müller, C. Global Crop Yields Can Be Lifted by Timely Adaptation of Growing Periods to Climate Change. Nat. Commun. 2022, 13, 7079. [Google Scholar] [CrossRef] [PubMed]
- Vaitkevičiūtė, G.; Aleliūnas, A.; Gibon, Y.; Armonienė, R. The Effect of Cold Acclimation, Deacclimation and Reacclimation on Metabolite Profiles and Freezing Tolerance in Winter Wheat. Front. Plant Sci. 2022, 13, 959118. [Google Scholar] [CrossRef] [PubMed]
- Rapacz, M.; Ergon, Å.; Höglind, M.; Jørgensen, M.; Jurczyk, B.; Østrem, L.; Rognli, O.A.; Tronsmo, A.M. Overwintering of Herbaceous Plants in a Changing Climate. Still More Questions than Answers. Plant Sci. 2014, 225, 34–44. [Google Scholar] [CrossRef]
- Le Gouis, J.; Oury, F.-X.; Charmet, G. How Changes in Climate and Agricultural Practices Influenced Wheat Production in Western Europe. J. Cereal Sci. 2020, 93, 102960. [Google Scholar] [CrossRef]
- Galiba, G.; Vágújfalvi, A.; Li, C.; Soltész, A.; Dubcovsky, J. Regulatory Genes Involved in the Determination of Frost Tolerance in Temperate Cereals. Plant Sci. 2009, 176, 12–19. [Google Scholar] [CrossRef]
- Gaudet, D.A.; Wang, Y.; Frick, M.; Puchalski, B.; Penniket, C.; Ouellet, T.; Robert, L.; Singh, J.; Laroche, A. Low Temperature Induced Defence Gene Expression in Winter Wheat in Relation to Resistance to Snow Moulds and Other Wheat Diseases. Plant Sci. 2011, 180, 99–110. [Google Scholar] [CrossRef]
- Skendžić, S.; Zovko, M.; Lešić, V.; Pajač Živković, I.; Lemić, D. Detection and Evaluation of Environmental Stress in Winter Wheat Using Remote and Proximal Sensing Methods and Vegetation Indices—A Review. Diversity 2023, 15, 481. [Google Scholar] [CrossRef]
- Fowler, D.B.; Limin, A.E. Breeding for Winter Hardiness in Cereals. Acta Agron. Hung. 1997, 45, 301–310. [Google Scholar]
- Fowler, D.B.; Limin, A.E.; Wang, S.; Ward, R.W. Relationship between Low-Temperature Tolerance and Vernalization Response in Wheat and Rye. Can. J. Plant Sci. 1996, 76, 37–42. [Google Scholar] [CrossRef]
- Laudencia-Chingcuanco, D.; Ganeshan, S.; You, F.; Fowler, B.; Chibbar, R.; Anderson, O. Genome-Wide Gene Expression Analysis Supports a Developmental Model of Low Temperature Tolerance Gene Regulation in Wheat (Triticum aestivum L.). BMC Genom. 2011, 12, 299. [Google Scholar] [CrossRef] [PubMed]
- Jaškūnė, K.; Armonienė, R.; Liatukas, Ž.; Statkevičiūtė, G.; Cesevičienė, J.; Brazauskas, G. Relationship between Freezing Tolerance and Leaf Growth during Acclimation in Winter Wheat. Agronomy 2022, 12, 859. [Google Scholar] [CrossRef]
- Vágújfalvi, A.; Galiba, G.; Dubcovsky, J.; Cattivelli, L. Two Loci on Wheat Chromosome 5A Regulate the Differential Cold-Dependent Expression of the Cor14b Gene in Frost-Tolerant and Frost-Sensitive Genotypes. Mol. Gen. Genet. MGG 2000, 263, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Båga, M.; Chodaparambil, S.V.; Limin, A.E.; Pecar, M.; Fowler, D.B.; Chibbar, R.N. Identification of Quantitative Trait Loci and Associated Candidate Genes for Low-Temperature Tolerance in Cold-Hardy Winter Wheat. Funct. Integr. Genom. 2007, 7, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wu, Y.; Liu, C.; Li, W.; St Amand, P.; Bernardo, A.; Wang, D.; Dong, L.; Yuan, X.; Zhang, H.; et al. High-Resolution Genome-Wide Association Study and Genomic Prediction for Disease Resistance and Cold Tolerance in Wheat. Theor. Appl. Genet. 2021, 134, 2857–2873. [Google Scholar] [CrossRef]
- Würschum, T.; Longin, C.F.H.; Hahn, V.; Tucker, M.R.; Leiser, W.L. Copy Number Variations of CBF Genes at the Fr-A2 Locus Are Essential Components of Winter Hardiness in Wheat. Plant J. 2017, 89, 764–773. [Google Scholar] [CrossRef]
