Development of Novel Monoclonal Antibodies to Wheat Alpha-Amylases Associated with Grain Quality Problems That Are Increasing with Climate Change
Abstract
1. Introduction
2. Results
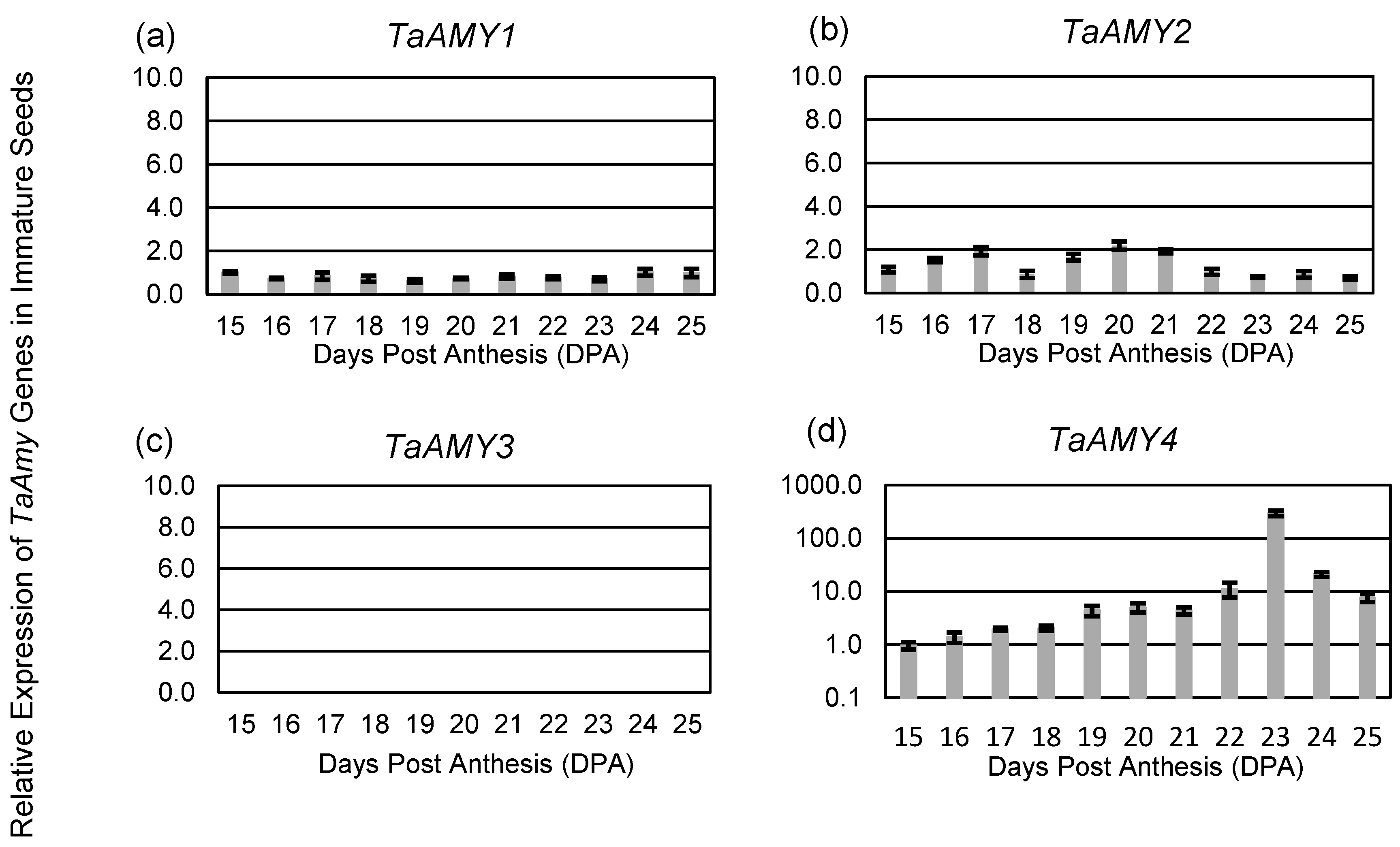
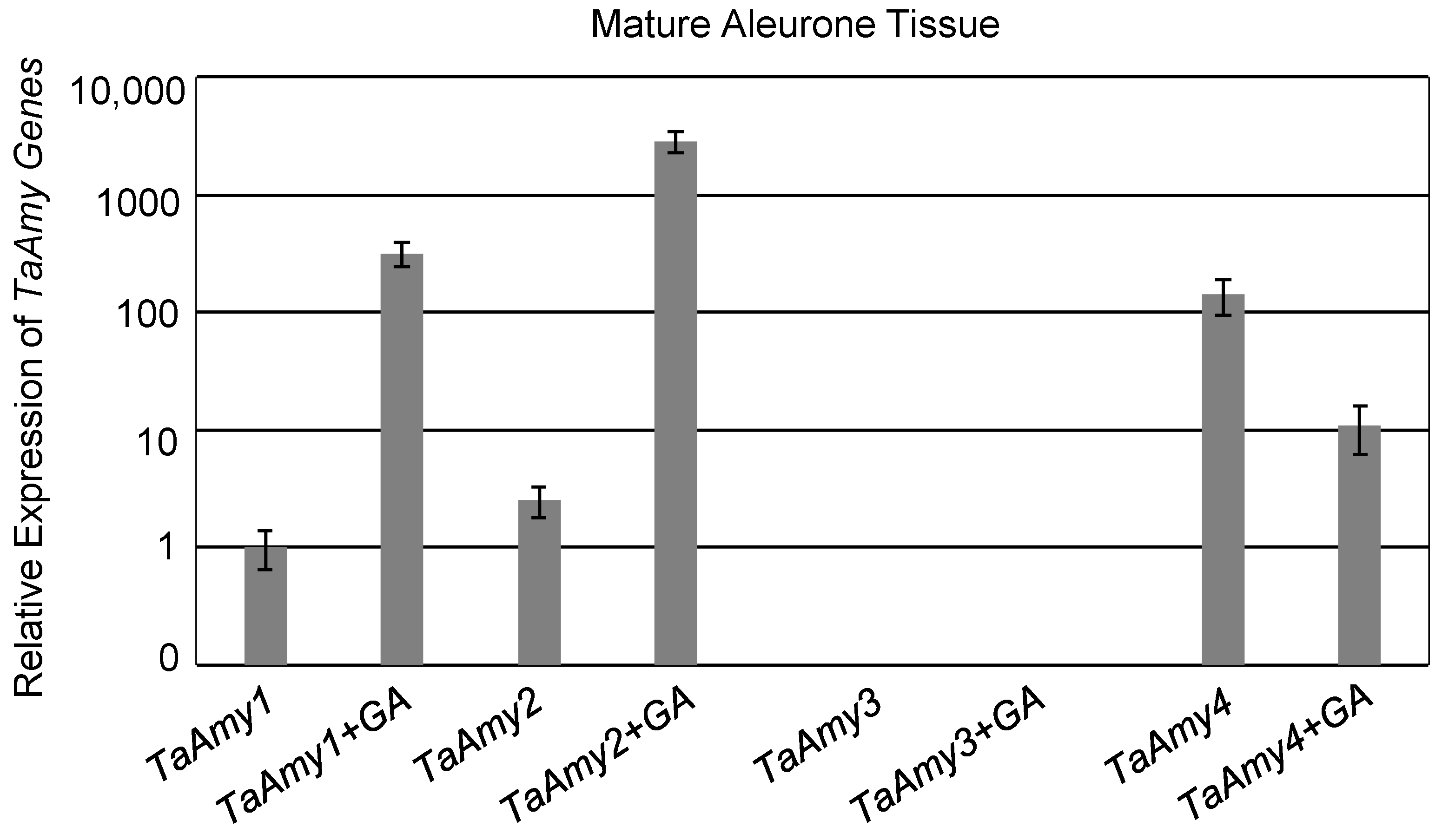
2.1. Expression of Analysis of Wheat (Triticum astevium; TA) α-Amylase (AMY) Genes
2.2. Monoclonal Antibody Development
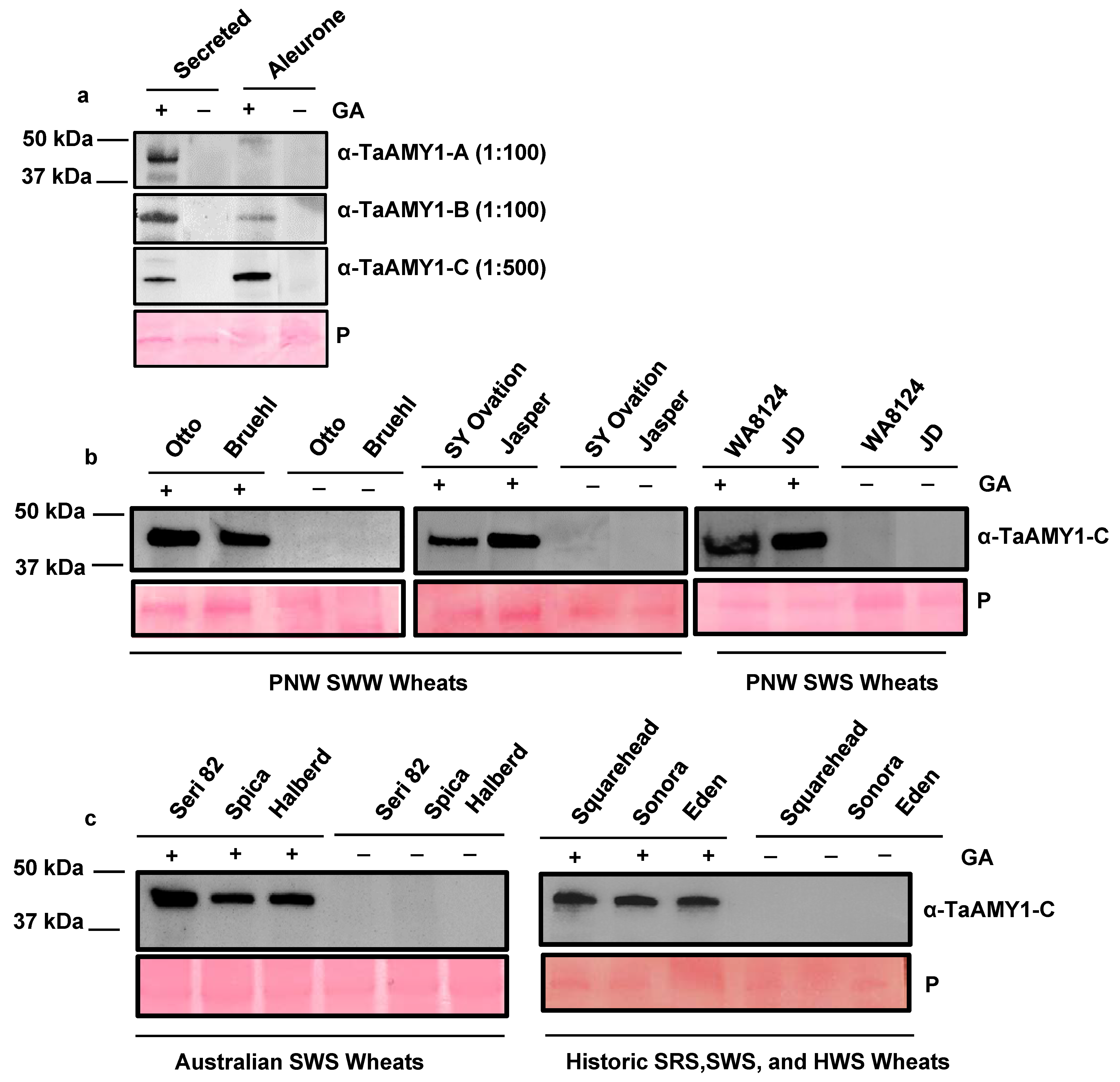
2.3. Validation of TaAMY Antibodies for α-Amylase Detection
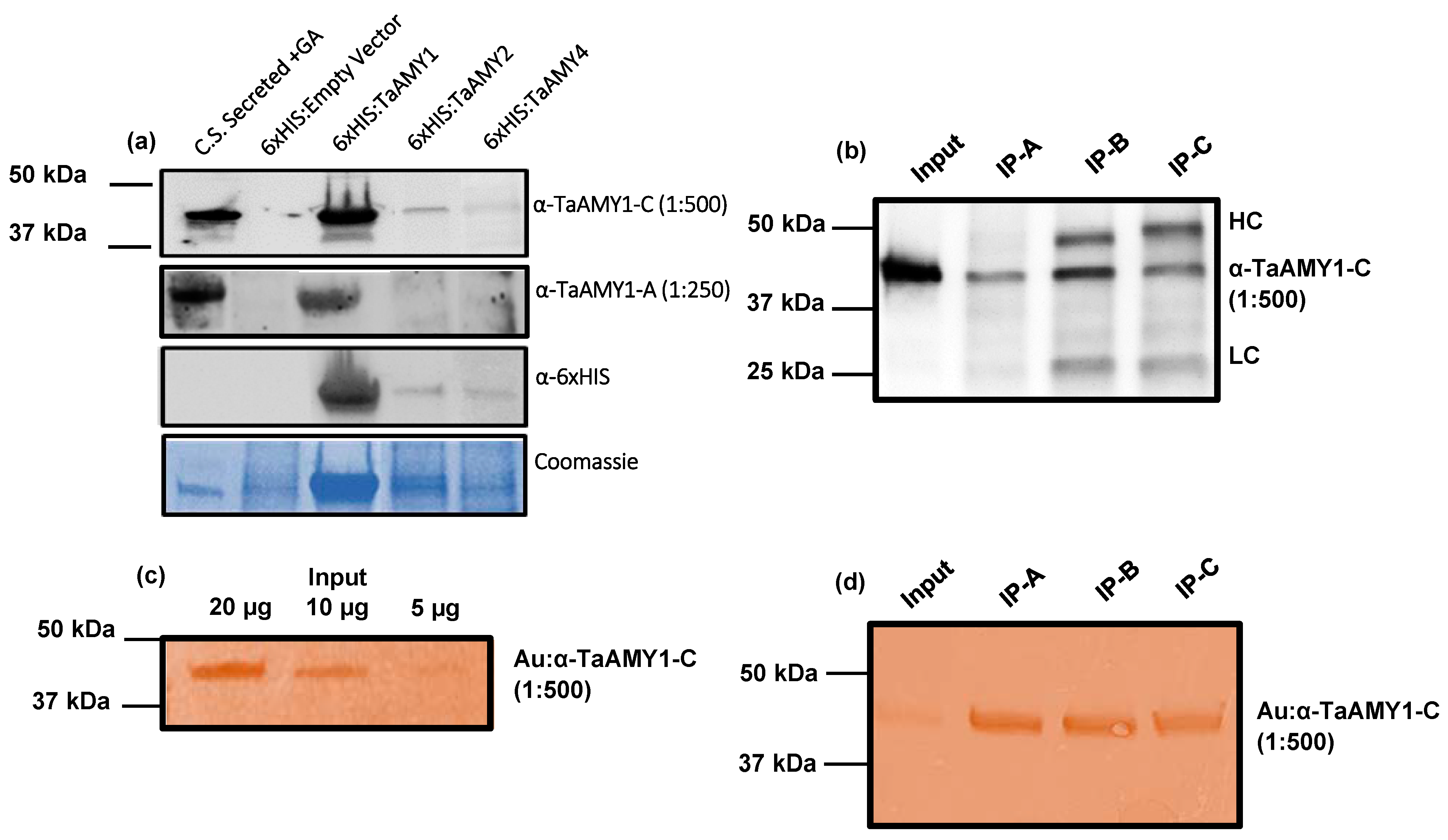
2.4. Validation of TaAMY Antibody Specificity
2.5. Evaluating the Use TaAMY Antibodies in Pairs
3. Discussion
3.1. Overview
3.2. Expression Analysis of Wheat α-Amylase Genes
3.3. Potential for an α-Amylase Immunoassay for Wheat
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Expression Analysis of α-Amylase Genes in Grain Development
4.3. Cloning of α-Amylase Genes and Recombinant Protein Expression
4.4. Statistical Analysis
4.5. Peptide Monoclonal Antibody Development
4.6. Induction of α-Amylase Enzymes from Aleurone Tissue
4.7. Monoclonal Antibody Validation and Peptide Sequencing
4.8. Alpha-Amylase Immunoprecipitation Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruger, J.E. Chapter 10: Enzymes of sprouted wheat and their possible technological significance. In Wheat: Production, Properties and Quality; Bushuk, W., Rasper, V.F., Eds.; Springer: Boston, MA, USA, 1994; pp. 143–153. [Google Scholar]
- Ross, A.S.; Bettge, A.D. Passing the Test on Wheat End-Use Quality; US Department of Agriculture: Washington, DC, USA, 2009; Volume Chapter 20, pp. 455–494. [Google Scholar]
- Kiszonas, A.M.; Engle, D.A.; Pierantoni, L.A.; Morris, C.F. Relationships between Falling Number, α-amylase activity, milling, cookie, and sponge cake quality of soft white wheat. Cereal Chem. 2018, 95, 373–385. [Google Scholar] [CrossRef]
