Genome-Wide Identification and Analysis of the EPF Gene Family in Sorghum bicolor (L.) Moench
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Analysis of the Sorghum EPF
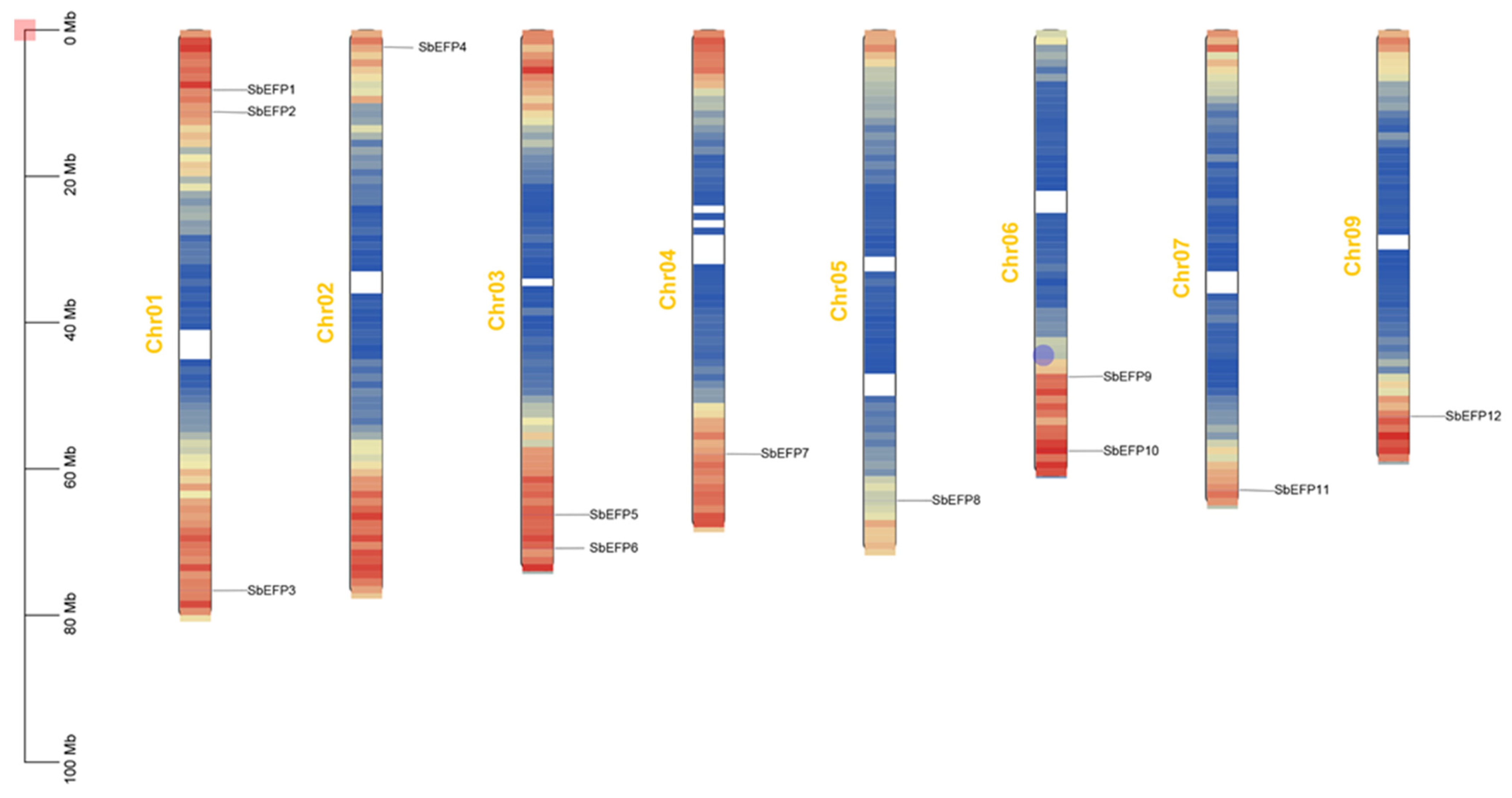
2.2. The Chromosomal Localization of the SbEPF Gene Family
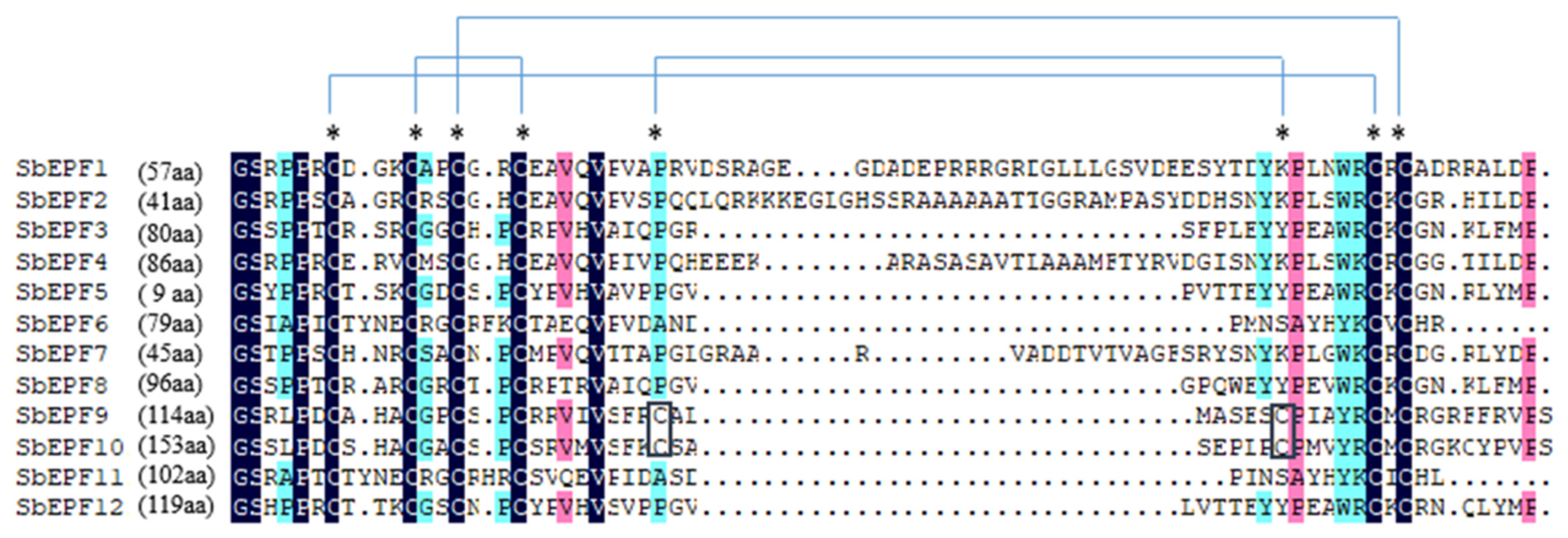
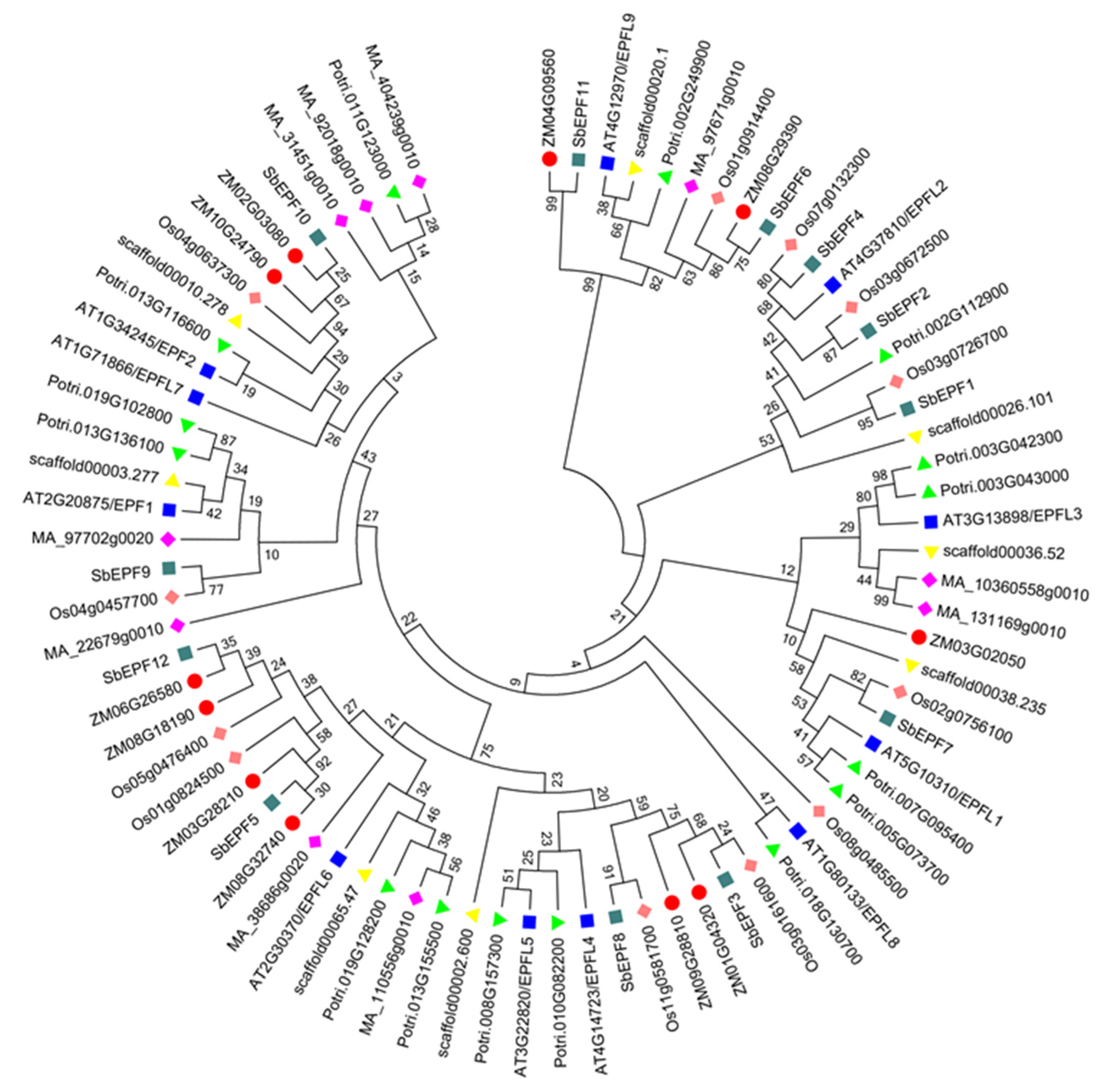
2.3. Evolutionary Analysis of Arabidopsis and Sorghum EPF
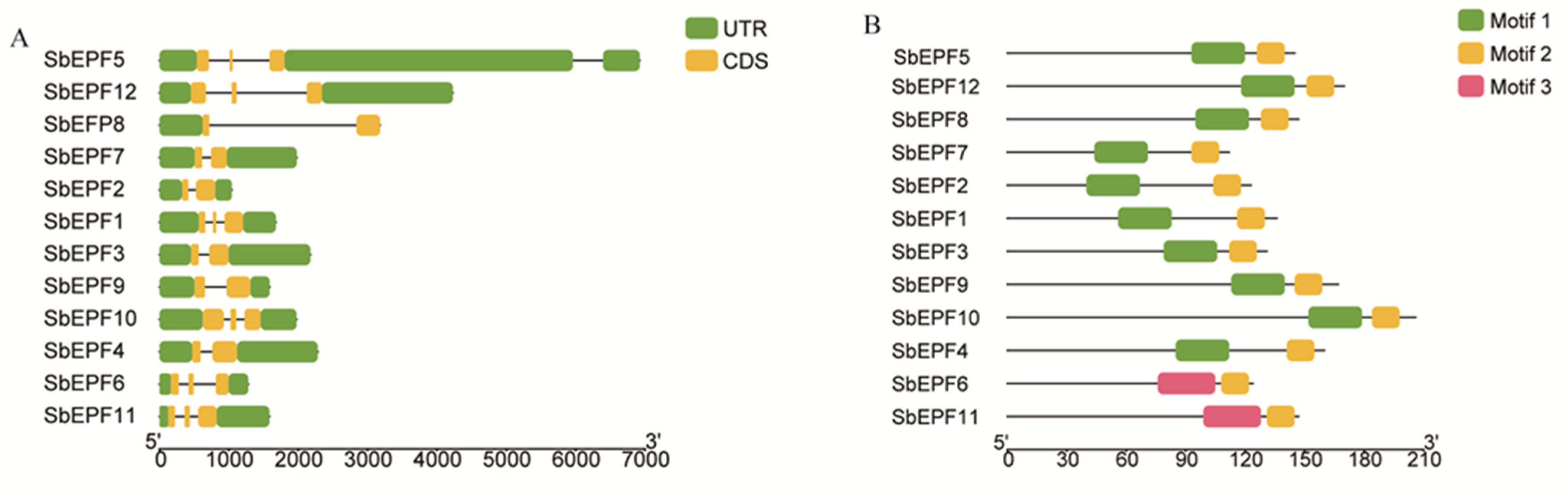
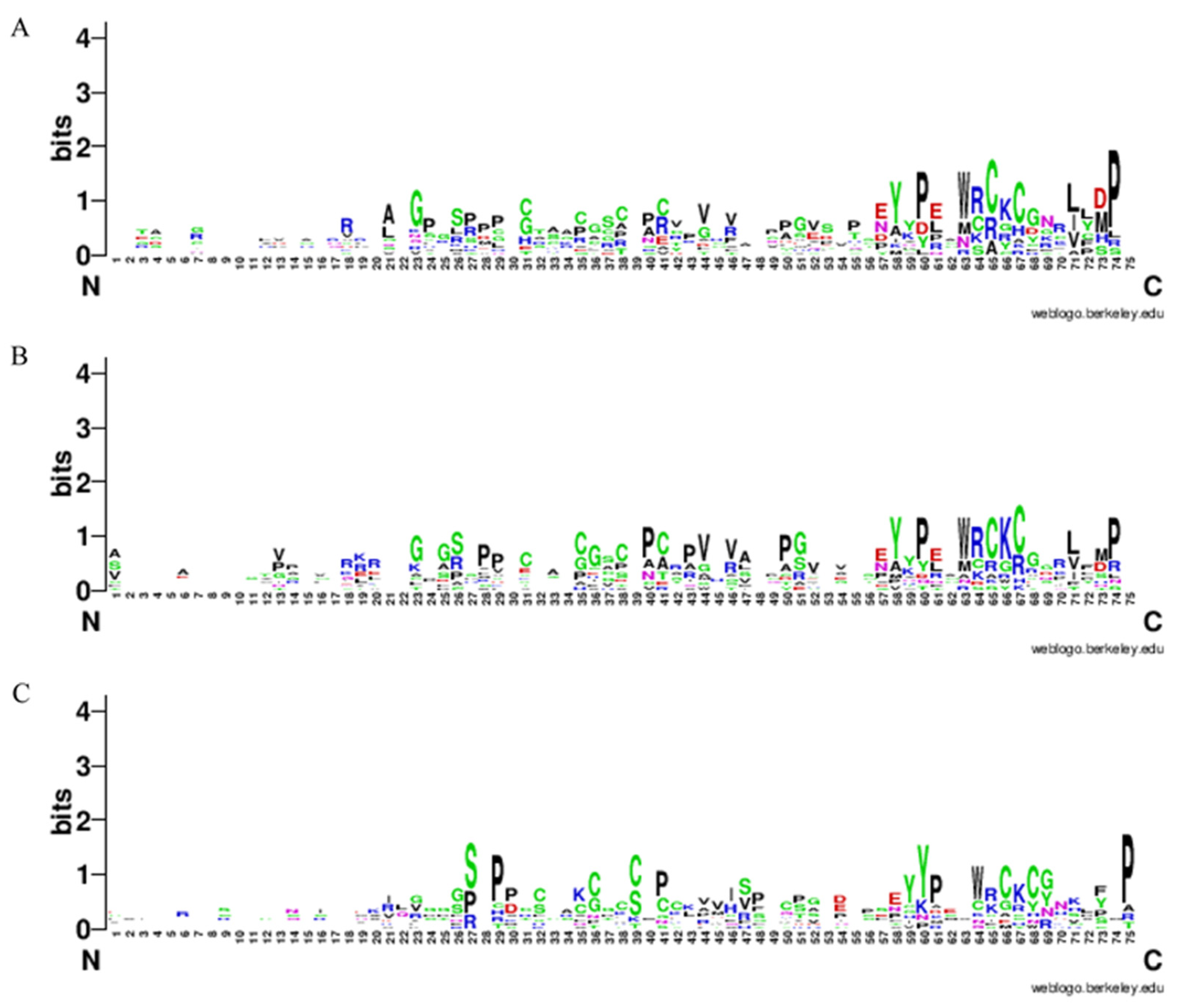
2.4. The Gene Structure and Protein Conserved Motifs Analysis of the Sorghum EPF Family Members
2.5. The EPF Genes’ Relationship between Sorghum and Arabidopsis
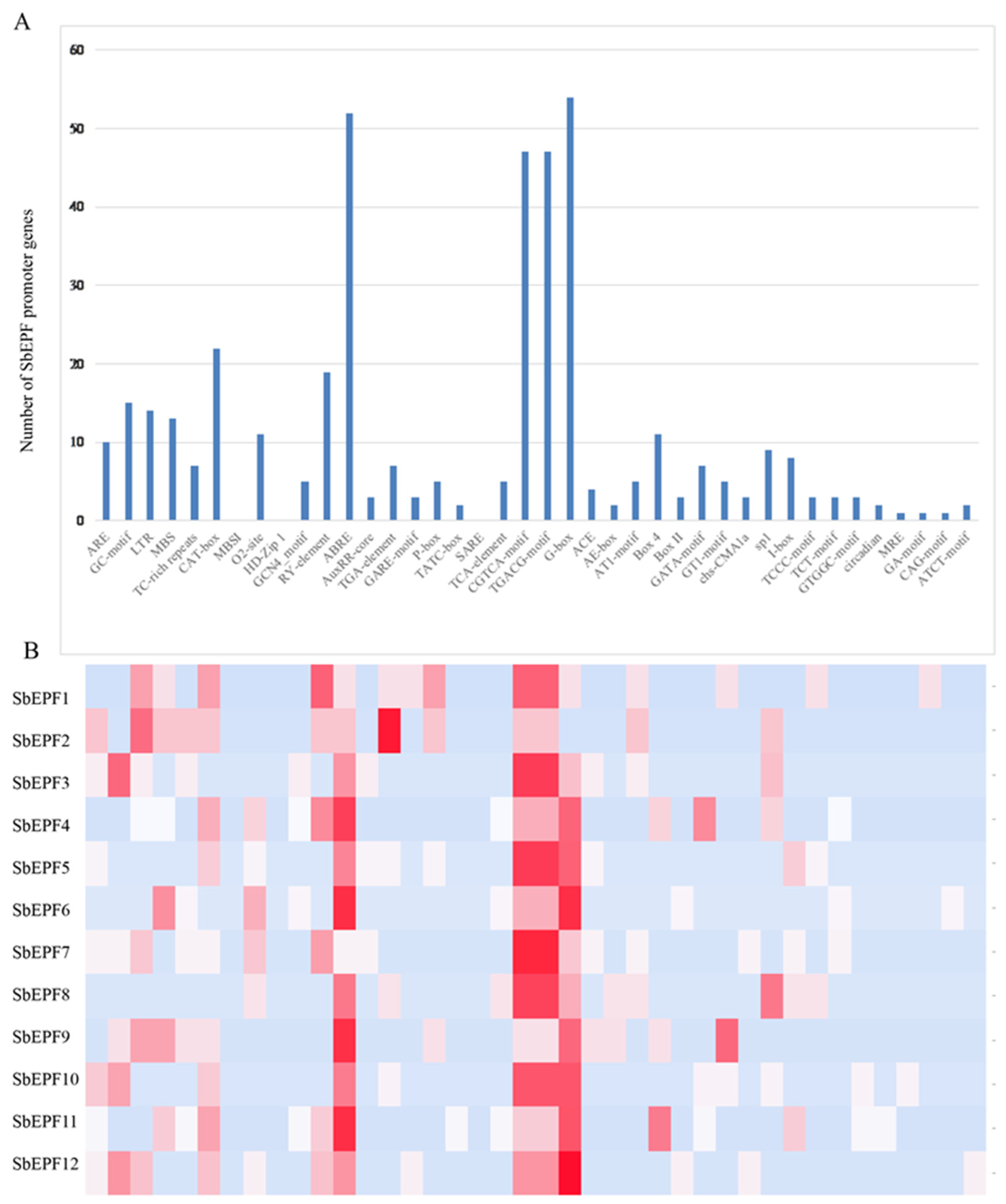
2.6. Analysis of the SbEPF Gene Family Promoter
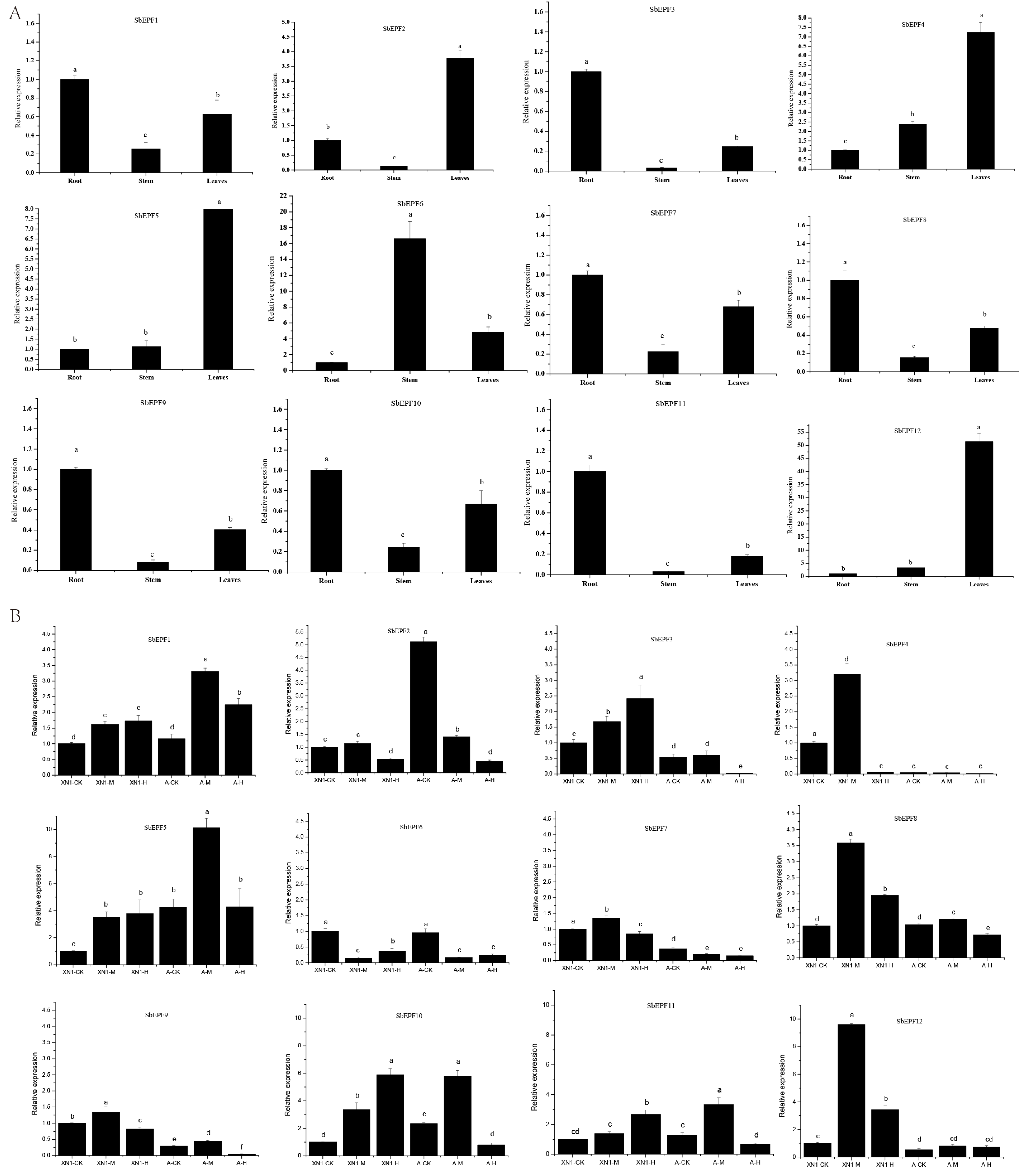
2.7. The Expression Profile of SbEPFs in Response to Various Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification and Characterization of SbEPF Genes in Sorghum
4.2. Evolutionary Relationships of EPF Family Genes
4.3. Analysis of SbEPF Genes and Protein Structures
4.4. Analysis of cis-Element in SbEPF Promoters
4.5. Plant Materials and Drought Sample Collections
4.6. RNA Extraction, Reverse Transcription, and qRT-PCR