- Babben, S.; Schliephake, E.; Janitza, P.; Berner, T.; Keilwagen, J.; Koch, M.; Arana-Ceballos, F.A.; Templer, S.E.; Chesnokov, Y.; Pshenichnikova, T.; et al. Association Genetics Studies on Frost Tolerance in Wheat (Triticum aestivum L.) Reveal New Highly Conserved Amino Acid Substitutions in CBF-A3, CBF-A15, VRN3 and PPD1 Genes. BMC Genom. 2018, 19, 409. [Google Scholar] [CrossRef]
- Soleimani, B.; Lehnert, H.; Babben, S.; Keilwagen, J.; Koch, M.; Arana-Ceballos, F.A.; Chesnokov, Y.; Pshenichnikova, T.; Schondelmaier, J.; Ordon, F.; et al. Genome Wide Association Study of Frost Tolerance in Wheat. Sci. Rep. 2022, 12, 5275. [Google Scholar] [CrossRef]
- Fowler, D.B.; N’diaye, A.; Laudencia-Chingcuanco, D.; Pozniak, C.J. Quantitative Trait Loci Associated with Phenological Development, Low-Temperature Tolerance, Grain Quality, and Agronomic Characters in Wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0152185. [Google Scholar] [CrossRef]
- Case, A.J.; Skinner, D.Z.; Garland-Campbell, K.A.; Carter, A.H. Freezing Tolerance-Associated Quantitative Trait Loci in the Brundage × Coda Wheat Recombinant Inbred Line Population. Crop Sci. 2014, 54, 982–992. [Google Scholar] [CrossRef]
- Hetterscheid, W.L.A.; Brandenburg, W.A. Culton versus Taxon: Conceptual Issues in Cultivated Plant Systematics. Taxon 1995, 44, 161–175. [Google Scholar] [CrossRef]
- Odilbekov, F.; Armonienė, R.; Koc, A.; Svensson, J.; Chawade, A. GWAS-Assisted Genomic Prediction to Predict Resistance to Septoria Tritici Blotch in Nordic Winter Wheat at Seedling Stage. Front. Genet. 2019, 10, 1224. [Google Scholar] [CrossRef] [PubMed]
- Alemu, A.; Brazauskas, G.; Gaikpa, D.S.; Henriksson, T.; Islamov, B.; Jørgensen, L.N.; Koppel, M.; Koppel, R.; Liatukas, Ž.; Svensson, J.T.; et al. Genome-Wide Association Analysis and Genomic Prediction for Adult-Plant Resistance to Septoria Tritici Blotch and Powdery Mildew in Winter Wheat. Front. Genet. 2021, 12, 1742. [Google Scholar] [CrossRef]
- Kumar, D.; Kushwaha, S.; Delvento, C.; Liatukas, Ž.; Vivekanand, V.; Svensson, J.T.; Henriksson, T.; Brazauskas, G.; Chawade, A. Affordable Phenotyping of Winter Wheat under Field and Controlled Conditions for Drought Tolerance. Agronomy 2020, 10, 882. [Google Scholar] [CrossRef]
- Bellucci, A.; Torp, A.M.; Bruun, S.; Magid, J.; Andersen, S.B.; Rasmussen, S.K. Association Mapping in Scandinavian Winter Wheat for Yield, Plant Height, and Traits Important for Second-Generation Bioethanol Production. Front. Plant Sci. 2015, 6, 1046. [Google Scholar] [CrossRef] [PubMed]
- Dotlačil, L.; Hermuth, J.; Stehno, Z.; Dvořáček, V.; Bradová, J.; Leišová, L. How Can Wheat Landraces Contribute to Present Breeding. Czech J. Genet. Plant Breed. 2010, 46, S70–S74. [Google Scholar] [CrossRef]
- Sthapit Kandel, J.; Huang, M.; Zhang, Z.; Skinner, D.Z.; See, D.R. Genetic Diversity of Clinal Freezing Tolerance Variation in Winter Wheat Landraces. Agronomy 2018, 8, 95. [Google Scholar] [CrossRef]
- Gupta, P.K.; Kulwal, P.L.; Jaiswal, V. Association Mapping in Plants in the Post-GWAS Genomics Era. In Advances in Genetics; Kumar, D., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 104, pp. 75–154. ISBN 0065-2660. [Google Scholar]
- Jaškūnė, K.; Kemešytė, V.; Aleliūnas, A.; Statkevičiūtė, G. Genome-Wide Markers for Seed Yield and Disease Resistance in Perennial Ryegrass. Crop J. 2022, 10, 508–514. [Google Scholar] [CrossRef]