- Mares, D.J.; Mrva, K. Wheat grain preharvest sprouting and late maturity alpha-amylase. Planta 2014, 240, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Cannon, A.E.; Marston, E.J.; Kiszonas, A.M.; Hauvermale, A.L.; See, D.R. Late-maturity α-amylase (LMA): Exploring the underlying mechanisms and end-use quality effects in wheat. Planta 2022, 255, 2. [Google Scholar] [CrossRef]
- Mares, D.; Mrva, K. Late-maturity α-amylase: Low falling number in wheat in the absence of preharvest sprouting. J. Cereal Sci. 2008, 47, 6–17. [Google Scholar] [CrossRef]
- Hu, Y.; Sjoberg, S.M.; Chen, C.; Hauvermale, A.L.; Morris, C.F.; Delwiche, S.R.; Cannon, A.E.; Steber, C.; Zhang, Z. As the number falls, alternatives to the Hagberg–Perten falling number method: A review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2105–2117. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, S. A rapid method for determining alpha-amylase activity. Cereal Chem. 1960, 37, 218–222. [Google Scholar]
- Perten, H. Application of falling number method for evaluating alpha-amylase activity. Cereal Chem. 1964, 41, 127–139. [Google Scholar]
- He, Y.; Lin, Y.L.; Chen, C.; Tsai, M.H.; Lin, A.H. Impacts of starch and the interactions between starch and other macromolecules on wheat falling number. Compr. Rev. Food Sci. Food Saf. 2019, 18, 641–654. [Google Scholar] [CrossRef]
- Daussant, J.; Renard, H.A. Development of different alpha-amylase isozymes, having high and low isoelectric points, during early stages of kernel development in wheat. J. Cereal Sci. 1987, 5, 13–21. [Google Scholar] [CrossRef]
- Gale, M.D.; Ainsworth, C.C. The relationship between alpha-amylase species found in developing and germinating wheat-grain. Biochem. Genet. 1984, 22, 1031–1036. [Google Scholar] [CrossRef]
- Derera, N.F. Preharvest Field Sprouting Cereals; CRC Press: Boca Raton, FL, USA, 1989. [Google Scholar] [CrossRef]