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.-K. Salt and Drought Stress Signal Transduction in Plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Mutava, R.N. Characterization of Grain Sorghum for Physiological and Yield Traits Associated with Drought Tolerance. Master Thesis, Kansas State University, Manhattan, KS, USA, 2009. [Google Scholar]
- Chadalavada, K.; Guna, K.; Kumari, B.R.; Kumar, T.S. Drought stress in sorghum: Impact on grain quality. ScienceDirect 2022, 5, 113–134. [Google Scholar]
- Gupta, A.; Andrés Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Gleick, P.H. The Worlds Water 2000–2001: The Biennial Report on Freshwater Resources; Island Press: Washington, DC, USA, 2000; p. 315. [Google Scholar]
- Abdel-Ghany, S.E.; Ullah, F.; Ben-Hur, A.; Reddy, A.S. Transcriptome Analysis of Drought-Resistant and Drought-Sensitive Sorghum (Sorghum bicolor) Genotypes in Response to PEG-Induced Drought Stress. Int. J. Mol. Sci. 2020, 21, 772. [Google Scholar] [CrossRef]
- Katsir, L.; Davies, K.A.; Bergmann, D.C.; Laux, T. Peptide Signaling in Plant Development. Curr. Biol. 2011, 21, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Takata, N.; Yokota, K.; Ohki, S.; Mori, M.; Taniguchi, T.; Kurita, M. Evolutionary Relationship and Structural Characterization of the EPF/EPFL Gene Family. PLoS ONE 2013, 8, e65183. [Google Scholar] [CrossRef]
- Lee, J.S.; Kuroha, T.; Hnilova, M.; Khatayevich, D.; Kanaoka, M.M.; McAbee, J.M.; Sarikaya, M.; Tamerler, C.; Torii, K.U. Direct interaction of ligand–receptor pairs specifying stomatal patterning. Gene Dev. 2012, 26, 126–136. [Google Scholar] [CrossRef]
