QTL Verification and Candidate Gene Screening of Fiber Quality and Lint Percentage in the Secondary Segregating Population of Gossypium hirsutum
Abstract
1. Introduction
2. Results and Analysis
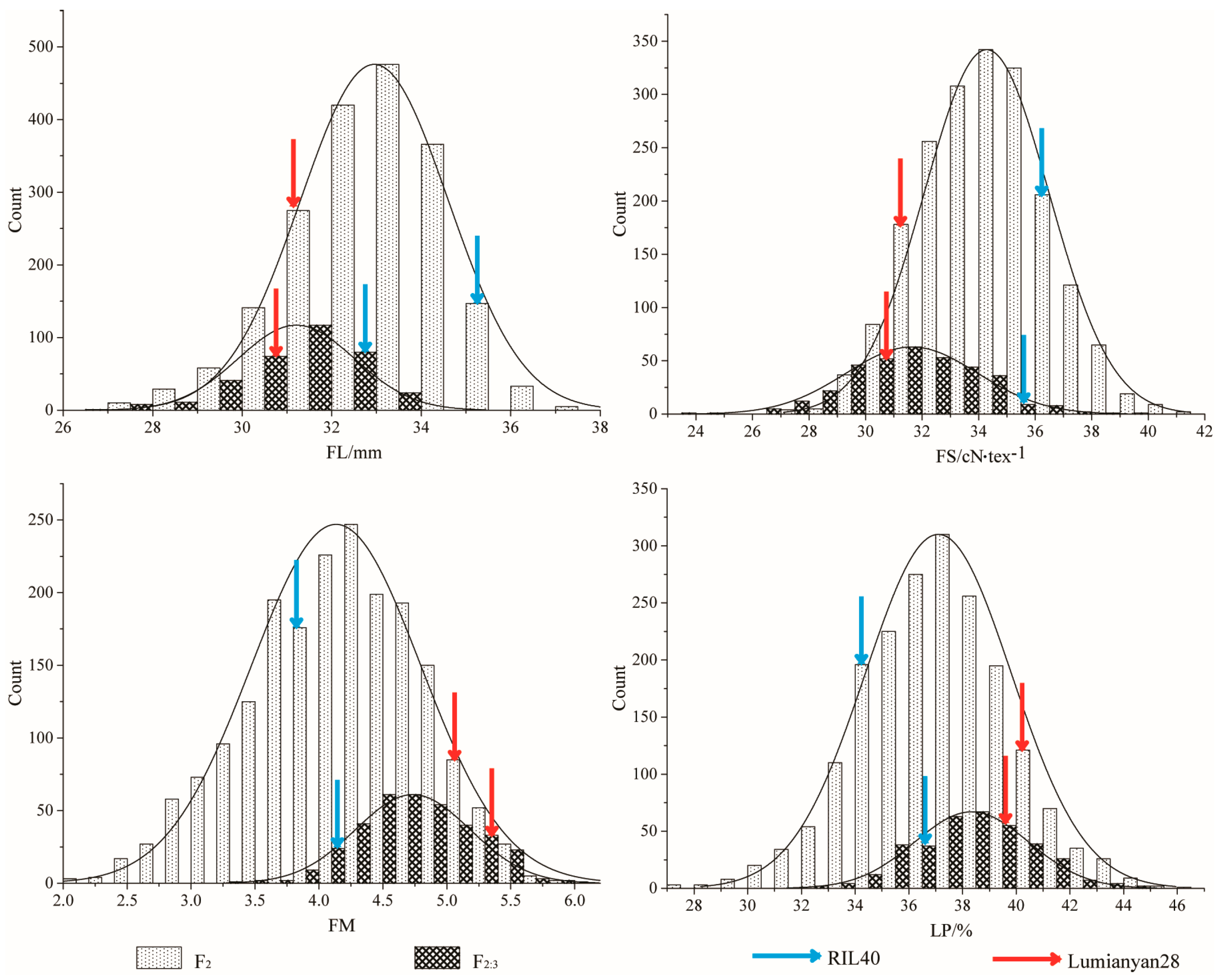
2.1. Phenotypic Statistics of Fiber Quality and Yield Traits of the Experimental Materials
2.2. Linkage Map Construction and QTL Mapping of the Target Loci
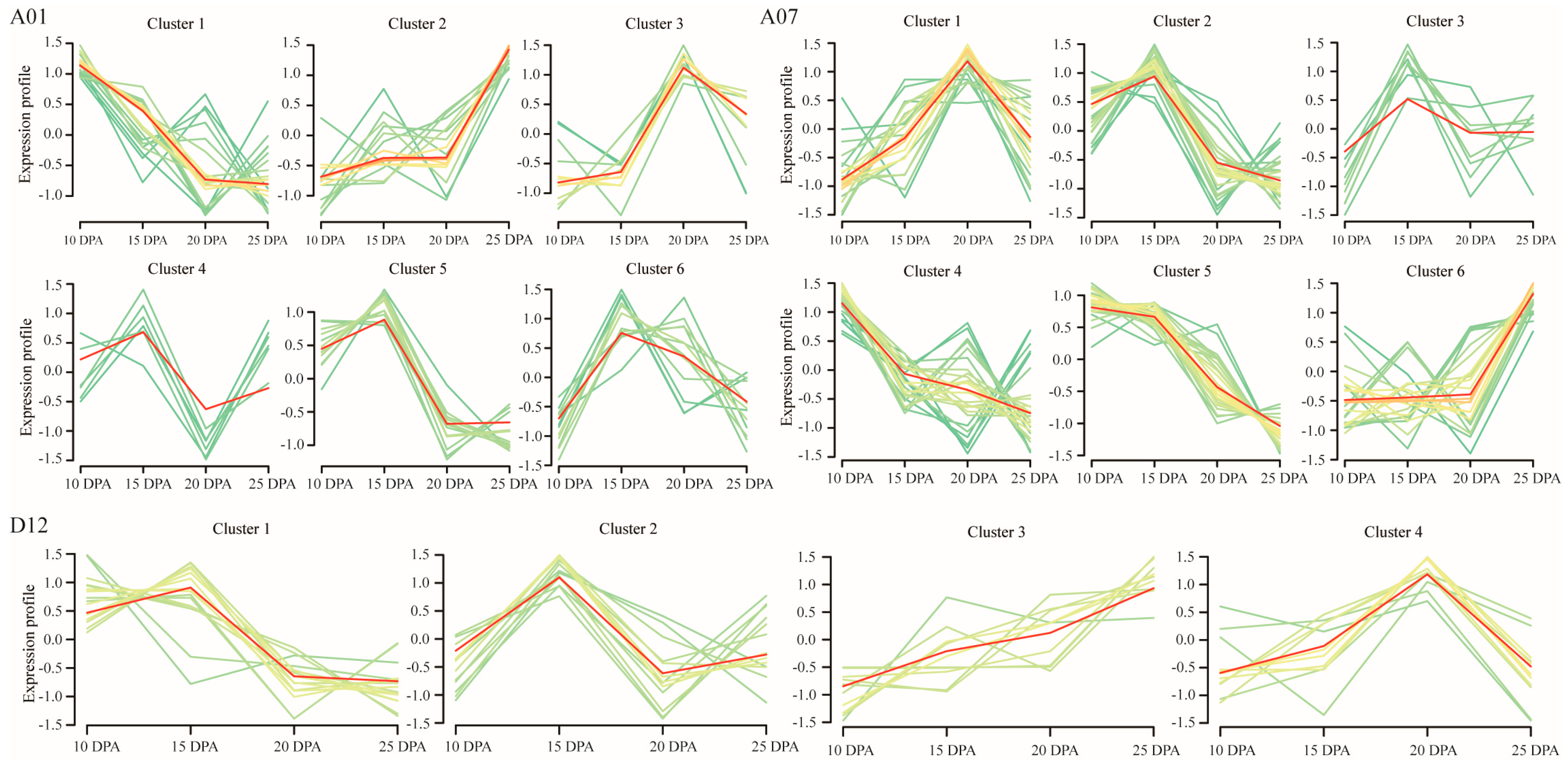
2.3. Screening and Analysis of DEGs from Genes within the QTL Intervals
3. Materials and Methods
3.1. Plant Materials and Phenotypic Measurement
3.2. Maker Development for Genotyping of the Secondary Population
3.3. QTL Mapping
3.4. Candidate Gene Screening Based on DEG Analysis between L28 and RIL40
3.5. qRT-PCR Experiment
4. Discussion
4.1. MAS Strategy in Breeding Practice
4.2. Function Validations of Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, Z.; Wang, X.; Liu, Z.; Gu, Q.; Zhang, Y.; Li, Z.; Ke, H.; Yang, J.; Wu, J.; Wu, L.; et al. Genome-wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol. J. 2017, 15, 982–996. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Wang, Q.; Hu, Y.; Jia, Y.; Chen, J.; Liu, B.; Zhang, Z.; Guan, X.; Chen, S.; Zhou, B.; et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 2017, 49, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Zeng, L.; Thyssen, G.N.; Delhom, C.D.; Kim, H.J.; Li, P.; Fang, D.D. Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor. Appl. Genet. 2016, 129, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, B.; Zhao, N.; Kong, J.; Hua, J. Fiber Quality Improvement in Upland Cotton (Gossypium hirsutum L.): Quantitative Trait Loci Mapping and Marker Assisted Selection Application. Front. Plant Sci. 2019, 10, 1585. [Google Scholar] [CrossRef]
