An Open-Source Package for Thermal and Multispectral Image Analysis for Plants in Glasshouse
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiment Setup
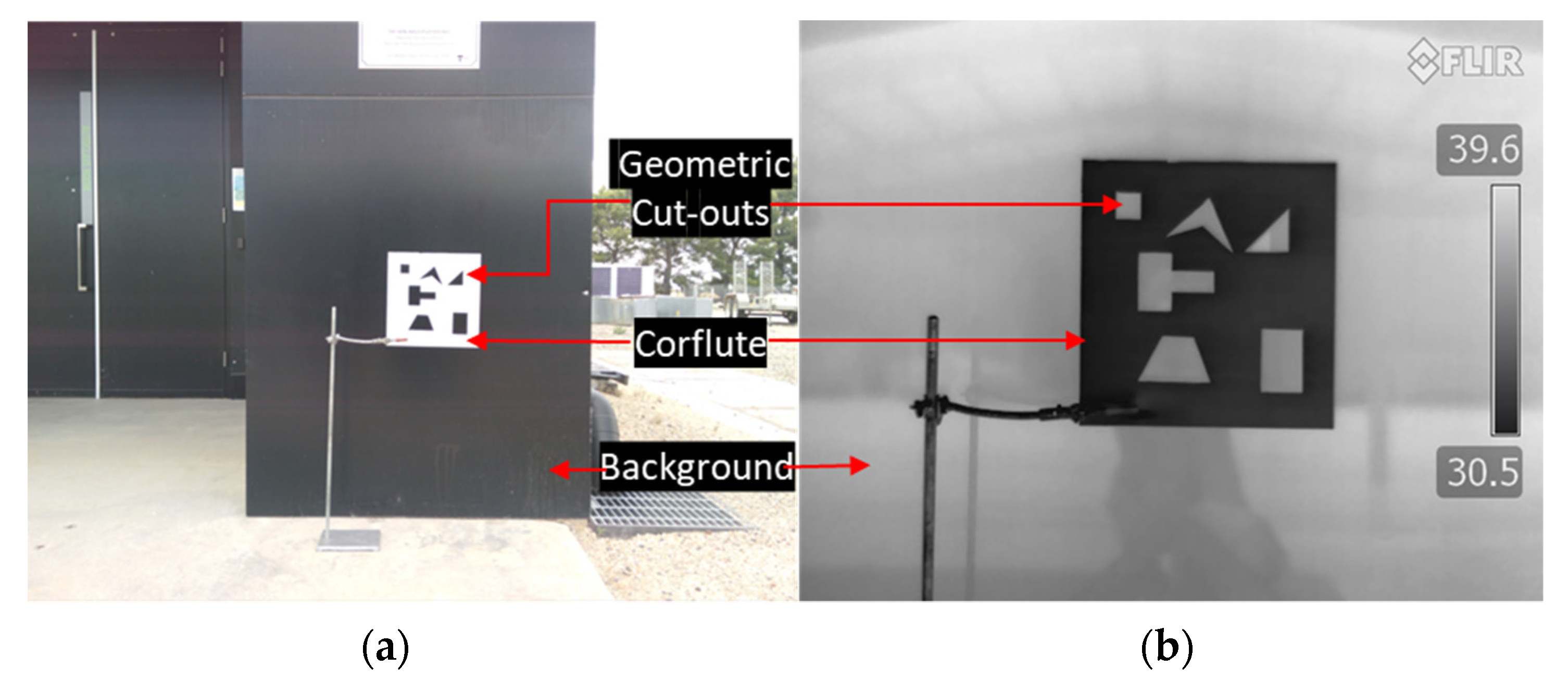
2.2. Integrated Sensor Platform and Imaging Setups
2.3. Image Processing
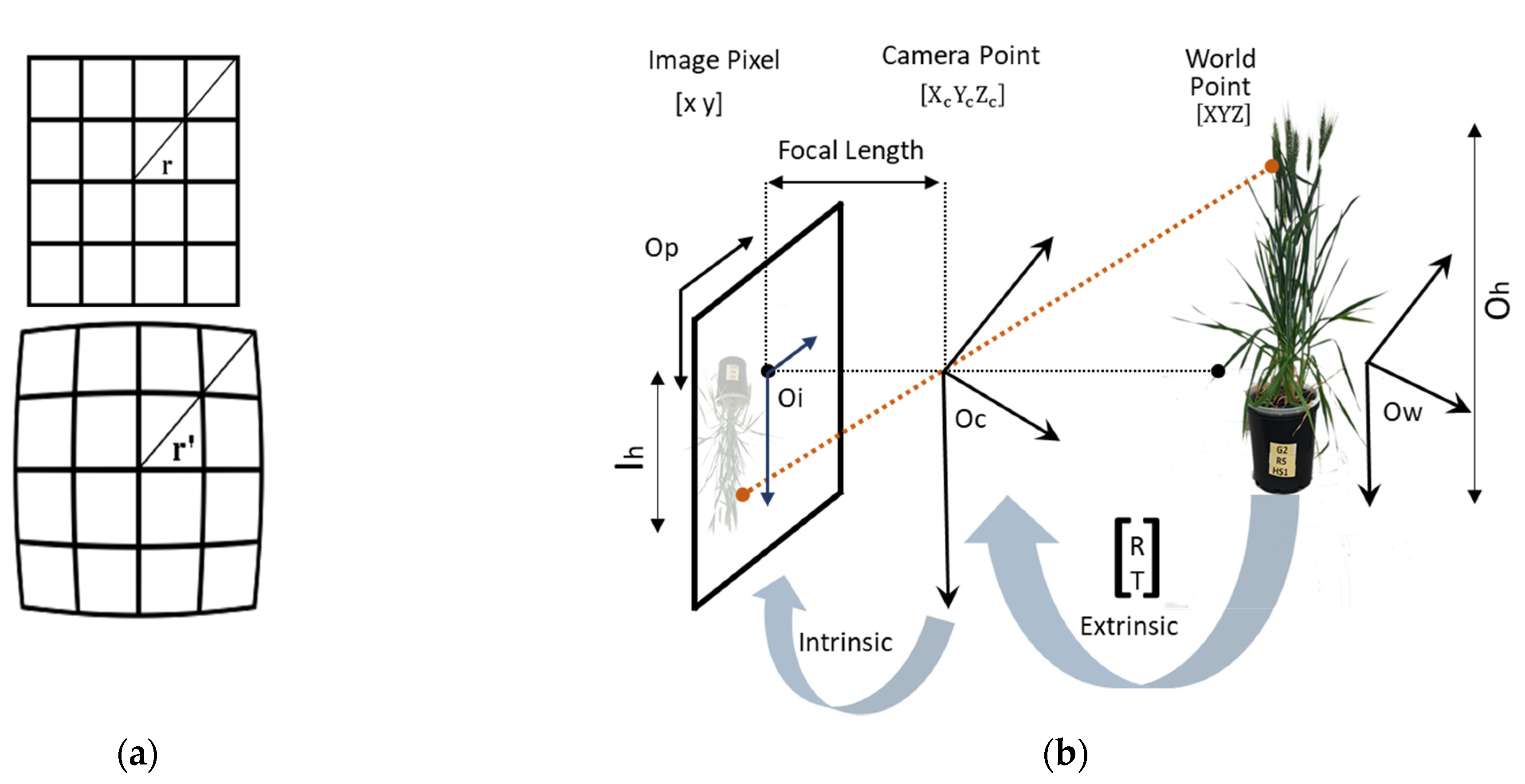
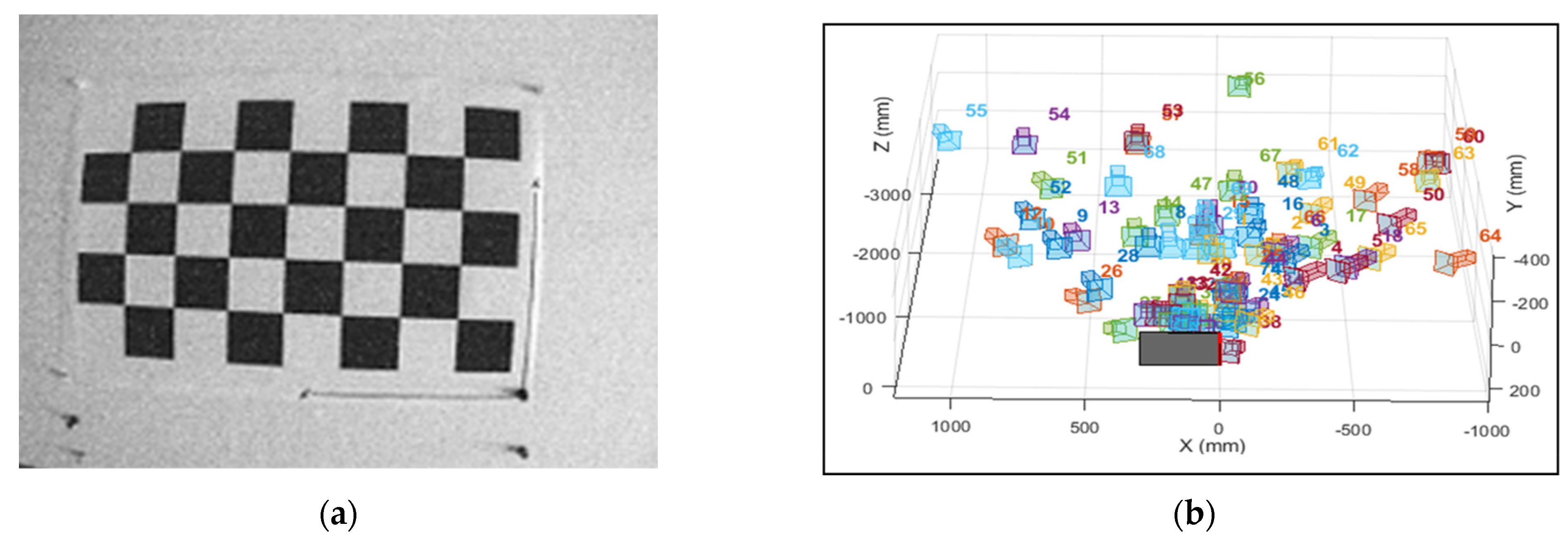
2.3.1. Correction of Radial Optical Distortions in Multispectral Images
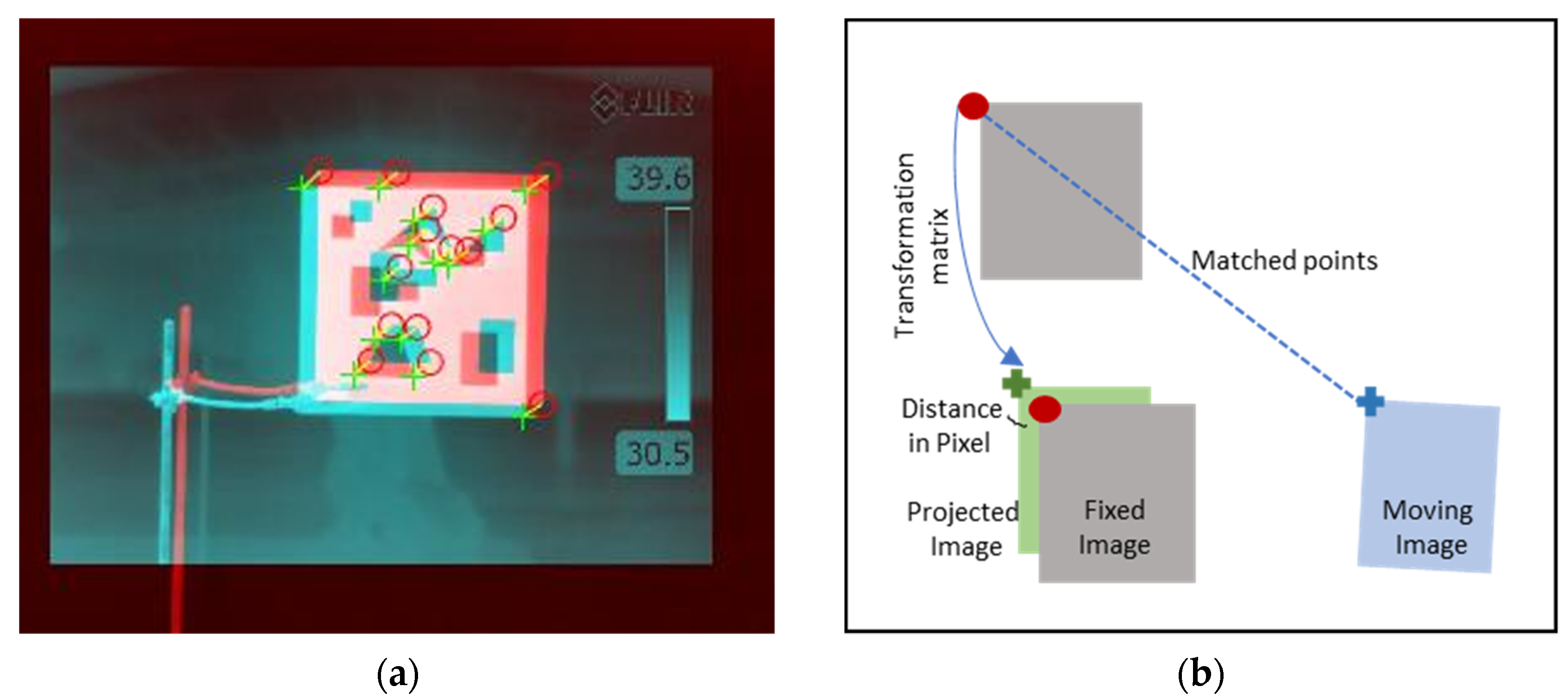
2.3.2. Registration of Optical, Multispectral, and Thermal Images
2.3.3. Radiometric Rescaling of Thermal Images
2.3.4. Gradient Removal and Illumination Correction of Multispectral Images
2.3.5. Segmentation to Separate the Plant from the Background
2.3.6. Vegetation Indices
3. Results
3.1. Correction of Radial Optical Distortion
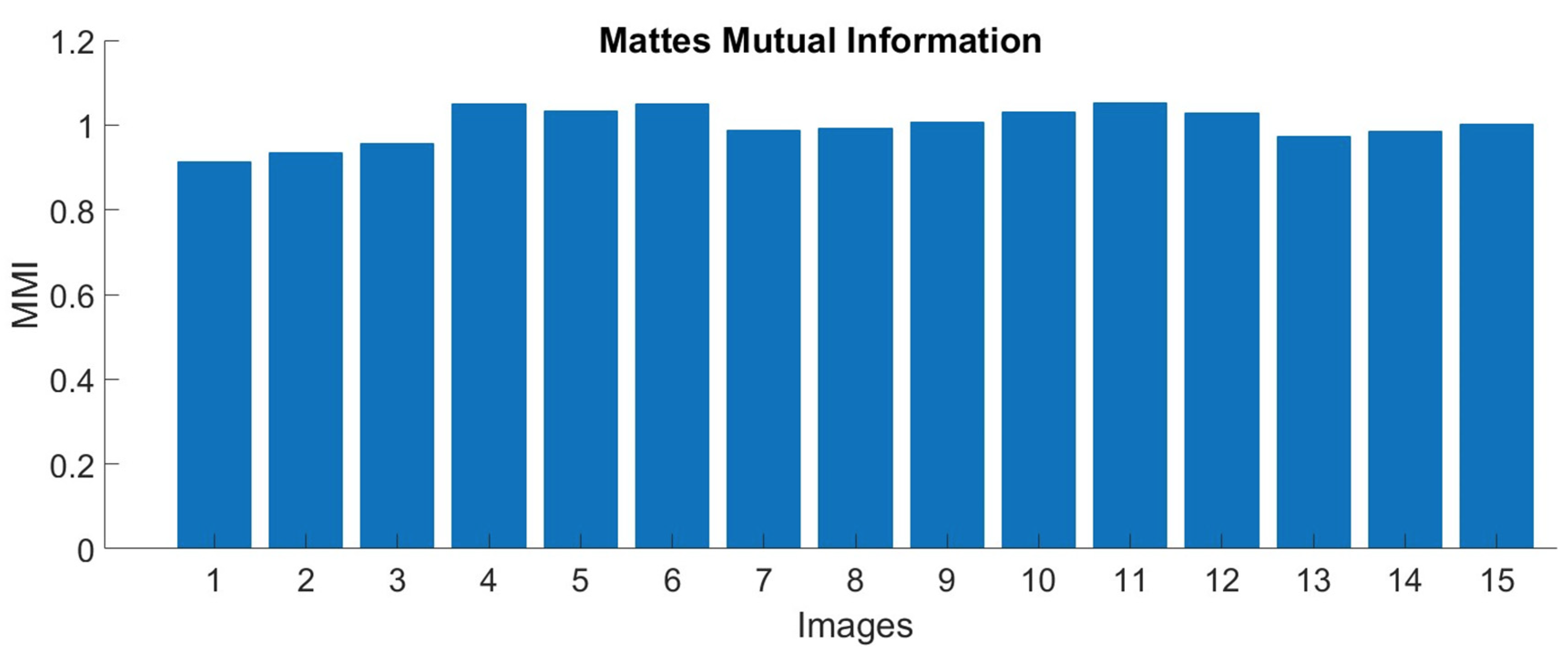
3.2. Image Registration
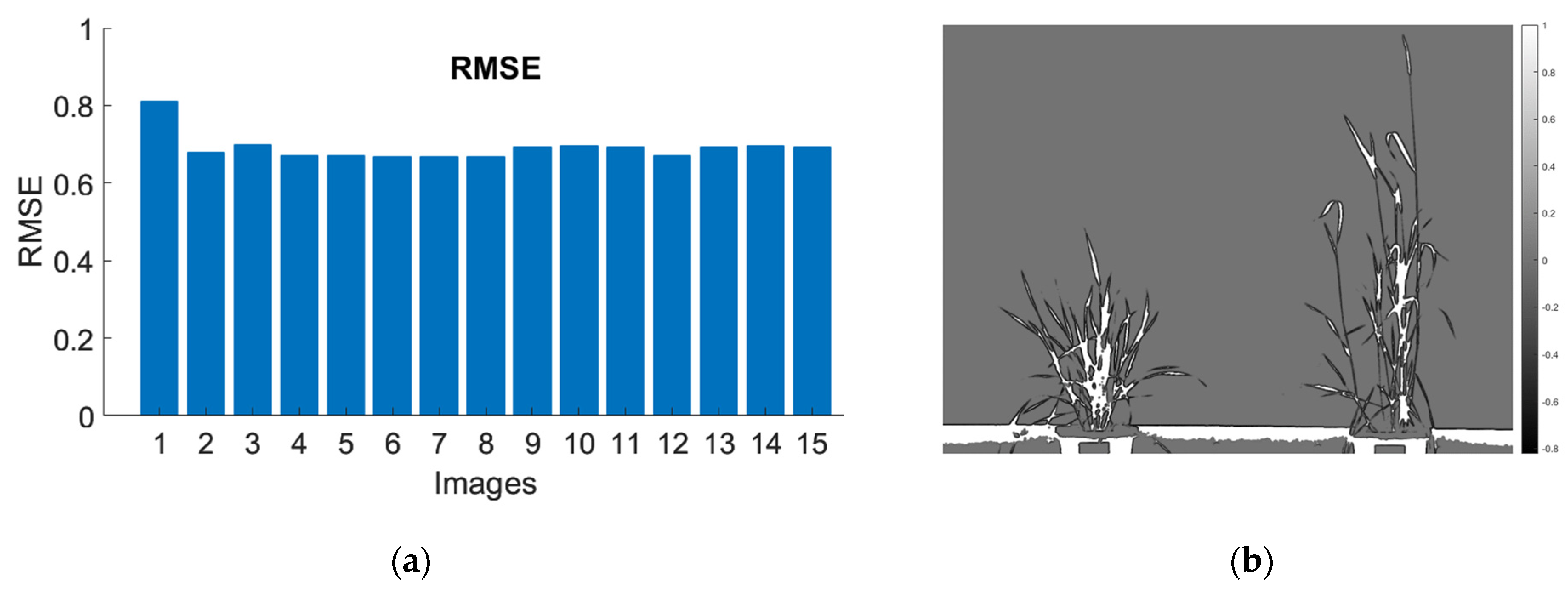
3.3. Segmentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, L.; Zhang, Q.; Huang, D. A Review of Imaging Techniques for Plant Phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.W.; Yang, L.; Yousaf, M.I.; Sami, A.; Mei, X.D.; Shah, L.; Rehman, S.; Xue, L.; Si, H.; Ma, C. Effects of Heat Stress on Growth, Physiology of Plants, Yield and Grain Quality of Different Spring Wheat (Triticum aestivum L.) Genotypes. Sustainability 2021, 13, 2972. [Google Scholar] [CrossRef]
- Mohanty, N. Photosynthetic characteristics and enzymatic antioxidant capacity of flag leaf and the grain yield in two cultivars of Triticum aestivum (L.) exposed to warmer growth conditions. J. Plant Physiol. 2003, 160, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Pineda, M.; Barón, M.; Pérez-Bueno, M. Thermal Imaging for Plant Stress Detection and Phenotyping. Remote Sens. 2020, 13, 68. [Google Scholar] [CrossRef]
- Xu, R.; Li, C.; Paterson, A. Multispectral imaging and unmanned aerial systems for cotton plant phenotyping. PLoS ONE 2019, 14, e0205083. [Google Scholar] [CrossRef]
- Waiphara, P.; Bourgenot, C.; Compton, L.J.; Prashar, A. Optical Imaging Resources for Crop Phenotyping and Stress Detection. Methods Mol. Biol. 2022, 2494, 255–265. [Google Scholar] [CrossRef]
