Abstract
Although zygotic embryogenesis is usually studied in the field of seed biology, great attention has been paid to the methods used to generate haploid embryos due to their applications in crop breeding. These mainly include two methods for haploid embryogenesis: in vitro microspore embryogenesis and in vivo haploid embryogenesis. Although microspore culture systems and maize haploid induction systems were discovered in the 1960s, little is known about the molecular mechanisms underlying haploid formation. In recent years, major breakthroughs have been made in in vivo haploid induction systems, and several key factors, such as the matrilineal (MTL), baby boom (BBM), domain of unknown function 679 membrane protein (DMP), and egg cell-specific (ECS) that trigger in vivo haploid embryo production in both the crops and Arabidopsis models have been identified. The discovery of these haploid inducers indicates that haploid embryogenesis is highly related to gamete development, fertilization, and genome stability in ealry embryos. Here, based on recent efforts to identify key players in haploid embryogenesis and to understand its molecular mechanisms, we summarize the different paths to haploid embryogenesis, and we discuss the mechanisms of haploid generation and its potential applications in crop breeding. Although these haploid-inducing factors could assist egg cells in bypassing fertilization to initiate embryogenesis or trigger genome elimination in zygotes after fertilization to form haploid embryos, the fertilization of central cells to form endosperms is a prerequisite step for haploid formation. Deciphering the molecular and cellular mechanisms for haploid embryogenesis, increasing the haploid induction efficiency, and establishing haploid induction systems in other crops are critical for promoting the application of haploid technology in crop breeding, and these should be addressed in further studies.
Although zygotic embryogenesis is the primary way to generate embryos in plant reproduction, there are still several alternative methods for forming haploid or diploid embryos. In addition to normal zygotic embryogenesis, in recent decades, a great deal of attention has been paid to the formation of haploid embryos due to their important applications in crop breeding. Haploid induction is an effective way to shorten breeding time and has been used in crop breeding for many years. Traditional breeding requires 7–8 generations to obtain ideal homozygous plants during the cross, while haploid technology can shorten the breeding time to 2–3 generations, which will greatly save on breeding time and reduce the cost of breeding [1]. Haploid embryogenesis is the core of haploid breeding technology.
According to the existing methods of generating haploid embryos, a haploid induction system can be divided into two categories: in vitro and in vivo methods. Microspore embryogenesis is the primary in vitro method used to induce haploid embryos. It was first developed in 1964 and has been employed in over 75 species. The in vitro methods for haploid embryogenesis are mainly achieved through the suitable in vitro cell culture system, which promotes haploid microspores to be reprogrammed and enter into embryogenic pathways to generate haploid embryos. However, these in vitro culture systems have not been established for many plant species, and the mechanisms of microspore embryogenesis remain largely unknown. The existing in vivo haploid induction systems can be further divided into CENH3-mediated haploid embryogenesis, parental factor-induced haploid embryogenesis, and transcriptional factor-triggered haploid embryogenesis. The haploid inducer line in maize was first discovered in 1959, but the mechanisms underlying haploid formation have long remained a mystery. The recent discoveries of several key molecular players in maize and Arabidopsis haploid induction have greatly aided us in understanding the mechanisms of haploid embryogenesis, demonstrating that defects in gamete development and fertilization are the fundamental mechanisms for haploid generation.
In this review, we summarize the recent advances in the methods for generating haploid embryogenesis, and we primarily focus on microspore embryogenesis, CENH3-mediated haploid embryogenesis, and parental factor-induced and transcription factor-triggered haploid embryogenesis. We also discuss the mechanisms underlying these methods and the potential applications of haploid embryogenesis in crop breeding.