- Rodriguez, M.V.; Barrero, J.M.; Corbineau, F.; Gubler, F.; Benech-Arnold, R.L. Dormancy in cereals (not too much, not so little): About the mechanisms behind this trait. Seed. Sci. Res. 2015, 25, 99–119. [Google Scholar] [CrossRef]
- Ritchie, S.; Gilroy, S. Gibberellins: Regulating genes and germination. New Phytol. 1998, 140, 363–383. [Google Scholar] [CrossRef] [PubMed]
- McCubbin, A.G.; Ritchie, S.M.; Swanson, S.J.; Gilroy, S. The calcium-dependent protein kinase HvCDPK1 mediates the gibberellic acid response of the barley aleurone through regulation of vacuolar function. Plant J. 2004, 39, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, S.; McCubbin, A.; Ambrose, G.; Kao, T.; Gilroy, S. The sensitivity of barley aleurone tissue to gibberellin is heterogenous and may be spatially determined. Plant Physiol. 1999, 120, 361–370. [Google Scholar] [CrossRef][Green Version]
- Lunn, G.D.; Kettlewell, P.S.; Major, B.J.; Scott, R.K. Effects of pericarp alpha-amylase activity on wheat (Triticum aestivum) Hagberg falling number. Ann. Appl. Biol. 2001, 138, 207–214. [Google Scholar] [CrossRef]
- Mieog, J.C.; Janeček, Š.; Ral, J.P. New insight in cereal starch degradation: Identification and structural characterization of four α-amylases in bread wheat. Amylase 2017, 1, 35–49. [Google Scholar] [CrossRef]
- Barrero, J.M.; Mrva, K.; Talbot, M.J.; White, R.G.; Taylor, J.; Gubler, F.; Mares, D.J. Genetic, hormonal, and physiological analysis of late maturity α-amylase in wheat. Plant Physiol. 2013, 161, 1265–1277. [Google Scholar] [CrossRef]
- Ral, J.P.; Whan, A.; Larroque, O.; Leyne, E.; Pritchard, J.; Dielen, A.S.; Howitt, C.A.; Morell, M.K.; Newberry, M. Engineering high α-amylase levels in wheat grain lowers F alling N umber but improves baking properties. Plant Biotechnol. J. 2016, 14, 364–376. [Google Scholar] [CrossRef]
- Newberry, M.; Zwart, A.B.; Whan, A.; Mieog, J.C.; Sun, M.; Leyne, E.; Pritchard, J.; Daneri-Castro, S.N.; Ibrahim, K.; Diepeveen, D.; et al. Does late maturity alpha-amylase impact wheat baking quality? Front. Plant Sci. 2018, 9, 1356. [Google Scholar] [CrossRef]