- Song, C.; Li, W.; Pei, X.; Liu, Y.; Ren, Z.; He, K.; Zhang, F.; Sun, K.; Zhou, X.; Ma, X.; et al. Dissection of the genetic variation and candidate genes of lint percentage by a genome-wide association study in upland cotton. Theor. Appl. Genet. 2019, 132, 1991–2002. [Google Scholar] [CrossRef]
- Huang, G.; Zhu, Y.X. Breeding cotton with superior fiber quality: Identification and utilization of multiple elite loci and exotic genetic resources. Sci. China Life Sci. 2021, 64, 1197–1198. [Google Scholar] [CrossRef]
- Said, J.I.; Song, M.; Wang, H.; Lin, Z.; Zhang, X.; Fang, D.D.; Zhang, J. A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol. Genet. Genom. 2015, 290, 1003–1025. [Google Scholar] [CrossRef]
- Pei, W.; Song, J.; Wang, W.; Ma, J.; Jia, B.; Wu, L.; Wu, M.; Chen, Q.; Qin, Q.; Zhu, H.; et al. Quantitative Trait Locus Analysis and Identification of Candidate Genes for Micronaire in an Interspecific Backcross Inbred Line Population of Gossypium hirsutum × Gossypium barbadense. Front. Plant Sci. 2021, 12, 763016. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef]
- Kushanov, F.N.; Turaev, O.S.; Ernazarova, D.K.; Gapparov, B.M.; Oripova, B.B.; Kudratova, M.K.; Rafieva, F.U.; Khalikov, K.K.; Erjigitov, D.S.; Khidirov, M.T.; et al. Genetic Diversity, QTL Mapping, and Marker-Assisted Selection Technology in Cotton (Gossypium spp.). Front. Plant Sci. 2021, 12, 779386. [Google Scholar] [CrossRef]
- Chavhan, R.; Sable, S.; Narwade, A.; Hinge, V.; Kalbande, B.; Mukherjee, A.; Chakrabarty, P.; Kadam, U.S. Multiplex molecular marker-assisted analysis of significant pathogens of cotton (Gossypium sp.). Biocatal. Agric. Biotechnol. 2023, 47, 102557. [Google Scholar] [CrossRef]