- Tsai, H.-Y.; Janss, L.L.; Andersen, J.R.; Orabi, J.; Jensen, J.D.; Jahoor, A.; Jensen, J. Genomic Prediction and GWAS of Yield, Quality and Disease-Related Traits in Spring Barley and Winter Wheat. Sci. Rep. 2020, 10, 3347. [Google Scholar] [CrossRef]
- Abou-Elwafa, S.F.; Shehzad, T. Genetic Diversity, GWAS and Prediction for Drought and Terminal Heat Stress Tolerance in Bread Wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2021, 68, 711–728. [Google Scholar] [CrossRef]
- Mäkinen, H.; Kaseva, J.; Trnka, M.; Balek, J.; Kersebaum, K.C.; Nendel, C.; Gobin, A.; Olesen, J.E.; Bindi, M.; Ferrise, R.; et al. Sensitivity of European Wheat to Extreme Weather. F. Crop. Res. 2018, 222, 209–217. [Google Scholar] [CrossRef]
- Olesen, J.E.; Børgesen, C.D.; Elsgaard, L.; Palosuo, T.; Rötter, R.P.; Skjelvåg, A.O.; Peltonen-Sainio, P.; Börjesson, T.; Trnka, M.; Ewert, F.; et al. Changes in Time of Sowing, Flowering and Maturity of Cereals in Europe under Climate Change. Food Addit. Contam. Part A 2012, 29, 1527–1542. [Google Scholar] [CrossRef]
- Gorash, A.; Armonienė, R.; Liatukas, Ž.; Brazauskas, G. The Relationship among Freezing Tolerance, Vernalization Requirement, Ppd Alleles and Winter Hardiness in European Wheat Cultivars. J. Agric. Sci. 2017, 155, 1353–1370. [Google Scholar] [CrossRef]
- Rapacz, M.; Macko-Podgórni, A.; Jurczyk, B.; Kuchar, L. Modeling Wheat and Triticale Winter Hardiness under Current and Predicted Winter Scenarios for Central Europe: A Focus on Deacclimation. Agric. For. Meteorol. 2022, 313, 108739. [Google Scholar] [CrossRef]
- Lupton, F.G.H. Wheat Breeding: Its Scientific Basis; Lupton, F.G.H., Ed.; Springer: Dordrecht, The Netherlands, 1987; pp. 51–70. ISBN 978-94-009-3131-2. [Google Scholar]
- Yang, C.J.; Ladejobi, O.; Mott, R.; Powell, W.; Mackay, I. Analysis of Historical Selection in Winter Wheat. Theor. Appl. Genet. 2022, 135, 3005–3023. [Google Scholar] [CrossRef] [PubMed]
- Haas, M.; Schreiber, M.; Mascher, M. Domestication and Crop Evolution of Wheat and Barley: Genes, Genomics, and Future Directions. J. Integr. Plant Biol. 2019, 61, 204–225. [Google Scholar] [CrossRef] [PubMed]
- Borlaug, N.E. Contributions of Conventional Plant Breeding to Food Production. Science 1983, 219, 689–693. [Google Scholar] [CrossRef]
- Evenson, R.E.; Gollin, D. Assessing the Impact of the Green Revolution, 1960 to 2000. Science 2003, 300, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Faris, J.D. Wheat Domestication: Key to Agricultural Revolutions Past and Future. In Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 439–464. ISBN 978-94-007-7572-5. [Google Scholar]
- Reif, J.C.; Zhang, P.; Dreisigacker, S.; Warburton, M.L.; van Ginkel, M.; Hoisington, D.; Bohn, M.; Melchinger, A.E. Wheat Genetic Diversity Trends during Domestication and Breeding. Theor. Appl. Genet. 2005, 110, 859–864. [Google Scholar] [CrossRef]
- Jol, A.; Füssel, H.-M. EEA Climate Change, Impacts and Vulnerability in Europe 2012; European Environment Agency: Copenhagen, Denmark, 2012; p. 59. [Google Scholar] [CrossRef]
- Twardosz, R.; Kossowska-Cezak, U. Exceptionally Cold and Mild Winters in Europe (1951–2010). Theor. Appl. Climatol. 2016, 125, 399–411. [Google Scholar] [CrossRef]
- Lejenäs, H. The Severe Winter in Europe 1941–42: The Large-Scale Circulation, Cut-off Lows, and Blocking. Bull. Am. Meteorol. Soc. 1989, 70, 271–281. [Google Scholar] [CrossRef]
- Brassley, P.; Segers, Y.; Van Molle, L. War, Agriculture, and Food: Rural Europe from the 1930s to the 1950s; Routledge: London, UK, 2012; ISBN 0415522161. [Google Scholar]
- FAOSTAT Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 17 October 2023).