- Neoh, G.K.; Dieters, M.J.; Tao, K.; Fox, G.P.; Nguyen, P.T.; Gilbert, R.G. Late-maturity alpha-amylase in wheat (Triticum aestivum) and its impact on fresh white sauce qualities. Foods 2021, 10, 201. [Google Scholar] [CrossRef]
- Ross, A.S.; Walker, C.E.; Booth, R.I.; Orth, R.A.; Wrigley, C.W. The rapid visco-analyzer—A new technique for the estimation of sprout damage. Cereal Foods World 1987, 32, 827–829. [Google Scholar]
- McKie, V.A.; McCleary, B.V. A rapid, automated method for measuring α-amylase in pre-harvest sprouted (sprout damaged) wheat. J. Cereal Sci. 2015, 64, 70–75. [Google Scholar] [CrossRef]
- Hauvermale, A.L.; Parveen, R.S.; Harris, T.J.; Tuttle, K.M.; Mikhaylenko, G.; Nair, S.; McCubbin, A.G.; Pumphrey, M.O.; Steber, C.M. Streamlined alpha-amylase assays for wheat preharvest sprouting and late maturity alpha-amylase detection. Agrosyst. Geosci. Environ. 2023, 6, e20327. [Google Scholar] [CrossRef]
- McCleary, B.V.; Sheehan, H. Measurement of cereal alpha-amylase: A new assay procedure. J. Cereal Sci. 1987, 6, 237–251. [Google Scholar] [CrossRef]
- Verity, J.C.K.; Hac, L.; Skerritt, J.H. Development of a field enzyme-linked immunosorbent assay (ELISA) for detection of α-amylase in preharvest-sprouted wheat. Cereal Chem. 1999, 76, 673–681. [Google Scholar] [CrossRef]
- Skerritt, J.H.; Heywood, R.H. A five-minute field test for on-farm detection of pre-harvest sprouting in wheat. Crop Sci. 2000, 40, 742–756. [Google Scholar] [CrossRef]
- Cheng, C.R.; Oldach, K.; Mrva, K.; Mares, D. Analysis of high pI α-Amy-1 gene family members expressed in late maturity α-amylase in wheat (Triticum aestivum L.). Mol. Breed. 2014, 33, 519–529. [Google Scholar] [CrossRef][Green Version]
- International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef]