- Bian, L.; Suo, J.; Situ, G.; Li, Z.; Fan, J.; Chen, F.; Dai, Q. Multispectral imaging using a single bucket detector. Sci. Rep. 2016, 6, 24752. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Huete, A.R.; Liu, H.Q.; Batchily, K.; Van Leeuwen, W. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Khaple, A.K.; Devagiri, G.M.; Veerabhadraswamy, N.; Babu, S.; Mishra, S.B. Chapter 6—Vegetation biomass and carbon stock assessment using geospatial approach. In Forest Resources Resilience and Conflicts; Kumar Shit, P., Pourghasemi, H.R., Adhikary, P.P., Bhunia, G.S., Sati, V.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 77–91. [Google Scholar]
- Banerjee, B.P.; Spangenberg, G.; Kant, S. Fusion of Spectral and Structural Information from Aerial Images for Improved Biomass Estimation. Remote Sens. 2020, 12, 3164. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Quantitative estimation of chlorophyll-a using reflectance spectra: Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.C.; Keydan, G.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30, 1248. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys. Res. Lett. 2006, 33, L11402. [Google Scholar] [CrossRef]
- Ju, C.-H.; Tian, Y.-C.; Yao, X.; Cao, W.-X.; Zhu, Y.; Hannaway, D. Estimating Leaf Chlorophyll Content Using Red Edge Parameters. Pedosphere 2010, 20, 633–644. [Google Scholar] [CrossRef]
- Melo, L.L.d.; Melo, V.G.M.L.d.; Marques, P.A.A.; Frizzone, J.A.; Coelho, R.D.; Romero, R.A.F.; Barros, T.H.d.S. Deep learning for identification of water deficits in sugarcane based on thermal images. Agric. Water Manag. 2022, 272, 107820. [Google Scholar] [CrossRef]
- Chandel, N.; Chakraborty, S.; Rajwade, Y.; Dubey, K.; Tiwari, M.K.; Jat, D. Identifying crop water stress using deep learning models. Neural Comput. Appl. 2021, 33, 5353–5367. [Google Scholar] [CrossRef]
- Schor, N.; Berman, S.; Dombrovsky, A.; Elad, Y.; Ignat, T.; Bechar, A. Development of a robotic detection system for greenhouse pepper plant diseases. Precis. Agric. 2017, 18, 394–409. [Google Scholar] [CrossRef]
- Ishimwe, R.; Abutaleb, K.; Ahmed, F. Applications of Thermal Imaging in Agriculture—A Review. Adv. Remote Sens. 2014, 3, 128–140. [Google Scholar] [CrossRef]