- FAO. The State of Food and Agriculture 1955; UN: Rome, Italy, 1955; ISBN 9789210472715. [Google Scholar]
- Diederichsen, A.; Solberg, S.Ø.; Jeppson, S. Morphological Changes in Nordic Spring Wheat (Triticum Aestivum L.) Landraces and Cultivars Released from 1892 to 1994. Genet. Resour. Crop Evol. 2012, 60, 569–585. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting Genetic Diversity from Landraces in Wheat Breeding for Adaptation to Climate Change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-Wide Comparative Diversity Uncovers Multiple Targets of Selection for Improvement in Hexaploid Wheat Landraces and Cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef]
- Mokanu, N.V.; Fayt, V.I. Differences in the Effects of Alleles of the Genes Vrd1 and Ppd-D1 with Respect to Winter Hardiness, Frost Tolerance and Yield in Winter Wheat. Cytol. Genet. 2008, 42, 384–390. [Google Scholar] [CrossRef]
- Sãulescu, N.N.; Braun, H.J. Cold Tolerance. In Application of Physiology in Wheat Breeding; Reynolds, M., Ortiz-Monaterio, J., McNab, A., Eds.; CIMMYT: Mexico City, Mexico, 2001; pp. 111–123. ISBN 970-648-077-3. [Google Scholar]
- IWGSC; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Souvorov, A.; Kapustin, Y.; Kiryutin, B.; Chetvernin, V.; Tatusova, T.; Lipman, D. Gnomon–NCBI Eukaryotic Gene Prediction Tool. Natl. Cent. Biotechnol. Inf. 2010, 25, 1–24. [Google Scholar]
- Clavijo, B.J.; Venturini, L.; Schudoma, C.; Accinelli, G.G.; Kaithakottil, G.; Wright, J.; Borrill, P.; Kettleborough, G.; Heavens, D.; Chapman, H. An Improved Assembly and Annotation of the Allohexaploid Wheat Genome Identifies Complete Families of Agronomic Genes and Provides Genomic Evidence for Chromosomal Translocations. Genome Res. 2017, 27, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Gundlach, H.; Spannagl, M.; Uauy, C.; Borrill, P.; Ramírez-González, R.H.; De Oliveira, R.; Mayer, K.F.X.; Paux, E.; Choulet, F.; et al. Impact of Transposable Elements on Genome Structure and Evolution in Bread Wheat. Genome Biol. 2018, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Chao, S.; Dubcovsky, J.; Dvorak, J.; Luo, M.-C.; Baenziger, S.P.; Matnyazov, R.; Clark, D.R.; Talbert, L.E.; Anderson, J.A.; Dreisigacker, S.; et al. Population- and Genome-Specific Patterns of Linkage Disequilibrium and SNP Variation in Spring and Winter Wheat (Triticum aestivum L.). BMC Genom. 2010, 11, 727. [Google Scholar] [CrossRef]
- Underwood, C.J.; Henderson, I.R.; Martienssen, R.A. Genetic and Epigenetic Variation of Transposable Elements in Arabidopsis. Curr. Opin. Plant Biol. 2017, 36, 135–141. [Google Scholar] [CrossRef]
- Monera, O.D.; Sereda, T.J.; Zhou, N.E.; Kay, C.M.; Hodges, R.S. Relationship of Sidechain Hydrophobicity and A-helical Propensity on the Stability of the Single-stranded Amphipathic A-helix. J. Pept. Sci. 1995, 1, 319–329. [Google Scholar] [CrossRef]
- Hirano, T.; Sato, M.H. Diverse Physiological Functions of FAB1 and Phosphatidylinositol 3,5-Bisphosphate in Plants. Front. Plant Sci. 2019, 10, 274. [Google Scholar] [CrossRef]
- Bak, G.; Lee, E.-J.; Lee, Y.; Kato, M.; Segami, S.; Sze, H.; Maeshima, M.; Hwang, J.-U.; Lee, Y. Rapid Structural Changes and Acidification of Guard Cell Vacuoles during Stomatal Closure Require Phosphatidylinositol 3,5-Bisphosphate. Plant Cell 2013, 25, 2202–2216. [Google Scholar] [CrossRef]