- Ritchie, S.; Gilroy, S. The regulation of the response of barley aleurone protoplasts to GA and ABA by elements of the phospholipid signal transduction pathway. Plant Physiol. 1996, 111, 701. [Google Scholar]
- Zhang, Q.; Li, C. Comparisons of Copy Number, Genomic Structure, and Conserved Motifs for α-Amylase Genes from Barley, Rice, and Wheat. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.; Deng, G.; Liang, J.; Zhang, H.; Li, Q.; Pan, Z.; Yu, M.; Long, H. Structural organization and functional divergence of high isoelectric point α-amylase genes in bread wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.). BMC Genet. 2019, 20, 25. [Google Scholar] [CrossRef]
- Farrell, A.D.; Kettlewell, P.S. The effect of temperature shock and grain morphology on alpha-amylase in developing wheat grain. Ann. Bot. 2008, 102, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Barrero, J.M.; Porfirio, L.; Hughes, T.; Chen, J.; Dillon, S.; Gubler, F.; Ral, J.P.F. Evaluation of the impact of heat on wheat dormancy, late maturity α-amylase and grain size under controlled conditions in diverse germplasm. Sci. Rep. 2020, 10, 17800. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Role of abscisic acid signaling in drought tolerance and preharvest sprouting under climate change. Clim. Change Plant Abiotic Stress Toler. 2013, 20, 521–554. [Google Scholar]
- Song, Y.; Linderholm, H.W.; Wang, C.; Tian, J.; Huo, Z.; Gao, P.; Song, Y.; Guo, A. The influence of excess precipitation on winter wheat under climate change in China from 1961 to 2017. Sci. Total Environ. 2019, 690, 189–196. [Google Scholar] [CrossRef]
- Steber, C.M. Avoiding problems in wheat with low falling numbers. Crops Soils 2017, 50, 22–25. [Google Scholar] [CrossRef]
- Patwa, N.; Penning, B.W. Genetics of a diverse soft winter wheat population for pre-harvest sprouting, agronomic, and flour quality traits. Front. Plant Sci. 2023, 14, 113780. [Google Scholar] [CrossRef]