- Fricke, W. Night-Time Transpiration—Favouring Growth? Trends Plant Sci. 2019, 24, 311–317. [Google Scholar] [CrossRef]
- Gil-Pérez, B.; Zarco-Tejada, P.; Correa-Guimaraes, A.; Relea-Gangas, E.; Gracia, L.M.; Hernández-Navarro, S.; Sanz Requena, J.F.; Berjón, A.; Martín-Gil, J. Remote sensing detection of nutrient uptake in vineyards using narrow-band hyperspectral imagery. Vitis 2010, 49, 167–173. [Google Scholar]
- Parihar, G.; Saha, S.; Giri, L.I. Application of infrared thermography for irrigation scheduling of horticulture plants. Smart Agric. Technol. 2021, 1, 100021. [Google Scholar] [CrossRef]
- Mutka, A.M.; Bart, R.S. Image-based phenotyping of plant disease symptoms. Front. Plant Sci. 2015, 5, 734. [Google Scholar] [CrossRef] [PubMed]
- Grant, O.; Chaves, M.; Jones, H. Optimizing thermal imaging as a technique for detecting stomatal closure induced by drought stress under greenhouse conditions. Physiol. Plant. 2006, 127, 507–518. [Google Scholar] [CrossRef]
- Leinonen, I.; Jones, H.G. Combining thermal and visible imagery for estimating canopy temperature and identifying plant stress. J. Exp. Bot. 2004, 55, 1423–1431. [Google Scholar] [CrossRef] [PubMed]
- Vieira, G.H.S.; Ferrarezi, R.S. Use of Thermal Imaging to Assess Water Status in Citrus Plants in Greenhouses. Horticulturae 2021, 7, 249. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, Y.; Shamaila, Z.; Zeng, A.; Song, J.; Liu, Y.; Wolfram, S.; Joachim, M.; He, X. Application of BP Neural Network in Predicting Winter Wheat Yield Based on Thermography Technology. Spectrosc. Spectr. Anal. 2013, 33, 1587–1592. [Google Scholar] [CrossRef]
- Bhandari, M.; Xue, Q.; Liu, S.; Stewart, B.A.; Rudd, J.C.; Pokhrel, P.; Blaser, B.; Jessup, K.; Baker, J. Thermal imaging to evaluate wheat genotypes under dryland conditions. Agrosystems Geosci. Environ. 2021, 4, e20152. [Google Scholar] [CrossRef]
- Berger, K.; Machwitz, M.; Kycko, M.; Kefauver, S.C.; Van Wittenberghe, S.; Gerhards, M.; Verrelst, J.; Atzberger, C.; van der Tol, C.; Damm, A.; et al. Multi-sensor spectral synergies for crop stress detection and monitoring in the optical domain: A review. Remote Sens. Environ. 2022, 280, 113198. [Google Scholar] [CrossRef]