- Richardson, L.G.L.; Torii, K.U. Take a deep breath: Peptide signalling in stomatal patterning and differentiation. J. Exp. Bot. 2013, 64, 5243–5251. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, S.; Dong, Y.; Zhao, Y.; Geng, A.; Xia, X.; Yin, W. PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. 2016, 14, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lin, J.; Zhu, M.; Zuo, H.; Shen, Y.; Li, J.; Wang k Li, P.; Tang, Q.; Liu, Z. Variations of stomata development in tea plant (Camellia sinensis) leaves in different light and temperature environments and genetic backgrounds. Hortic. Res. 2022, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Huang, Y.; Liu, L.; Zhang, M.; Chai, M.; Xi, X.; Aslam, M.; Wang, L.; Ma, S.; Su, H. Signaling by the EPFL-ERECTA family coordinates female germline specification through the BZR1 family in Arabidopsis. Plant Cell 2023, 35, 1455–1473. [Google Scholar] [CrossRef]
- Hughes, J.; Hepworth, C.; Dutton, C.; Dunn, J.A.; Hunt, L.; Stephens, J.; Waugh, R.; Cameron, D.D.; Gray, J.E. Reducing Stomatal Density in Barley Improves Drought Tolerance without Impacting on Yield. Plant Physiol. 2017, 174, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.; Hunt, L.; Afsharinafar, M.; Meselmani, M.A.; Mitchell, A.; Howells, R.; Wallington, E.; Fleming, A.J.; Gray, J.E. Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 2019, 70, 4737–4747. [Google Scholar] [CrossRef]