- Jurczyk, B.; Grzesiak, M.; Pociecha, E.; Wlazło, M.; Rapacz, M. Diverse Stomatal Behaviors Mediating Photosynthetic Acclimation to Low Temperatures in Hordeum vulgare. Front. Plant Sci. 2019, 9, 963. [Google Scholar] [CrossRef]
- Bricker, J.; Garrick, M.D. An Isoleucine-Valine Substitution in the β Chain of Rabbit Hemoglobin. Biochim. Biophys. Acta Protein Struct. 1974, 351, 437–441. [Google Scholar] [CrossRef]
- Båga, M.; Bahrani, H.; Larsen, J.; Hackauf, B.; Graf, R.J.; Laroche, A.; Chibbar, R.N. Association Mapping of Autumn-Seeded Rye (Secale cereale L.) Reveals Genetic Linkages between Genes Controlling Winter Hardiness and Plant Development. Sci. Rep. 2022, 12, 5793. [Google Scholar] [CrossRef] [PubMed]
- Erath, W.; Bauer, E.; Fowler, D.B.; Gordillo, A.; Korzun, V.; Ponomareva, M.; Schmidt, M.; Schmiedchen, B.; Wilde, P.; Schön, C.-C. Exploring New Alleles for Frost Tolerance in Winter Rye. Theor. Appl. Genet. 2017, 130, 2151–2164. [Google Scholar] [CrossRef] [PubMed]
- He, G.H.Y.; Helbing, C.C.; Wagner, M.J.; Sensen, C.W.; Riabowol, K. Phylogenetic Analysis of the ING Family of PHD Finger Proteins. Mol. Biol. Evol. 2005, 22, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Kruse, E.B.; Carle, S.W.; Wen, N.; Skinner, D.Z.; Murray, T.D.; Garland-Campbell, K.A.; Carter, A.H. Genomic Regions Associated with Tolerance to Freezing Stress and Snow Mold in Winter Wheat. G3 Genes Genomes Genet. 2017, 7, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Armonienė, R.; Liatukas, Ž.; Brazauskas, G. Evaluation of Freezing Tolerance of Winter Wheat (Triticum aestivum L.) under Controlled Conditions and in the Field. Zemdirbyste 2013, 100, 417–424. [Google Scholar] [CrossRef][Green Version]
- Helgadóttir, Á.; Aavola, R.; Isolahti, M.; Marum, P.; Persson, C.; Aleliūnas, A.; Brazauskas, G.; Krisjánsdóttir, T.A.; Asp, T.; Rognli, O.A. Adaptability and Phenotypic Stability of Lolium perenne L. Cultivars of Diverse Origin Grown at the Margin of the Species Distribution. J. Agron. Crop Sci. 2018, 204, 493–504. [Google Scholar] [CrossRef]
- Mishra, M.; Kanwar, P.; Singh, A.; Pandey, A.; Kapoor, S.; Pandey, G.K. Plant Omics: Genome-Wide Analysis of ABA Repressor1 (ABR1) Related Genes in Rice during Abiotic Stress and Development. Omi. A J. Integr. Biol. 2013, 17, 439–450. [Google Scholar] [CrossRef]
- Tang, R.-J.; Luan, S. Regulation of Calcium and Magnesium Homeostasis in Plants: From Transporters to Signaling Network. Curr. Opin. Plant Biol. 2017, 39, 97–105. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. Cold Stress Signaling Networks in Arabidopsis. J. Plant Biol. 2013, 56, 69–76. [Google Scholar] [CrossRef]
- Shu, S.; Ye, K. Structural and Functional Analysis of Ribosome Assembly Factor Efg1. Nucleic Acids Res. 2018, 46, 2096–2106. [Google Scholar] [CrossRef]
- Vaz-Drago, R.; Custódio, N.; Carmo-Fonseca, M. Deep Intronic Mutations and Human Disease. Hum. Genet. 2017, 136, 1093–1111. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Srivastava, R. Plant Biosynthetic Engineering Through Transcription Regulation: An Insight into Molecular Mechanisms During Environmental Stress. In Biosynthetic Technology and Environmental Challenges; Varjani, S.J., Parameswaran, B., Kumar, S., Khare, S.K., Eds.; Springer: Singapore, 2018; pp. 51–72. ISBN 978-981-10-7434-9. [Google Scholar]