- Galieni, A.; D’Ascenzo, N.; Stagnari, F.; Pagnani, G.; Xie, Q.; Pisante, M. Past and Future of Plant Stress Detection: An Overview From Remote Sensing to Positron Emission Tomography. Front. Plant Sci. 2021, 11, 1975. [Google Scholar] [CrossRef]
- Stutsel, B.; Johansen, K.; Malbéteau, Y.M.; McCabe, M.F. Detecting Plant Stress Using Thermal and Optical Imagery From an Unoccupied Aerial Vehicle. Front. Plant Sci. 2021, 12, 2225. [Google Scholar] [CrossRef]
- Bai, G.F.; Blecha, S.; Ge, Y.; Walia, H.; Phansak, P. Characterizing Wheat Response to Water Limitation Using Multispectral and Thermal Imaging. Trans. ASABE 2017, 60, 1457–1466. [Google Scholar] [CrossRef]
- Cucho-Padin, G.; Rinza Díaz, J.; Ninanya Tantavilca, J.; Loayza, H.; Roberto, Q.; Ramirez, D. Development of an Open-Source Thermal Image Processing Software for Improving Irrigation Management in Potato Crops (Solanum tuberosum L.). Sensors 2020, 20, 472. [Google Scholar] [CrossRef]
- Bulanon, D.M.; Burks, T.F.; Alchanatis, V. Image fusion of visible and thermal images for fruit detection. Biosyst. Eng. 2009, 103, 12–22. [Google Scholar] [CrossRef]
- Rosenqvist, E.; Großkinsky, D.K.; Ottosen, C.-O.; van de Zedde, R. The Phenotyping Dilemma—The Challenges of a Diversified Phenotyping Community. Front. Plant Sci. 2019, 10, 163. [Google Scholar] [CrossRef]
- Jiménez-Bello, M.; Ballester, C.; Castel, J.; Intrigliolo, D. Development and validation of an automatic thermal imaging process for assessing plant water status. Agric. Water Manag. 2011, 98, 1497–1504. [Google Scholar] [CrossRef]
- Kelcey, J.; Lucieer, A. Sensor Correction of a 6-Band Multispectral Imaging Sensor for UAV Remote Sensing. Remote Sens. 2012, 4, 1462–1493. [Google Scholar] [CrossRef]
- Lu, H.; Fan, T.; Ghimire, P.; Deng, L. Experimental Evaluation and Consistency Comparison of UAV Multispectral Minisensors. Remote Sens. 2020, 12, 2542. [Google Scholar] [CrossRef]
- Stephen23. Natural-Order Filename Sort. MATLAB Central File Exchange. 2022. Available online: https://www.mathworks.com/matlabcentral/fileexchange/47434-natural-order-filename-sort (accessed on 6 November 2022).