- Mohammed, U.; Caine, R.S.; Atkinson, J.A.; Harrison, E.L.; Wells, D.; Chater, C.C.; Gray, J.E.; Swarup, R.; Murchie, E.H. Rice plants overexpressing OsEPF1 show reduced stomatal density and increased root cortical aerenchyma formation. Sci. Rep. 2019, 9, 5584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xie, T.; Zhang, C.; Li, J.; Wang, Z.; Li, H.; Liu, X.; Yin, L.; Wang, S.; Zhang, S.; et al. Overexpression of the potato StEPF2 gene confers enhanced drought tolerance in Arabidopsis. J. Exp. Bot. 2020, 14, 479–490. [Google Scholar] [CrossRef]
- Bessho-Uehara, K.; Wang, D.R.; Furuta, T.; Minami, A.; Nagai, K.; Gamuyao, R.; Asano, K.; Angeles-Shim, R.B.; Shimizu, Y.; Ayano, M.; et al. Loss of function at RAE2, apreviously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl. Acad. Sci. USA 2016, 113, 8969–8974. [Google Scholar] [CrossRef]
- Jin, J.; Hua, L.; Zhu, Z.; Tan, L.; Zhao, X.; Zhang, W.; Liu, F.; Fu, Y.; Cai, H.; Sun, X.; et al. GAD1 Encodes a Secreted Peptide That Regulates Grain Number, Grain Length and Awn Development in Rice Domestication. Plant Cell 2016, 10, 2453–2463. [Google Scholar] [CrossRef]
- Kawamoto, N.; Del Carpio, D.P.; Hofmann, A.; Mizuta, Y.; Kurihara, D.; Higashiyama, T.; Uchida, N.; Torii, K.U.; Colombo, L.; Growth, G.; et al. A Peptide Pair Coordinates Regular Ovule Initiation Patterns with Seed Number and Fruit Size. Curr. Biol. 2009, 30, 4352–4361. [Google Scholar] [CrossRef]