- Sun, J.; Sun, Y.; Ahmed, R.I.; Ren, A.; Xie, M. Research Progress on Plant RING-Finger Proteins. Genes 2019, 10, 973. [Google Scholar] [CrossRef]
- Dong, C.-H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.-K. The Negative Regulator of Plant Cold Responses, HOS1, Is a RING E3 Ligase That Mediates the Ubiquitination and Degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Meng, Q.; Zhang, H.; Huang, J. Knock-down of a RING Finger Gene Confers Cold Tolerance. Bioengineered 2016, 7, 39–45. [Google Scholar] [CrossRef]
- Kadowaki, T.; Kadowaki, H.; Accili, D.; Taylor, S.I. Substitution of Lysine for Asparagine at Position 15 in the Alpha-Subunit of the Human Insulin Receptor. A Mutation That Impairs Transport of Receptors to the Cell Surface and Decreases the Affinity of Insulin Binding. J. Biol. Chem. 1990, 265, 19143–19150. [Google Scholar] [CrossRef]
- Gaffney, D.; Pullinger, C.R.; O’Reilly, D.S.J.; Hoffs, M.S.; Cameron, I.; Vass, J.K.; Kulkarni, M.V.; Kane, J.P.; Schumaker, V.N.; Watts, G.F.; et al. Influence of an Asparagine to Lysine Mutation at Amino Acid 3516 of Apolipoprotein B on Low-Density Lipoprotein Receptor Binding. Clin. Chim. Acta 2002, 321, 113–121. [Google Scholar] [CrossRef]
- Fradgley, N. NIAB Global and UK Subset Wheat Pedigree Files. 9 Decemebr 2021 Update. Available online: https://figshare.com/articles/dataset/NIAB_global_and_UK_subset_wheat_pedigree_files_09_12_21_update/17153363 (accessed on 17 October 2023).
- Shaw, P.D.; Graham, M.; Kennedy, J.; Milne, I.; Marshall, D.F. Helium: Visualization of Large Scale Plant Pedigrees. BMC Bioinform. 2014, 15, 259. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. [Google Scholar]
- Hansen, V.T.; Grimenes, A.A. Meteorologiske Data for Ås 2009; NMBU: Ås, Norway, 2010; ISBN 978-82-7636-023-3. [Google Scholar]
- Hansen, V.T.; Grimenes, A.A. Meteorologiske Data for Ås 2010; NMBU: Ås, Norway, 2011; ISBN 978-82-7636-024-0. [Google Scholar]
- Yao, E.; Blake, V.C.; Cooper, L.; Wight, C.P.; Michel, S.; Cagirici, H.B.; Lazo, G.R.; Birkett, C.L.; Waring, D.J.; Jannink, J.-L.; et al. GrainGenes: A Data-Rich Repository for Small Grains Genetics and Genomics. Database 2022, 2022, baac034. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genomics. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Blake, V.C.; Birkett, C.; Matthews, D.E.; Hane, D.L.; Bradbury, P.; Jannink, J. The Triticeae Toolbox: Combining Phenotype and Genotype Data to Advance Small-Grains Breeding. Plant Genome 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 17 October 2023).
- de Mendiburu, F.; Yaseen, M. Agricolae: Statistical Procedures for Agricultural Research. R Package Version 1.4.0. Available online: https://myaseen208.com/agricolae//index.html (accessed on 17 October 2023).
- Gauch, H.G.; Qian, S.; Piepho, H.-P.; Zhou, L.; Chen, R. Consequences of PCA Graphs, SNP Codings, and PCA Variants for Elucidating Population Structure. PLoS ONE 2019, 14, e0218306. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. Available online: https://cran.r-project.org/package=factoextra (accessed on 17 October 2023).