- Zang, Y.; Hu, Y.; Xu, C.; Wu, S.; Wang, Y.; Ning, Z.; Han, Z.; Si, Z.; Shen, W.; Zhang, Y.; et al. GhUBX controlling helical growth results in production of stronger cotton fiber. iScience 2021, 24, 102930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shen, C.; Zhu, D.; Le, Y.; Wang, N.; Li, Y.; Zhang, X.; Lin, Z. Fine-mapping and candidate gene analysis of qFL-c10-1 controlling fiber length in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2022, 135, 4483–4494. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hou, J.; Chen, L.; Li, Q.; Fang, X.; Wang, J.; Hao, Y.; Yang, P.; Wang, W.; Zhang, D.; et al. Natural variation of GhSI7 increases seed index in cotton. Theor. Appl. Genet. 2022, 135, 3661–3672. [Google Scholar] [CrossRef]
- Fang, X.; Liu, X.; Wang, X.; Wang, W.; Liu, D.; Zhang, J.; Liu, D.; Teng, Z.; Tan, Z.; Liu, F.; et al. Fine-mapping qFS07.1 controlling fiber strength in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2017, 130, 795–806. [Google Scholar] [CrossRef]
- Liu, R.; Gong, J.; Xiao, X.; Zhang, Z.; Li, J.; Liu, A.; Lu, Q.; Shang, H.; Shi, Y.; Ge, Q.; et al. GWAS Analysis and QTL Identification of Fiber Quality Traits and Yield Components in Upland Cotton Using Enriched High-Density SNP Markers. Front. Plant Sci. 2018, 9, 1067. [Google Scholar] [CrossRef]
- Paterson, A.H.; Brubaker, C.L.; Wendel, J.F. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Report. 1993, 11, 122–127. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. In Selected Works in Mathematics and Statistics; Springer: New Delhi, India, 2016; pp. 125–130. [Google Scholar]
- Wang, S.; Basten, C.; Zeng, Z. Windows QTL Cartographer v2.5. 2012. Available online: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 1 August 2012).
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 8 March 2016).