- Ningwei, Q.; Arslan, J. ‘China’s Wheat Growers Face Disaster after Heavy Rain Batters Crop. Reuters. Available online: https://www.reuters.com/markets/commodities/chinas-wheat-growers-face-disaster-after-heavy-rain-batters-crop-2023-06-07/ (accessed on 7 June 2023).
- Chang, C.; Zhang, H.; Lu, J.; Si, H.; Ma, C. Genetic Improvement of Wheat with Pre-Harvest Sprouting Resistance in China. Genes 2023, 14, 837. [Google Scholar] [CrossRef]
- Farrell, A.D.; Kettlewell, P.S.; Simmonds, J.; Flintham, J.E.; Snape, J.W.; Werner, P.; Jack, P.L. Control of late maturity alpha-amylase in wheat by the dwarfing gene Rht-D1b and genes on the 1B/1R translocation. Mol. Breed. 2013, 32, 425–436. [Google Scholar] [CrossRef]
- Derkx, A.; Baumann, U.; Cheong, J.; Mrva, K.; Sharma, N.; Pallotta, M.; Mares, D. A major locus on wheat chromosome 7B associated with late-maturity α-amylase encodes a putative ent-copalyl diphosphate synthase. Front. Plant Sci. 2021, 12, 637685. [Google Scholar] [CrossRef] [PubMed]
- Kondhare, K.R.; Kettlewell, P.S.; Farrell, A.D.; Hedden, P.; Monaghan, J.M. Effects of exogenous abscisic acid and gibberellic acid on pre-maturity α-amylase formation in wheat grains. Euphytica 2012, 188, 51–60. [Google Scholar] [CrossRef]
- Kondhare, K.R.; Kettlewell, P.S.; Farrell, A.D.; Hedden, P.; Monaghan, J.M. The role of sensitivity to abscisic acid and gibberellin in pre-maturity α-amylase formation in wheat grains. J. Cereal Sci. 2013, 58, 472–478. [Google Scholar] [CrossRef]
- Kondhare, K.R.; Hedden, P.; Kettlewell, P.S.; Farrell, A.D.; Monaghan, J.M. Use of the hormone-biosynthesis inhibitors fluridone and paclobutrazol to determine the effects of altered abscisic acid and gibberellin levels on pre-maturity α-amylase formation in wheat grains. J. Cereal Sci. 2014, 60, 210–216. [Google Scholar] [CrossRef]