- Jhan, J.-P.; Rau, J.; Haala, N.; Cramer, M. Investigation of Parallax Issues for Multi-Lens Multispectral Camera Band Co-Registration. ISPRS—Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, XLII-2/W6, 157–163. [Google Scholar] [CrossRef]
- Choi, K.; Lam, E.; Wong, K. Automatic source camera identification using the intrinsic lens radial distortion. Opt. Express 2006, 14, 11551–11565. [Google Scholar] [CrossRef]
- Wu, F.; Wang, X. Correction of image radial distortion based on division model. Opt. Eng. 2017, 56, 013108. [Google Scholar] [CrossRef]
- Heikkilä, J.; Silvén, O. A four-step camera calibration procedure with implicit image correction. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Juan, Puerto Rico, 17–19 June 1997; pp. 1106–1112. [Google Scholar]
- Zhang, Z. A Flexible New Technique for Camera Calibration. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1330–1334. [Google Scholar] [CrossRef]
- Wang, G.; Zheng, H.; Zhang, X. A Robust Checkerboard Corner Detection Method for Camera Calibration Based on Improved YOLOX. Front. Phys. 2022, 9, 828. [Google Scholar] [CrossRef]
- Torr, P.H.S.; Zisserman, A. MLESAC: A New Robust Estimator with Application to Estimating Image Geometry. Comput. Vis. Image Underst. 2000, 78, 138–156. [Google Scholar] [CrossRef]
- Banerjee, B.; Raval, S.; Cullen, P.J. Alignment of UAV-hyperspectral bands using keypoint descriptors in a spectrally complex environment. Remote Sens. Lett. 2018, 9, 524–533. [Google Scholar] [CrossRef]
- Chui, H.; Win, L.; Schultz, R.; Duncan, J.S.; Rangarajan, A. A unified non-rigid feature registration method for brain mapping. Med. Image Anal. 2003, 7, 113–130. [Google Scholar] [CrossRef]
- Myronenko, A.; Song, X. Intensity-based image registration by minimizing residual complexity. IEEE Trans. Med. Imaging 2010, 29, 1882–1891. [Google Scholar] [CrossRef]
- Aghajani, K.; Yousefpour, R.; Shirpour, M.; Manzuri, M.T. Intensity based image registration by minimizing the complexity of weighted subtraction under illumination changes. Biomed. Signal Process. Control 2016, 25, 35–45. [Google Scholar] [CrossRef]
- Aylward, S.; Jomier, J.; Barre, S.; Davis, B.; Ibanez, L. Optimizing ITK’s Registration Methods for Multi-processor, Shared-Memory Systems. Insight J. 2007. [Google Scholar] [CrossRef]
- Keikhosravi, A.; Li, B.; Liu, Y.; Eliceiri, K.W. Intensity-based registration of bright-field and second-harmonic generation images of histopathology tissue sections. Biomed. Opt. Express 2020, 11, 160–173. [Google Scholar] [CrossRef]