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Chen, J.; Zhang, Z.; Liu, F.; Li, J.; Zhao, T.; Hu, Y.; Zhang, T.; Fang, L. COTTONOMICS: A comprehensive cotton multi-omics database. Database 2022, 2022, baac080. [Google Scholar] [CrossRef]
- Kumar, L.; E Futschik, M. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Jamshed, M.; Shi, Y.; Liu, A.; Gong, J.; Wang, S.; Zhang, J.; Sun, F.; Jia, F.; et al. Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population. Plant Biotechnol. J. 2020, 18, 239–253. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, A.; Li, J.; Zhang, J.; Li, S.; Zhang, J.; Ma, L.; He, R.; Song, W.; Guo, L.; et al. Examining two sets of introgression lines across multiple environments reveals background-independent and stably expressed quantitative trait loci of fiber quality in cotton. Theor. Appl. Genet. 2020, 133, 2075–2093. [Google Scholar] [CrossRef]
- Li, L.; Lu, K.; Chen, Z.; Mu, T.; Hu, Z.; Li, X. Dominance, Overdominance and Epistasis Condition the Heterosis in Two Heterotic Rice Hybrids. Genetics 2008, 180, 1725–1742. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.-D.; Zhang, J.-H.; Wang, S.-F.; Gong, W.-K.; Shi, Y.-Z.; Liu, A.-Y.; Li, J.-W.; Gong, J.-W.; Shang, H.-H.; Yuan, Y.-L. QTL mapping for fiber quality traits across multiple generations and environments in upland cotton. Mol. Breed. 2012, 30, 569–582. [Google Scholar] [CrossRef]
- Kong, W.; Deng, X.; Yang, J.; Zhang, C.; Sun, T.; Ji, W.; Zhong, H.; Fu, X.; Li, Y. High-resolution bin-based linkage mapping uncovers the genetic architecture and heterosis-related loci of plant height in indica–japonica derived populations. Plant J. 2022, 110, 814–827. [Google Scholar] [CrossRef]
- Yang, P.; Sun, X.; Liu, X.; Wang, W.; Hao, Y.; Chen, L.; Liu, J.; He, H.; Zhang, T.; Bao, W.; et al. Identification of Candidate Genes for Lint Percentage and Fiber Quality Through QTL Mapping and Transcriptome Analysis in an Allotetraploid Interspecific Cotton CSSLs Population. Front. Plant Sci. 2022, 13, 882051. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Gong, J.; Zhang, J.; Zhang, Z.; Shi, Y.; Li, J.; Liu, A.; Gong, W.; Ge, Q.; Deng, X. Quantitative trait loci and transcriptome analysis reveal genetic basis of fiber quality traits in CCRI70 RIL population of Gossypium hirsutum. Front. Plant Sci. 2021, 12, 753755. [Google Scholar] [CrossRef]
- Zhang, K.; Kuraparthy, V.; Fang, H.; Zhu, L.; Sood, S.; Jones, D.C. High-density linkage map construction and QTL analyses for fiber quality, yield and morphological traits using CottonSNP63K array in upland cotton (Gossypium hirsutum L.). BMC Genom. 2019, 20, 889. [Google Scholar] [CrossRef]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G.; et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, J.; Chen, Y.; Zhang, C.; Gong, J.; Song, Z.; Zhou, J.; Wang, J.; Zhao, C.; Jiao, M.; et al. Identification of candidate genes for key fibre-related QTLs and derivation of favourable alleles in Gossypium hirsutum recombinant inbred lines with G. barbadense introgressions. Plant Biotechnol. J. 2020, 18, 707–720. [Google Scholar] [CrossRef]