- Kondhare, K.R.; Farrell, A.D.; Kettlewell, P.S.; Hedden, P.; Monaghan, J.M. Pre-maturity α-amylase in wheat: The role of abscisic acid and gibberellins. J. Cereal Sci. 2015, 63, 95–108. [Google Scholar] [CrossRef]
- Mares, D.; Derkx, A.; Cheong, J.; Zaharia, I.; Asenstorfer, R.; Mrva, K. Gibberellins in developing wheat grains and their relationship to late maturity α-amylase (LMA). Planta 2022, 255, 119. [Google Scholar] [CrossRef]
- Liu, C.; Tuttle, K.M.; Campbell, K.A.G.; Pumphrey, M.O.; Steber, C.M. Investigating conditions that induce late maturity alpha-amylase (LMA) using Northwestern US spring wheat (Triticum aestivum L.). Seed Sci. Res. 2021, 31, 169–177. [Google Scholar] [CrossRef]
- Carter, A.H.; Jones, S.S.; Balow, K.A.; Shelton, G.B.; Burke, A.B.; Lyon, S.; Morris, C.F. Registration of ‘Jasper’ soft white winter wheat. J. Plant Regist. 2017, 11, 263–268. [Google Scholar] [CrossRef]
- Carter, A.H.; Jones, S.S.; Lyon, S.R.; Balow, K.A.; Shelton, G.B.; Higginbotham, R.W.; Chen, X.M.; Engle, D.A.; Baik, B.; Guy, S.O.; et al. Registration of ‘Otto’ wheat. J. Plant Regist. 2013, 7, 195–200. [Google Scholar] [CrossRef]
- Jones, S.S.; Murray, T.D.; Lyon, S.R.; Morris, C.F.; Line, R.F. Registration of ‘Bruehl’ wheat (Registrations of Cultivars). Crop Sci. 2001, 41, 2006–2008. [Google Scholar] [CrossRef]
- Tan, M.K. Molecular Marker Development for LMA Determinant in Spica and Cranbrook; Value Added Wheat CRC: Sydney, Australia, 2004. [Google Scholar]
- Mares, D.J.; Mrva, K. Mapping quantitative trait loci associated with variation in grain dormancy in Australian wheat. Aust. J. Agric. Res. 2001, 52, 1257–1265. [Google Scholar] [CrossRef]
- Oñate-Sánchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 2008, 1, 93. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.K.; Steber, C.M. Transcriptional mechanisms associated with seed dormancy and dormancy loss in the gibberellin-insensitive sly1-2 mutant of Arabidopsis thaliana. PLoS ONE 2017, 12, e0179143. [Google Scholar] [CrossRef] [PubMed]