- Dey, N. Uneven Illumination Correction of Digital Images: A Survey of the State-of-the-Art. Optik 2019, 183, 483–495. [Google Scholar] [CrossRef]
- Banerjee, B.P.; Joshi, S.; Thoday-Kennedy, E.; Pasam, R.K.; Tibbits, J.; Hayden, M.; Spangenberg, G.; Kant, S. High-throughput phenotyping using digital and hyperspectral imaging-derived biomarkers for genotypic nitrogen response. J. Exp. Bot. 2020, 71, 4604–4615. [Google Scholar] [CrossRef]
- Mishra, P.; Lohumi, S.; Ahmad Khan, H.; Nordon, A. Close-range hyperspectral imaging of whole plants for digital phenotyping: Recent applications and illumination correction approaches. Comput. Electron. Agric. 2020, 178, 105780. [Google Scholar] [CrossRef]
- Roth, L.; Aasen, H.; Walter, A.; Liebisch, F. Extracting leaf area index using viewing geometry effects—A new perspective on high-resolution unmanned aerial system photography. ISPRS J. Photogramm. Remote Sens. 2018, 141, 161–175. [Google Scholar] [CrossRef]
- Weyler, J.; Magistri, F.; Seitz, P.; Behley, J.; Stachniss, C. In-Field Phenotyping Based on Crop Leaf and Plant Instance Segmentation. In Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2022; pp. 2968–2977. [Google Scholar]
- Mario, S.; Madec, S.; David, E.; Velumani, K.; Lozano, R.; Weiss, M.; Frederic, B. SegVeg: Segmenting RGB images into green and senescent vegetation by combining deep and shallow methods. Plant Phenomics 2022, 2022, 9803570. [Google Scholar]
- Thanh, D.; Thanh, L.; Dvoenko, S.; Prasath, S.; San, N. Adaptive Thresholding Segmentation Method for Skin Lesion with Normalized Color Channels of NTSC and YCbCr. In Proceedings of the 14th International Conference on Pattern Recognition and Information Processing (PRIP’2019), Minsk, Belarus, 21–23 May 2019; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Fang, H.; Liang, S. Leaf Area Index Models. In Reference Module in Earth Systems and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Zhao, H.; Yang, C.; Guo, W.; Zhang, L.; Zhang, D. Automatic Estimation of Crop Disease Severity Levels Based on Vegetation Index Normalization. Remote Sens. 2020, 12, 1930. [Google Scholar] [CrossRef]
- Morales, A.; Guerra Hernández, R.; Horstrand, P.; Diaz, M.; Jimenez, A.; Melián, J.; Lopez, S.; Lopez, J. A Multispectral Camera Development: From the Prototype Assembly until Its Use in a UAV System. Sensors 2020, 20, 6129. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Kakarla, S.; Roberts, P. Detection of target spot and bacterial spot diseases in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precis. Agric. 2020, 21, 955–978. [Google Scholar] [CrossRef]