- Zhou, J.R.; Li, J.; Lin, J.X.; Xu, H.M.; Chu, N.; Wang, Q.N.; Gao, S.J. Genome-wide characterization of cys-tathionine-β-synthase domain-containing proteins in sugarcane reveals their role in defense responses under multiple stressors. Front. Plant Sci. 2022, 13, 985653. [Google Scholar] [CrossRef]
- Shin, J.S.; So, W.M.; Kim, S.Y.; Noh, M.; Hyoung, S.; Yoo, K.S.; Shin, J.S. CBSX3-Trxo-2 regulates ROS generation of mitochondrial complex II (succinate dehydrogenase) in Arabidopsis. Plant Sci. 2020, 294, 110458. [Google Scholar] [CrossRef]
- Yoo, K.S.; Ok, S.H.; Jeong, B.C.; Jung, K.W.; Cui, M.H.; Hyoung, S.; Lee, M.R.; Song, H.K.; Shin, J.S. Single cystathionine β-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell 2011, 23, 3577–3594. [Google Scholar] [CrossRef]
- Jung, K.W.; Kim, Y.Y.; Yoo, K.S.; Ok, S.H.; Cui, M.H.; Jeong, B.C.; Yoo, S.D.; Jeung, J.U.; Shin, J.S. A cystathionine-β-synthase domain-containing protein, CBSX2, regulates endothecial secondary cell wall thickening in anther development. Plant Cell Physiol. 2013, 54, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Tomar, S.; Subba, A.; Bala, M.; Singh, A.K.; Pareek, A.; Singla-Pareek, S.L. Genetic Conservation of CBS Domain Containing Protein Family in Oryza Species and Their Association with Abiotic Stress Responses. Int. J. Mol. Sci. 2022, 23, 1687. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Li, Y.; Li, F.; Wang, Z. Genome-wide characterization and expression analysis of cystathionine β-synthase genes in plant development and abiotic stresses of cotton (Gossypium spp.). Int. J. Biol. Macromol. 2021, 193, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Negi, J.; Young, J.; Israelsson, M.; Schroeder, J.I.; Iba, K. Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat. Cell Biol. 2006, 8, 391–397. [Google Scholar] [CrossRef]
- Matrosova, A.; Bogireddi, H.; Mateo-Peñas, A.; Hashimoto-Sugimoto, M.; Iba, K.; Schroeder, J.I.; Israelsson-Nordström, M. The HT1 protein kinase is essential for red light-induced stomatal opening and genetically interacts with OST1 in red light and CO2-induced stomatal movement responses. New Phytol. 2015, 208, 1126–1137. [Google Scholar] [CrossRef]
- Hõrak, H.; Sierla, M.; Tõldsepp, K.; Wang, C.; Wang, Y.S.; Nuhkat, M.; Valk, E.; Pechter, P.; Merilo, E.; Salojärvi, J.; et al. A Dominant Mutation in the HT1 Kinase Uncovers Roles of MAP Kinases and GHR1 in CO2-Induced Stomatal Closure. Plant Cell 2016, 28, 2493–2509. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, P.; Zhang, X.; Ren, D.; Yang, S. BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 2011, 67, 1081–1093. [Google Scholar] [CrossRef]
- Wierzba, M.P.; Tax, F.E. An Allelic Series of bak1 Mutations Differentially Alter bir1 Cell Death, Immune Response, Growth, and Root Development Phenotypes in Arabidopsis thaliana. Genetics 2016, 202, 689–702. [Google Scholar] [CrossRef][Green Version]
- Halter, T.; Imkampe, J.; Mazzotta, S.; Wierzba, M.; Postel, S.; Bücherl, C.; Kiefer, C.; Stahl, M.; Chinchilla, D.; Wang, X.; et al. The Leucine-Rich Repeat Receptor Kinase BIR2 Is a Negative Regulator of BAK1 in Plant Immunity. Curr. Biol. 2014, 24, 134–143. [Google Scholar] [CrossRef]