- Bacheva, A.V.; Gotmanova, N.N.; Belogurov, A.A.; Kudriaeva, A.A. Control of Genome through Variative Nature of Histone-Modifying Ubiquitin Ligases. Biochemistry 2021, 86, S71–S95. [Google Scholar] [CrossRef]
- Hong, J.H.; Savina, M.; Du, J.; Devendran, A.; Kannivadi Ramakanth, K.; Tian, X.; Sim, W.S.; Mironova, V.V.; Xu, J. A Sacrifice-for-Survival Mechanism Protects Root Stem Cell Niche from Chilling Stress. Cell 2017, 170, 102–113.e14. [Google Scholar] [CrossRef]

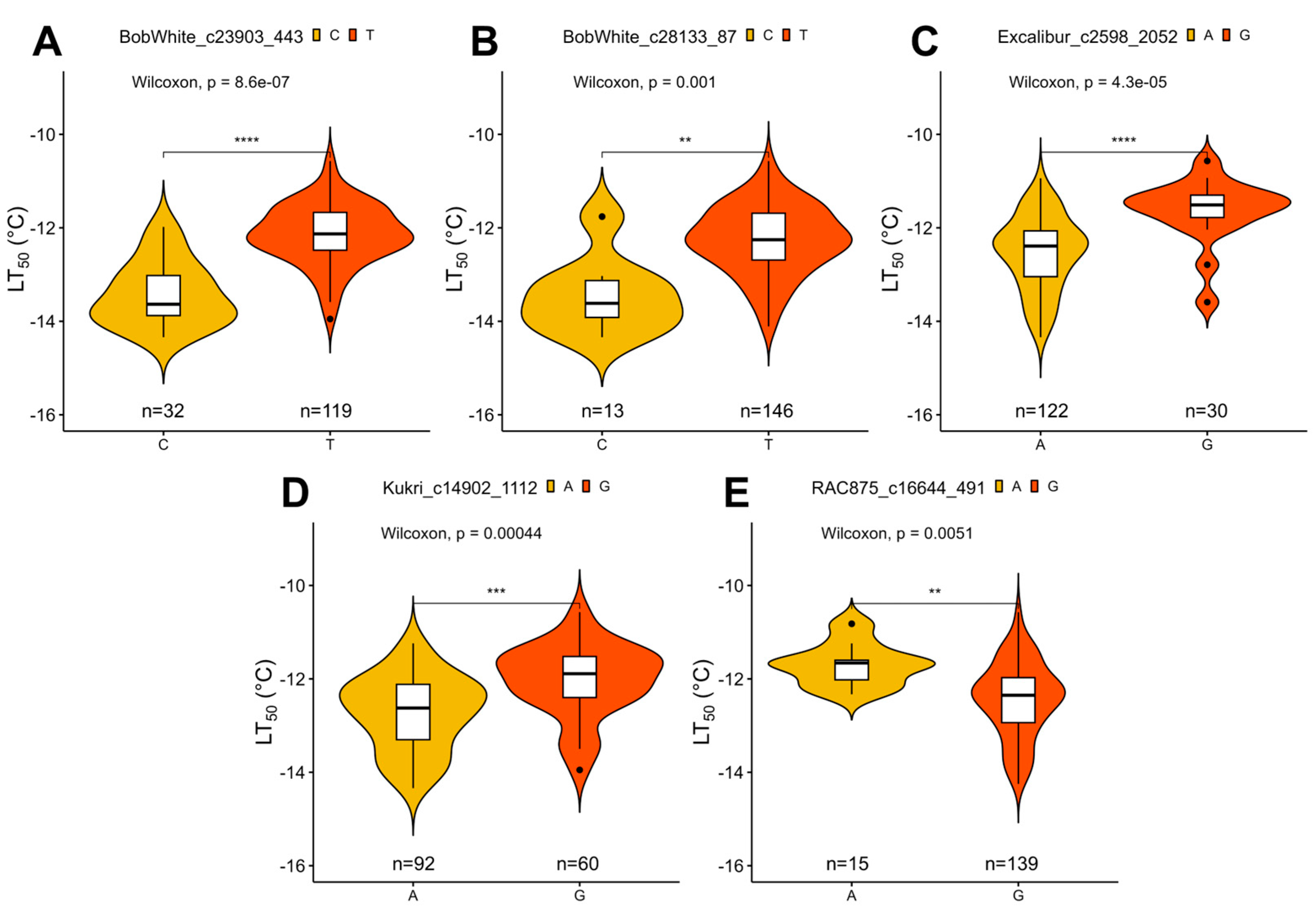
| SNP Marker | Chromosome | Physical Position | MAF | Alleles | GWAS Model | Effect |
|---|---|---|---|---|---|---|
| BobWhite_c23903_443 | 5B | 548427978 | 0.23 | T/C | BLINK **** | 0.33 |
| FarmCPU *** | 0.28 | |||||
| BobWhite_c28133_87 | 2A | 102022208 | 0.08 | T/C | BLINK **** | 0.44 |
| FarmCPU *** | 0.37 | |||||
| Excalibur_c2598_2052 | 5A | 519951564 | 0.21 | A/G | BLINK **** | 0.28 |
| FarmCPU **** | 0.29 | |||||
| Kukri_c14902_1112 | 2B | 206373209 | 0.4 | A/G | BLINK ** | 0.19 |
| FarmCPU * | 0.17 | |||||
| RAC875_c16644_491 | 7A | 18887907 | 0.11 | G/A | FarmCPU ** | −0.33 |
| SNP Marker | Chromosome | Physical Position | MAF | Alleles | GWAS Model | Effect |
|---|---|---|---|---|---|---|
| Eidum 2008–2009 | ||||||
| Excalibur_c43822_370 | 3B | 648752466 | 0.11 | C/T | BLINK **** | 17.67 |
| FarmCPU **** | 11.68 | |||||
| GLM * | 14.39 | |||||
| MLM * | 16.48 | |||||
| MLMM **** | 22.07 | |||||
| wsnp_Ex_c607_1204908 | 5B | 10438244 | 0.03 | T/C | BLINK ** | −12.49 |
| FarmCPU **** | −11.08 | |||||
| MLMM **** | −13.93 | |||||
| Kukri_c57491_156 | 2B | 440825074 | 0.22 | T/C | FarmCPU *** | 6.60 |
| Tdurum_contig47476_528 | UN | 51199268 | 0.10 | C/T | GLM * | 14.94 |
| MLM* | 16.36 | |||||