- Drisya, J.; Kumar, D.S.; Roshni, T. Chapter 27—Spatiotemporal Variability of Soil Moisture and Drought Estimation Using a Distributed Hydrological Model. In Integrating Disaster Science and Management; Samui, P., Kim, D., Ghosh, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 451–460. [Google Scholar]
- He, J.; Zhang, N.; Xi, S.; Lu, J.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.; Tian, Y. Estimating Leaf Area Index with a New Vegetation Index Considering the Influence of Rice Panicles. Remote Sens. 2019, 11, 1809. [Google Scholar] [CrossRef]
- Borges, M.V.V.; de Oliveira Garcia, J.; Batista, T.S.; Silva, A.N.M.; Baio, F.H.R.; da Silva Junior, C.A.; de Azevedo, G.B.; de Oliveira Sousa Azevedo, G.T.; Teodoro, L.P.R.; Teodoro, P.E. High-throughput phenotyping of two plant-size traits of Eucalyptus species using neural networks. J. For. Res. 2022, 33, 591–599. [Google Scholar] [CrossRef]
- Boiarskii, B.; Hasegawa, H. Comparison of NDVI and NDRE Indices to Detect Differences in Vegetation and Chlorophyll Content. J. Mech. Contin. Math. Sci. 2019, 4, 20–29. [Google Scholar] [CrossRef]
- Roujean, J.-L.; Breon, F.-M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E.; D’Alessio, P. A broad-band leaf chlorophyll vegetation index at the canopy scale. Precis. Agric. 2008, 9, 303–319. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Renieblas, G.P.; Nogués, A.T.; González, A.M.; Gómez-Leon, N.; Del Castillo, E.G. Structural similarity index family for image quality assessment in radiological images. J. Med. Imaging 2017, 4, 035501. [Google Scholar] [CrossRef] [PubMed]
- Grande, J.C. Principles of Image Analysis. Metallogr. Microstruct. Anal. 2012, 1, 227–243. [Google Scholar] [CrossRef]
- Drap, P.; Lefèvre, J. An Exact Formula for Calculating Inverse Radial Lens Distortions. Sensors 2016, 16, 807. [Google Scholar] [CrossRef]
- Das Choudhury, S.; Bashyam, S.; Qiu, Y.; Samal, A.; Awada, T. Holistic and component plant phenotyping using temporal image sequence. Plant Methods 2018, 14, 35. [Google Scholar] [CrossRef]
- Saleh, Z.H.; Apte, A.P.; Sharp, G.C.; Shusharina, N.P.; Wang, Y.; Veeraraghavan, H.; Thor, M.; Muren, L.P.; Rao, S.S.; Lee, N.Y.; et al. The distance discordance metric-a novel approach to quantifying spatial uncertainties in intra- and inter-patient deformable image registration. Phys. Med. Biol. 2014, 59, 733–746. [Google Scholar] [CrossRef]
- Memon, J.; Sami, M.; Khan, R.A.; Uddin, M. Handwritten Optical Character Recognition (OCR): A Comprehensive Systematic Literature Review (SLR). IEEE Access 2020, 8, 142642–142668. [Google Scholar] [CrossRef]