- Großeholz, R.; Feldman-Salit, A.; Wanke, F.; Schulze, S.; Glöckner, N.; Kemmerling, B.; Harter, K.; Kummer, U. Specifying the role of BAK1-interacting receptor-like kinase 3 in brassinosteroid signaling. J. Integr. Plant Biol. 2020, 62, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, C.; Ren, H.; Zhao, T.; Li, Q.; Wang, S.; Zhang, Y.; Xiao, F.; Wang, X. BAK1 Mediates Light Intensity to Phosphorylate and Activate Catalases to Regulate Plant Growth and Development. Int. J. Mol. Sci. 2020, 21, 1437. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Jourquin, J.; Njo, M.F.; Nguyen, L.; Beeckman, T.; Fernandez, A.I. The Phloem Intercalated with Xylem-Correlated 3 Receptor-Like Kinase Constitutively Interacts with Brassinosteroid Insensitive 1-Associated Receptor Kinase 1 and Is Involved in Vascular Development in Arabidopsis. Front. Plant Sci. 2021, 12, 706633. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Yang, D.; Ha, Y.; Lee, J.Y.; Kim, J.Y.; Oh, M.H.; Nam, K.H. Open stomata 1 exhibits dual serine/threonine and tyrosine kinase activity in regulating abscisic acid signaling. J. Exp. Bot. 2021, 72, 5494–5507. [Google Scholar] [CrossRef]
- Thomas, B.R.; Romero, G.O.; Nevins, D.J.; Rodriguez, R.L. New perspectives on the endo-beta-glucanases of glycosyl hydrolase Family 17. Int. J. Biol. Macromol. 2000, 27, 139–144. [Google Scholar] [CrossRef]
- Santos, C.; Martins, D.C.; González-Bernal, M.J.; Rubiales, D.; Vaz Patto, M.C. Integrating Phenotypic and Gene Expression Linkage Mapping to Dissect Rust Resistance in Chickling Pea. Front. Plant Sci. 2022, 13, 837613. [Google Scholar] [CrossRef]
- Jung, J.Y.; Min, C.W.; Jang, J.W.; Gupta, R.; Kim, J.H.; Kim, Y.H.; Cho, S.W.; Song, Y.H.; Jo, I.H.; Rakwal, R.; et al. Proteomic Analysis Reveals a Critical Role of the Glycosyl Hydrolase 17 Protein in Panax ginseng Leaves under Salt Stress. Int. J. Mol. Sci. 2023, 24, 3693. [Google Scholar] [CrossRef]
- Pervaiz, T.; Liu, T.; Fang, X.; Ren, Y.; Li, X.; Liu, Z.; Fiaz, M.; Fang, J.; Shangguan, L. Identification of GH17 gene family in Vitis vinifera and expression analysis of GH17 under various adversities. Physiol. Mol. Biol. Plants An. Int. J. Funct. Plant Biol. 2021, 27, 1423–1436. [Google Scholar] [CrossRef]
- Felipe, J.E.L.; Lachica, J.A.P.; Dela Cueva, F.M.; Laurel, N.R.; Alcasid, C.E.; Sison, M.L.J.; Valencia, L.D.C.; Ocampo, E.T.M. Validation and molecular analysis of β-1,3-GLU2 SNP marker associated with resistance to Colletotrichum gloeosporioides in mango (Mangifera indica L.). Physiol. Mol. Plant Pathol. 2022, 118, 101804. [Google Scholar] [CrossRef]
- Millet, N.; Latgé, J.-P.; Mouyna, I. Members of Glycosyl-Hydrolase Family 17 of A. fumigatus Differentially Affect Morphogenesis. J. Fungi 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Gě, Q.; Cūi, Y.; Lǐ, J.; Gōng, J.; Lú, Q.; Lǐ, P.; Shí, Y.; Shāng, H.; Liú, À.; Dèng, X.; et al. Disequilibrium evolution of the Fructose-1,6-bisphosphatase gene family leads to their functional biodiversity in Gossypium species. BMC Genom. 2020, 21, 379. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, A.; Gě, Q.; Zhāng, Q.; Lǐ, J.; Gōng, J.; Liú, R.; Shí, Y.; Shāng, H.; Liú, À.; Iqbal, M.S.; et al. Multi-responses of O-methyltransferase genes to salt stress and fiber development of Gossypium species. BMC Plant Biol. 2021, 21, 37. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, S.; Sun, D.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; Chen, Z. CONSTANS-Like 9 (OsCOL9) Interacts with Receptor for Activated C-Kinase 1(OsRACK1) to Regulate Blast Resistance through Salicylic Acid and Ethylene Signaling Pathways. PLoS ONE 2016, 11, e0166249. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gu, F.; Dong, S.; Liu, W.; Wang, H.; Chen, Z.; Wang, J. CONSTANS-like 9 (COL9) delays the flowering time in Oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 2016, 479, 173–178. [Google Scholar] [CrossRef]
- Qin, W.; Yu, Y.; Jin, Y.; Wang, X.; Liu, J.; Xi, J.; Li, Z.; Li, H.; Zhao, G.; Hu, W.; et al. Genome-Wide Analysis Elucidates the Role of CONSTANS-like Genes in Stress Responses of Cotton. Int. J. Mol. Sci. 2018, 19, 2658. [Google Scholar] [CrossRef]
- Stenmark, H.; Olkkonen, V.M. The Rab GTPase family. Genome Biol. 2001, 2, REVIEWS3007. [Google Scholar] [CrossRef]
- Briegas, B.; Corbacho, J.; Parra-Lobato, M.C.; Paredes, M.A.; Labrador, J.; Gallardo, M.; Gomez-Jimenez, M.C. Transcriptome and Hormone Analyses Revealed Insights into Hormonal and Vesicle Trafficking Regulation among Olea europaea Fruit Tissues in Late Development. Int. J. Mol. Sci. 2020, 21, 4819. [Google Scholar] [CrossRef]
- Lawson, T.; Lycett, G.W.; Mayes, S.; Ho, W.K.; Chin, C.F. Transcriptome-wide identification and characterization of the Rab GTPase family in mango. Mol. Biol. Rep. 2020, 47, 4183–4197. [Google Scholar] [CrossRef]
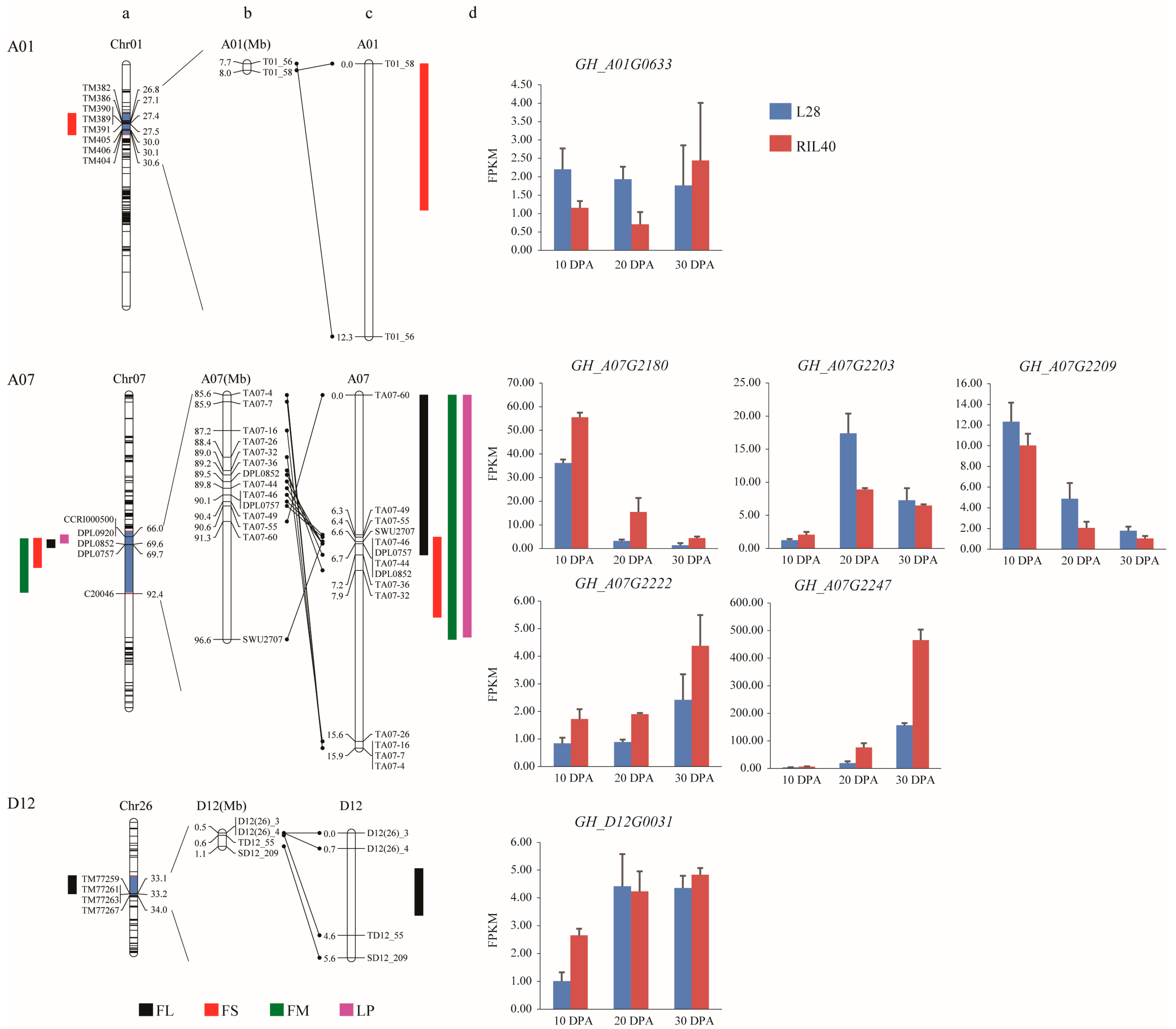

| Chromosome | QTL Compositions | Physical Position (Mb) | |||
|---|---|---|---|---|---|
| FS | FL | FM | LP | ||
| A01 | qFS-chr01-2 | 7.63–8.04 | |||
| A07 | qFS-chr07-2 | qFL-chr07-2 | qFM-chr07-1 | qLP-chr07-3 | 89.53–90.08 |
| D12 | qFL-chr26-1 | 0.46–0.52 | |||
| Parent Materials | Population | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Year | L28 | RIL40 | |AVDP| | Generation | Range | Min | Max | Average | SD | Skewness | Kurtosis |
| FL/mm | 2019 | 31.39 | 35.44 ** | 4.06 | F2 | 10.35 | 27.22 | 37.57 | 32.95 | 1.65 | −0.42 | 0.15 |
| 2020 | 30.90 | 32.94 * | 2.04 | F2:3 | 7.37 | 26.62 | 33.99 | 31.21 | 1.30 | −0.65 | 0.49 | |
| FS/cN∙tex−1 | 2019 | 31.28 | 36.49 ** | 5.20 | F2 | 14.30 | 27.02 | 41.32 | 34.28 | 2.21 | 0.06 | −0.13 |
| 2020 | 30.18 | 35.05 * | 4.87 | F2:3 | 15.68 | 23.61 | 39.29 | 31.64 | 2.34 | 0.01 | 0.32 | |
| FM | 2019 | 5.05 | 3.79 ** | 1.26 | F2 | 3.88 | 2.14 | 6.02 | 4.13 | 0.66 | −0.25 | −0.31 |
| 2020 | 5.37 | 4.01 * | 1.36 | F2:3 | 2.58 | 3.35 | 5.93 | 4.73 | 0.45 | −0.02 | −0.22 | |
| LP/% | 2019 | 40.21 | 34.20 ** | 6.01 | F2 | 19.20 | 27.45 | 46.65 | 37.10 | 2.72 | −0.05 | 0.28 |
| 2020 | 39.36 | 36.16 * | 3.20 | F2:3 | 11.86 | 32.97 | 44.83 | 38.34 | 2.12 | 0.09 | −0.14 | |
| Trait | Generation | FL/mm | FS/cN∙tex−1 | FM | LP/% | ||||
|---|---|---|---|---|---|---|---|---|---|
| F2 | F2:3 | F2 | F2:3 | F2 | F2:3 | F2 | F2:3 | ||
| FL/mm | F2 | 1 | |||||||
| F2:3 | 0.593 ** | 1 | |||||||
| FS/cN∙tex−1 | F2 | 0.252 ** | - | 1 | |||||
| F2:3 | - | 0.300 ** | 0.328 ** | 1 | |||||
| FM | F2 | −0.348 ** | - | −0.028 | - | 1 | |||
| F2:3 | - | −0.434** | - | 0.163 ** | 0.594 ** | 1 | |||
| LP/% | F2 | −0.351 ** | - | −0.189 ** | - | 0.508 ** | - | 1 | |