| Kukri_rep_c85536_598 | UN | 11380 | 0.11 | T/C | GLM * | −13.51 |
| MLM * | −15.16 | |||||
| RFL_Contig3621_1157 | 4A | 742335140 | 0.08 | G/A | MLM * | −15.35 |
| Tdurum_contig47476_495 | UN | 51199301 | 0.08 | G/A | MLM * | −15.28 |
| RAC875_c88279_291 | 6B | 246361296 | 0.16 | T/C | MLMM **** | −9.19 |
| Eidum 2010–2011 | ||||||
| Tdurum_contig50731_961 | 5B | 617710009 | 0.49 | A/C | BLINK *** | 14.54 |
| Vollebekk 2008–2009 | ||||||
| wsnp_JD_c10233_10936535 | 3B | 424792590 | 0.13 | C/A | BLINK *** | −26.47 |
| MLMM * | −26.02 | |||||
| BobWhite_c18566_106 | 6B | 3883636 | 0.29 | C/A | FarmCPU * | −13.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaitkevičiūtė, G.; Chawade, A.; Lillemo, M.; Liatukas, Ž.; Aleliūnas, A.; Armonienė, R. Genome-Wide Association Analysis of Freezing Tolerance and Winter Hardiness in Winter Wheat of Nordic Origin. Plants 2023, 12, 4014. https://doi.org/10.3390/plants12234014
Vaitkevičiūtė G, Chawade A, Lillemo M, Liatukas Ž, Aleliūnas A, Armonienė R. Genome-Wide Association Analysis of Freezing Tolerance and Winter Hardiness in Winter Wheat of Nordic Origin. Plants. 2023; 12(23):4014. https://doi.org/10.3390/plants12234014
Chicago/Turabian StyleVaitkevičiūtė, Gabija, Aakash Chawade, Morten Lillemo, Žilvinas Liatukas, Andrius Aleliūnas, and Rita Armonienė. 2023. "Genome-Wide Association Analysis of Freezing Tolerance and Winter Hardiness in Winter Wheat of Nordic Origin" Plants 12, no. 23: 4014. https://doi.org/10.3390/plants12234014
APA StyleVaitkevičiūtė, G., Chawade, A., Lillemo, M., Liatukas, Ž., Aleliūnas, A., & Armonienė, R. (2023). Genome-Wide Association Analysis of Freezing Tolerance and Winter Hardiness in Winter Wheat of Nordic Origin. Plants, 12(23), 4014. https://doi.org/10.3390/plants12234014