- Guo, T.; Lu, Z.-Q.; Xiong, Y.; Shan, J.-X.; Ye, W.-W.; Dong, N.-Q.; Kan, Y.; Yang, Y.-B.; Zhao, H.-Y.; Yu, H.-X.; et al. Optimization of rice panicle architecture by specifically suppressing ligand-receptor pairs. Nat. Commun. 2023, 14, 1640. [Google Scholar] [CrossRef]
- Caine, R.S.; Yin, X.; Sloan, J.; Harrison, E.L.; Mohammed, U.; Fulton, T.; Biswal, A.K.; Dionora, J.; Chater, C.C.; Coe, R.A.; et al. with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 2018, 221, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Han, S.; Li, Z.; Huang, M.; Niu, M.-X.; Yu, X.; Liu, C.; Wang, H.-L.; Yin, W.; Xia, X.; et al. PdEPFL6 reduces stomatal density to improve drought tolerance in poplar. Ind. Crop Prod. 2022, 182, 114873. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Han, S.; Jiao, Z.; Niu, M.X.; Yu, X.; Huang, M.; Liu, C.; Wang, H.L.; Zhou, Y.; Mao, W.; Wang, X.; et al. Genome-Wide Comprehensive Analysis of GASA Gene Family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef]
- Jiang, Q.; Yang, J.; Wang, Q.; Zhou, K.; Mao, K.; Ma, F. Overexpression of MdEPF2 improves water use efficiency and reduces oxidative stress in tomato. Environ. Exp. Bot. 2019, 162, 321–332. [Google Scholar] [CrossRef]