- Zoller, M.J.; Smith, M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1983; Volume 100, pp. 468–500. [Google Scholar]
- Tabor, S.; Richardson, C.C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. USA 1987, 84, 4767–4771. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, K.M.; Martinez, S.A.; Schramm, E.C.; Takebayashi, Y.; Seo, M.; Steber, C.M. Grain dormancy loss is associated with changes in ABA and GA sensitivity and hormone accumulation in bread wheat, Triticum aestivum (L.). Seed Sci. Res. 2015, 25, 179–193. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hauvermale, A.L.; Matzke, C.; Bohaliga, G.; Pumphrey, M.O.; Steber, C.M.; McCubbin, A.G. Development of Novel Monoclonal Antibodies to Wheat Alpha-Amylases Associated with Grain Quality Problems That Are Increasing with Climate Change. Plants 2023, 12, 3798. https://doi.org/10.3390/plants12223798
Hauvermale AL, Matzke C, Bohaliga G, Pumphrey MO, Steber CM, McCubbin AG. Development of Novel Monoclonal Antibodies to Wheat Alpha-Amylases Associated with Grain Quality Problems That Are Increasing with Climate Change. Plants. 2023; 12(22):3798. https://doi.org/10.3390/plants12223798
Chicago/Turabian StyleHauvermale, Amber L., Courtney Matzke, Gamila Bohaliga, Mike O. Pumphrey, Camille M. Steber, and Andrew G. McCubbin. 2023. "Development of Novel Monoclonal Antibodies to Wheat Alpha-Amylases Associated with Grain Quality Problems That Are Increasing with Climate Change" Plants 12, no. 22: 3798. https://doi.org/10.3390/plants12223798
APA StyleHauvermale, A. L., Matzke, C., Bohaliga, G., Pumphrey, M. O., Steber, C. M., & McCubbin, A. G. (2023). Development of Novel Monoclonal Antibodies to Wheat Alpha-Amylases Associated with Grain Quality Problems That Are Increasing with Climate Change. Plants, 12(22), 3798. https://doi.org/10.3390/plants12223798