- Pereyra Irujo, G. IRimage: Open source software for processing images from infrared thermal cameras. PeerJ Comput. Sci. 2022, 8, e977. [Google Scholar] [CrossRef]
- Wang, S.; Sun, G.; Zheng, B.; Du, Y. A Crop Image Segmentation and Extraction Algorithm Based on Mask RCNN. Entropy 2021, 23, 1160. [Google Scholar] [CrossRef]
- Sodjinou, S.G.; Mohammadi, V.; Sanda Mahama, A.T.; Gouton, P. A deep semantic segmentation-based algorithm to segment crops and weeds in agronomic color images. Inf. Process. Agric. 2022, 9, 355–364. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, X.; Qi, J.; Yang, S. Automatic instance segmentation of orchard canopy in unmanned aerial vehicle imagery using deep learning. Front. Plant Sci. 2022, 13, 1041791. [Google Scholar] [CrossRef]
- Munz, S.; Reiser, D. Approach for Image-Based Semantic Segmentation of Canopy Cover in Pea-Oat Intercropping. Agriculture 2020, 10, 354. [Google Scholar] [CrossRef]


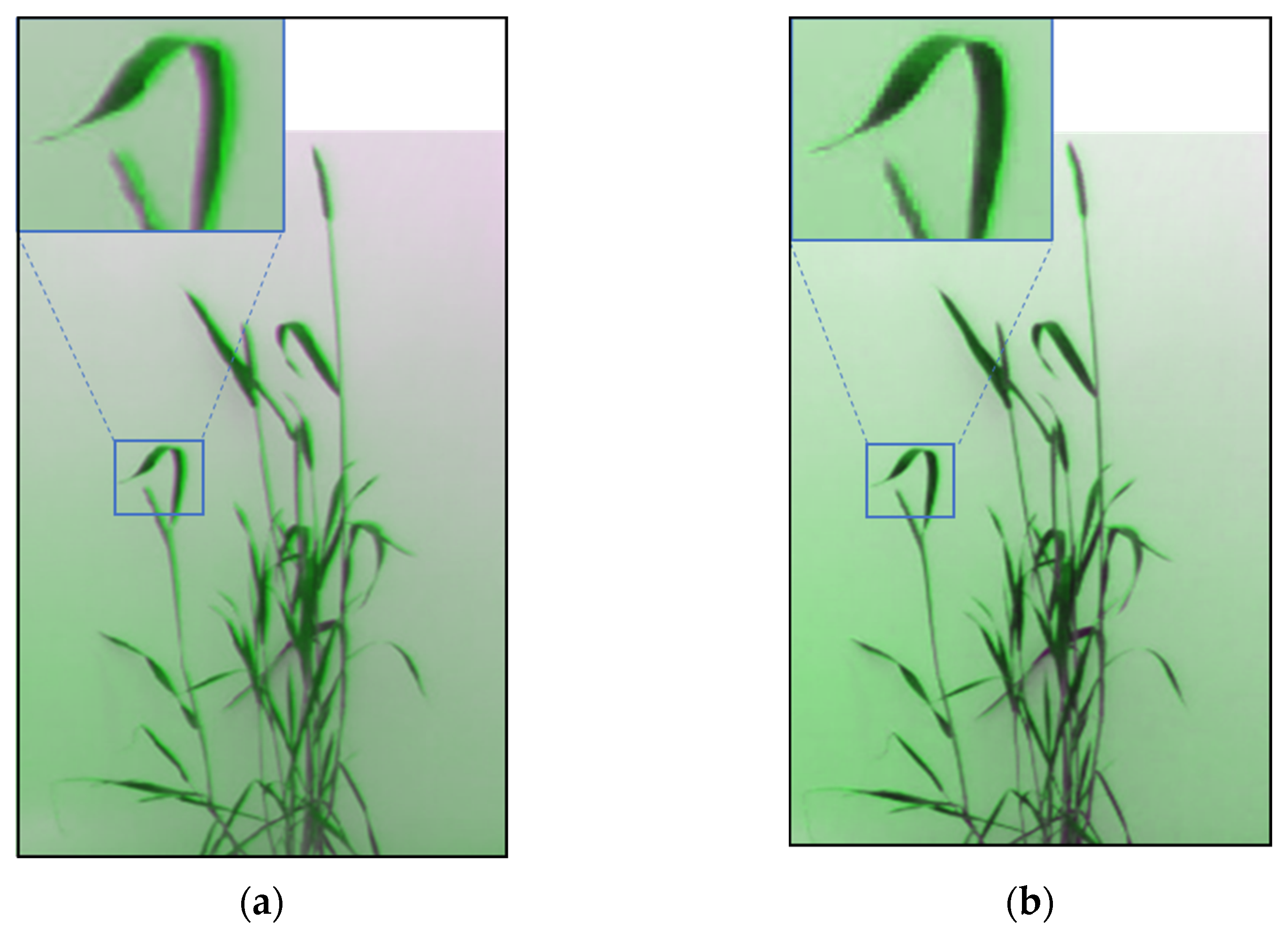
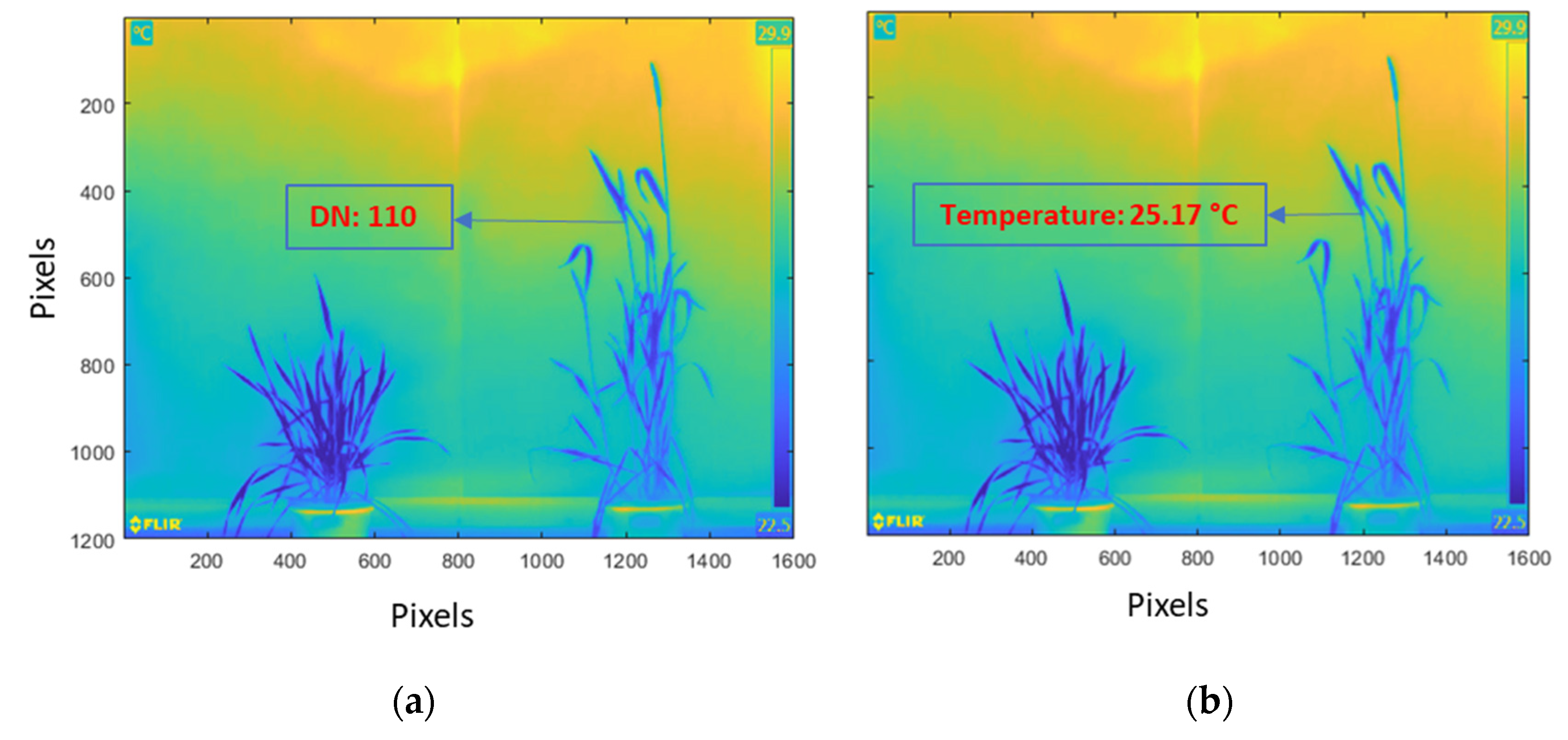


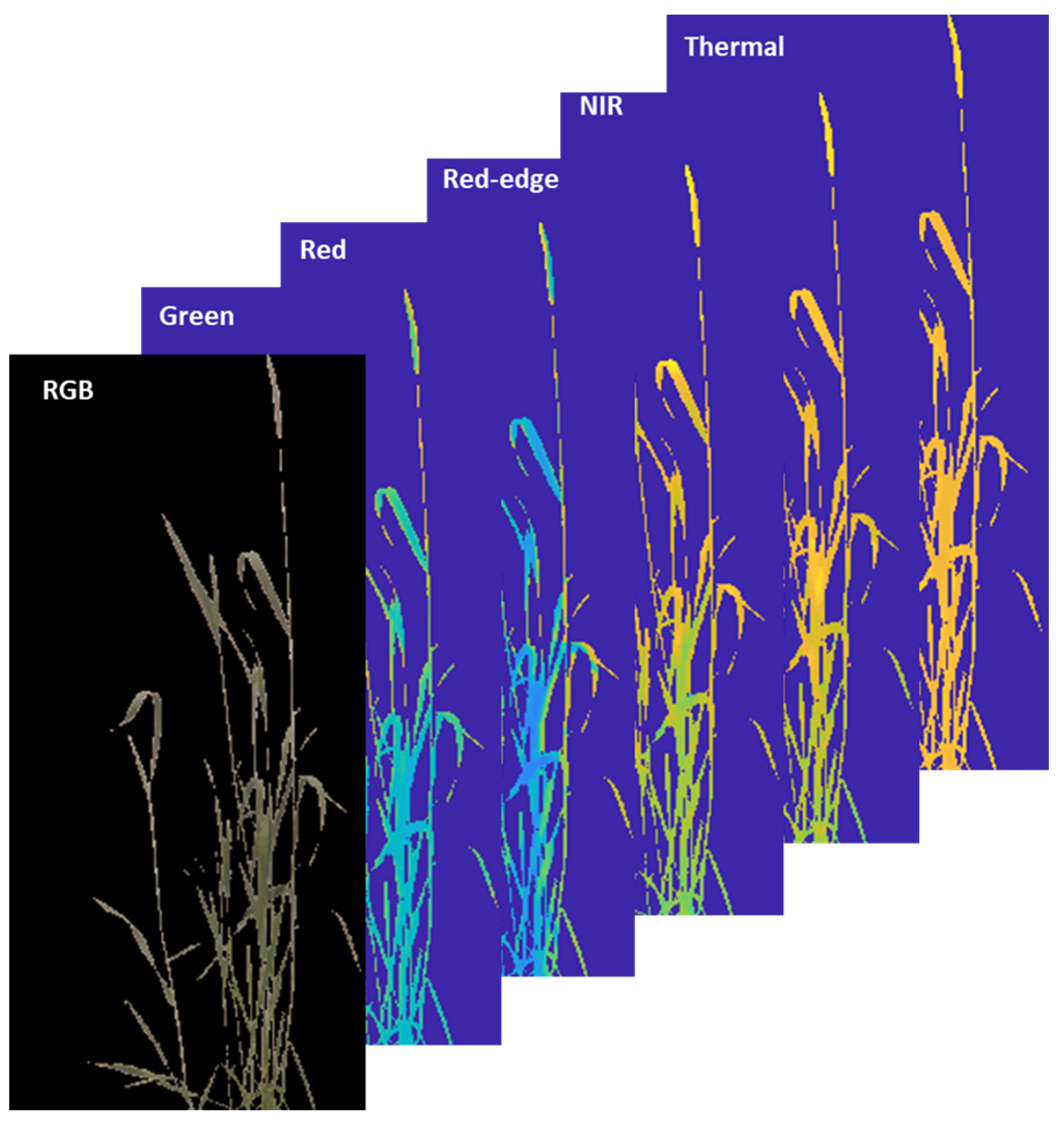
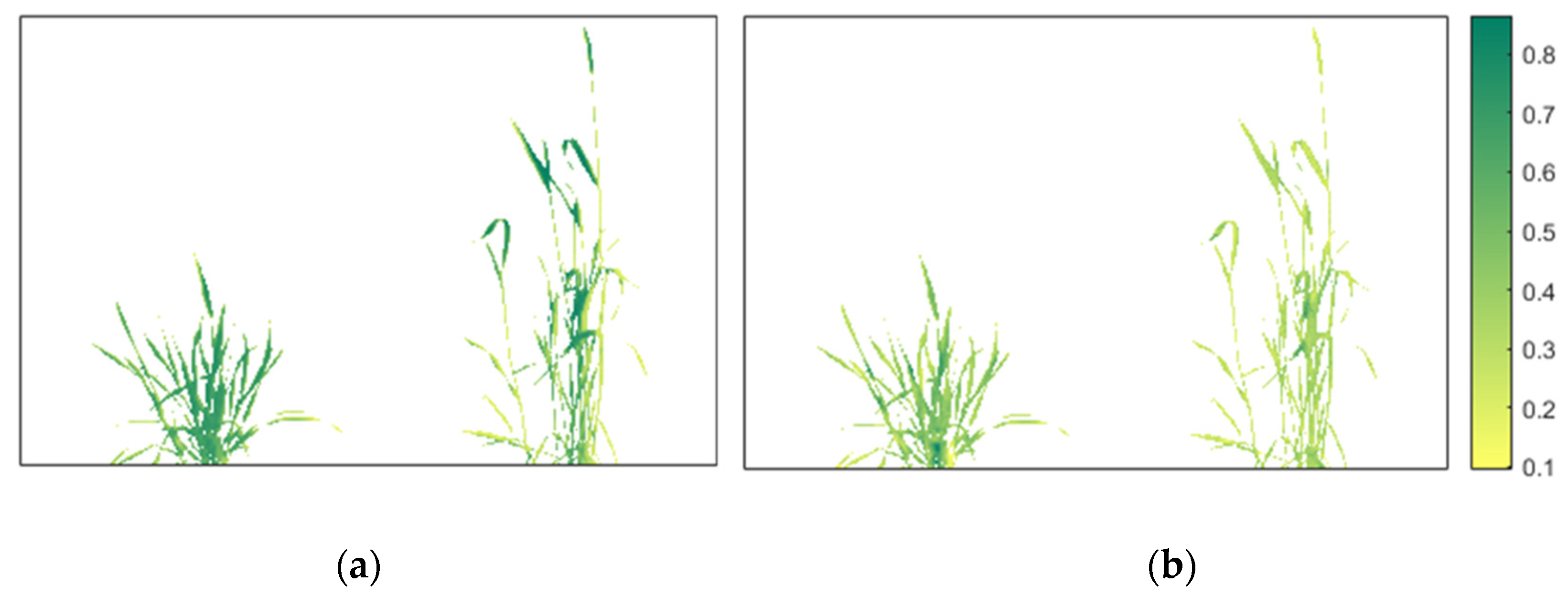

| Rigid | Nonrigid |
|---|---|
| Indices | Equations | References |
|---|---|---|
| Normalized Difference Vegetation Index (NDVI) | [8] | |
| Normalized Difference Red Edge (NDRE) | [12] | |
| Chlorophyll Index red edge (CIre) | [14] | |
| Triangle Vegetation Index (TVI) | 0.5(120( | [66] |
| Renormalized Difference Vegetation Index (RDVI) | [68] | |
| Chlorophyll Vegetation Index (CVI) | [69] | |
| Chlorophyll Index green (CIg) | [70] |
| Bands | Red | Red-Edge | NIR | Green |
|---|---|---|---|---|
| Mean reprojection error (pixels) | 0.29 | 0.60 | 0.21 | 0.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, N.; Banerjee, B.P.; Hayden, M.; Kant, S. An Open-Source Package for Thermal and Multispectral Image Analysis for Plants in Glasshouse. Plants 2023, 12, 317. https://doi.org/10.3390/plants12020317
Sharma N, Banerjee BP, Hayden M, Kant S. An Open-Source Package for Thermal and Multispectral Image Analysis for Plants in Glasshouse. Plants. 2023; 12(2):317. https://doi.org/10.3390/plants12020317
Chicago/Turabian StyleSharma, Neelesh, Bikram Pratap Banerjee, Matthew Hayden, and Surya Kant. 2023. "An Open-Source Package for Thermal and Multispectral Image Analysis for Plants in Glasshouse" Plants 12, no. 2: 317. https://doi.org/10.3390/plants12020317
APA StyleSharma, N., Banerjee, B. P., Hayden, M., & Kant, S. (2023). An Open-Source Package for Thermal and Multispectral Image Analysis for Plants in Glasshouse. Plants, 12(2), 317. https://doi.org/10.3390/plants12020317