- Ohki, S.; Takeuchi, M.; Mori, M. The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2011, 2, 512. [Google Scholar] [CrossRef]
- Caine, R.S.; Chater, C.C.; Kamisugi, Y.; Cuming, A.C.; Beerling, D.J.; Gray, J.E.; Fleming, A.J. An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 2016, 143, 3306–3314. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Sugano, S.S.; Hara-Nishimura, I. Positive and negative peptide signals control stomatal density. Cell Mol. Life Sci. 2011, 68, 2081–2088. [Google Scholar] [CrossRef]
- Ho CM, K.; Paciorek, T.; Abrash, E.; Bergmann, D.C. Modulators of Stomatal Lineage Signal Transduction Alter Membrane Contact Sites and Reveal Specialization among ERECTA Kinases. Dev. Cell 2016, 38, 345–357. [Google Scholar]
- Herrmann, A.; Torii, K.U. Shouting out loud: Signaling modules in the regulation of stomatal development. Plant Physiol. 2021, 185, 765–780. [Google Scholar] [CrossRef]
- Uchida, N.; Lee, J.S.; Horst, R.J.; Lai, H.H.; Kajita, R.; Kakimoto, T.; Tasaka, M.; Torii, K.U. Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 2012, 109, 6337–6342. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Tasaka, M. Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. J. Exp. Bot. 2013, 64, 5335–5343. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Shi, Y.; Wang, J.; Wang, Z.; Zhang, X.; Jia, X.; Du, Q.; Niu, J.; Liu, B.; Du, R.; et al. Integration of transcriptome and metabolome analyses reveals sorghum roots responding to cadmium stress through regulation of the flavonoid biosynthesis pathway. Front. Plant Sci. 2023, 14, 1144265. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Li, B.; Iwakawa, H.; Pan, Y.; Tang, X.; Ling-Hu, Q.; Liu, Y.; Sheng, S.; Feng, L.; Zhang, H.; et al. Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 2020, 581, 89–93. [Google Scholar] [CrossRef]
| Name | Gene Model ID (V3.1.1) | CDS Length (bp) | Protein Length (aa) | Molecular Weight (KDa) | Protein Isoelectric Point (pI) | Chr. no. | Formula |
|---|---|---|---|---|---|---|---|
| SbEPF1 | Sobic.001G106500 | 408 | 135 | 14.788 | 7.61 | 01 | C633H1025N197O198S7 |
| SbEPF2 | Sobic.001G140400 | 369 | 122 | 12.905 | 9.81 | 01 | C555H906N178O161S8 |
| SbEPF3 | Sobic.001G496400 | 393 | 130 | 13.932 | 9.24 | 01 | C622H964N176O169S10 |
| SbEPF4 | Sobic.002G025300 | 480 | 159 | 17.445 | 9.04 | 02 | C747H1227N231O224S13 |
| SbEPF5 | Sobic.003G339600 | 435 | 144 | 15.893 | 9.28 | 03 | C684H1069N213O188S19 |
| SbEPF6 | Sobic.003G399800 | 372 | 123 | 13.486 | 8.78 | 03 | C586H938N170O171S12 |
| SbEPF7 | Sobic.004G229700 | 335 | 111 | 11.784 | 9.24 | 04 | C504H813N153O153S10 |
| SbEPF8 | Sobic.005G166500 | 441 | 146 | 15.689 | 11.51 | 05 | C672H1105N221O183S15 |
| SbEPF9 | Sobic.006G104400 | 501 | 166 | 18.062 | 9.22 | 06 | C778H1239N243O226S14 |