| F2:3 | - | −0.453** | - | −0.008 | - | 0.597 ** | 0.583 ** | 1 | |
| Chromosome | Trait | QTL | Generation | Position (cM) | Marker Interval | LOD | Additive | Dominant | R2/% | Physical Interval |
|---|---|---|---|---|---|---|---|---|---|---|
| A01 | FS | qFS-A01-1 | F2:3 | 1.01 | T01_58-T01_56 | 3.40 | 0.10 | −1.12 | 1.45 | 7.73–8.00 |
| A07 | FL | qFL-A07-1 | F2 | 6.31 | TA07-49-TA07-36 | 38.84 | 0.55 | 0.47 | 2.31 | 88.95–91.29 |
| F2:3 | 0.01 | TA07-60-TA07-32 | 14.98 | 0.51 | 0.70 | 8.79 | ||||
| FS | qFS-A07-1 | F2 | 8.01 | TA07-49-TA07-32 | 42.53 | 0.89 | 0.34 | 5.79 | 88.95–90.63 | |
| FM | qFM-A07-1 | F2 | 6.61 | TA07-49-TA07-36 | 71.33 | −0.35 | −0.09 | 11.34 | 88.95–91.29 | |
| F2:3 | 8.91 | TA07-60-TA07-32 | 29.85 | −0.29 | −0.04 | 31.87 | ||||
| LP | qLP-A07-1 | F2 | 6.61 | TA07-49-TA07-36 | 73.40 | −1.53 | −0.29 | 12.67 | 88.95–91.29 | |
| F2:3 | 8.91 | TA07-60-TA07-32 | 26.57 | −1.30 | −0.40 | 27.36 | ||||
| D12 | FL | qFL-D12-1 | F2 | 2.71 | D12(26)_3-TD12_55 | 4.44 | 0.17 | −1.26 | 0.03 | 0.48–0.59 |
| Gene ID | 10 DPA * | 20 DPA | 30 DPA | Gene Name | Arabidopsis ID | ArabDesc | |||
|---|---|---|---|---|---|---|---|---|---|
| FDR | Log2FC | FDR | Log2FC | FDR | Log2FC | ||||
| GH_A01G0633 | 0.01 | −1.05 | 0.01 | −1.51 | - | - | CBSX5 | AT5G53750 | CBS domain-containing protein |
| GH_A07G2180 | - | - | 0.00 | 2.20 | - | - | NA | AT3G13130 | transmembrane protein |
| GH_A07G2203 | - | - | 0.00 | −1.01 | - | - | COL9 | AT3G07650 | CONSTANS-like 9 |
| GH_A07G2209 | - | - | 0.01 | −1.29 | - | - | RABB1C | AT4G17170 | RAB GTPase homolog B1C |
| GH_A07G2222 | - | - | 0.00 | 1.05 | - | - | BIR1 | AT5G48380 | BAK1-interacting receptor-like kinase 1 |
| GH_A07G2247 | - | - | 0.00 | 1.94 | 0.00 | 1.33 | GHL17 | AT3G07320 | O-Glycosyl hydrolases family 17 protein |
| GH_D12G0031 | 0.00 | 1.27 | - | - | - | - | HT1 | AT3G22750 | Protein kinase superfamily protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Zhu, M.; Shi, Y.; Li, J.; Gong, J.; Xiao, X.; Chen, Q.; Yuan, Y.; Gong, W. QTL Verification and Candidate Gene Screening of Fiber Quality and Lint Percentage in the Secondary Segregating Population of Gossypium hirsutum. Plants 2023, 12, 3737. https://doi.org/10.3390/plants12213737
Liu R, Zhu M, Shi Y, Li J, Gong J, Xiao X, Chen Q, Yuan Y, Gong W. QTL Verification and Candidate Gene Screening of Fiber Quality and Lint Percentage in the Secondary Segregating Population of Gossypium hirsutum. Plants. 2023; 12(21):3737. https://doi.org/10.3390/plants12213737
Chicago/Turabian StyleLiu, Ruixian, Minghui Zhu, Yongqiang Shi, Junwen Li, Juwu Gong, Xianghui Xiao, Quanjia Chen, Youlu Yuan, and Wankui Gong. 2023. "QTL Verification and Candidate Gene Screening of Fiber Quality and Lint Percentage in the Secondary Segregating Population of Gossypium hirsutum" Plants 12, no. 21: 3737. https://doi.org/10.3390/plants12213737
APA StyleLiu, R., Zhu, M., Shi, Y., Li, J., Gong, J., Xiao, X., Chen, Q., Yuan, Y., & Gong, W. (2023). QTL Verification and Candidate Gene Screening of Fiber Quality and Lint Percentage in the Secondary Segregating Population of Gossypium hirsutum. Plants, 12(21), 3737. https://doi.org/10.3390/plants12213737