| SbEPF10 | Sobic.006G233600 | 618 | 205 | 21.524 | 9.07 | 06 | C916H1513N279O276S21 |
| SbEPF11 | Sobic.007G197500 | 441 | 146 | 15.566 | 8.83 | 07 | C658H1076N208O207S11 |
| SbEPF12 | Sobic.009G173200 | 510 | 169 | 18.181 | 9.64 | 09 | C799H1268N236O226S12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Z.; Wang, J.; Shi, Y.; Wang, Z.; Zhang, J.; Du, Q.; Liu, B.; Jia, X.; Niu, J.; Gu, C.; et al. Genome-Wide Identification and Analysis of the EPF Gene Family in Sorghum bicolor (L.) Moench. Plants 2023, 12, 3912. https://doi.org/10.3390/plants12223912
Jiao Z, Wang J, Shi Y, Wang Z, Zhang J, Du Q, Liu B, Jia X, Niu J, Gu C, et al. Genome-Wide Identification and Analysis of the EPF Gene Family in Sorghum bicolor (L.) Moench. Plants. 2023; 12(22):3912. https://doi.org/10.3390/plants12223912
Chicago/Turabian StyleJiao, Zhiyin, Jinping Wang, Yannan Shi, Zhifang Wang, Jing Zhang, Qi Du, Bocheng Liu, Xinyue Jia, Jingtian Niu, Chun Gu, and et al. 2023. "Genome-Wide Identification and Analysis of the EPF Gene Family in Sorghum bicolor (L.) Moench" Plants 12, no. 22: 3912. https://doi.org/10.3390/plants12223912
APA StyleJiao, Z., Wang, J., Shi, Y., Wang, Z., Zhang, J., Du, Q., Liu, B., Jia, X., Niu, J., Gu, C., & Lv, P. (2023). Genome-Wide Identification and Analysis of the EPF Gene Family in Sorghum bicolor (L.) Moench. Plants, 12(22), 3912. https://doi.org/10.3390/plants12223912