1. Microspore Embryogenesis
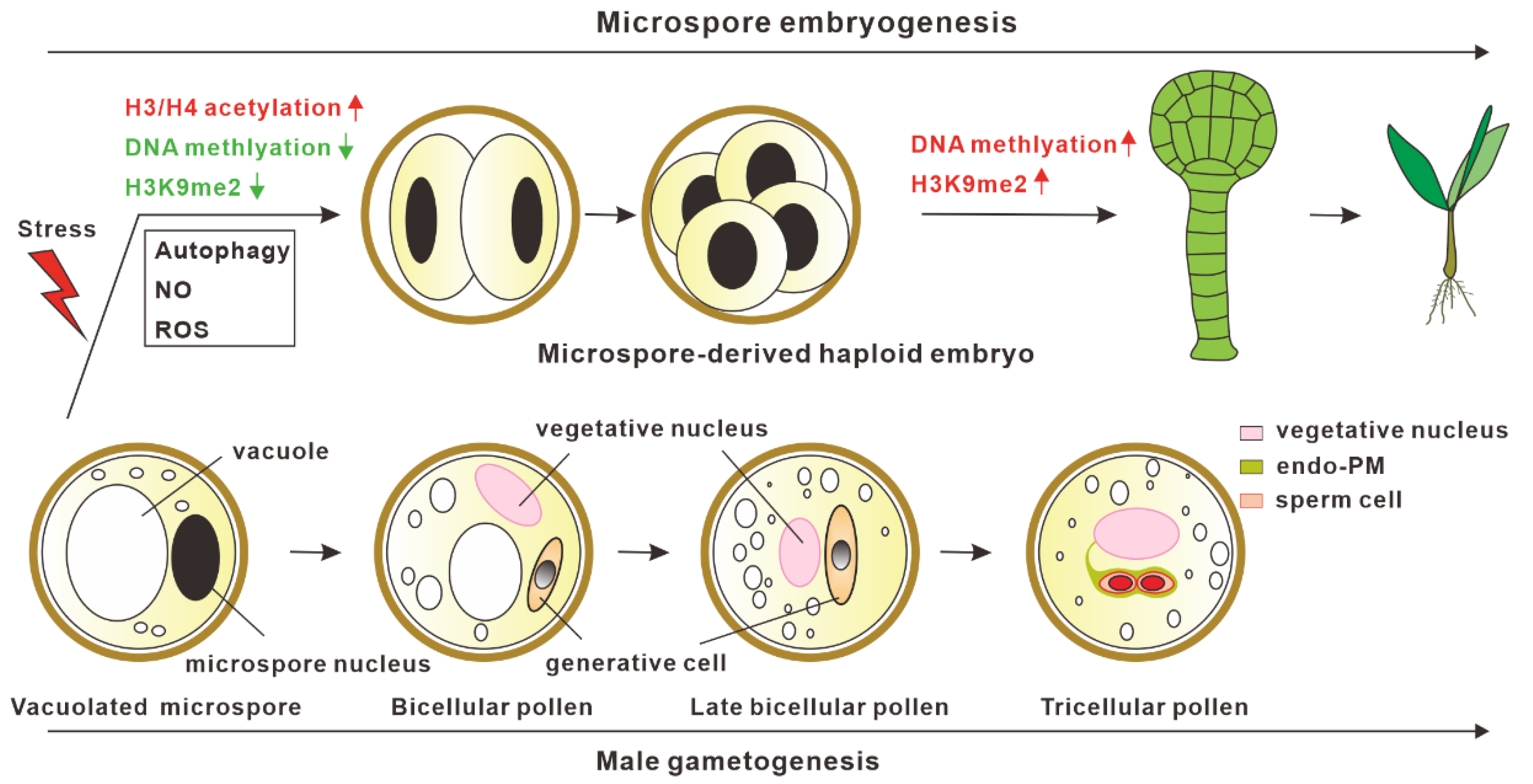
Microspore embryogenesis is a type of embryogenesis in which haploid microspores after stress treatment undergo cell reprogramming and shift into embryogenic pathways to generate haploid embryos [2,3,4,5,6,7,8]. The haploid embryos then can be automatically doubled or chemically treated to produce double haploids, thus reducing the time required to obtain homozygous plants [9,10]. Among these microspore embryogenesis systems, exine-dehisced Brassica napus microspores treated by physical stress can induce polarization and develop into typical embryos with the differentiation of an embryo proper and suspensor [11]. However, microspore embryogenesis induction systems have not been well established in many other plants, such as Arabidopsis thaliana and Solanum lycopersicum, indicating that the embryogenic potential of microspores or signals for triggering cell fate reprogramming into embryogenic pathways may vary in different species.
In microspore embryogenesis, the reprogramming of microspores into the embryo developmental pathway after stress treatment is the critical step for haploid embryo generation. Epigenetic mechanisms, including H3/ H4 deacetylation, DNA methylation, and H3K9me2, are reported to be involved in microspore embryogenesis [12]. The inhibition of histone deacetylases (HDAC) activities by the chemical inhibitor Trichostatin A (TSA) can efficiently promote microspore embryogenesis in both B. napus and A. thaliana. Suppression of HDAC activities by TSA leads to the hyperacetylation of histones H3 and H4, which results in the upregulation of genes related to cell wall remodeling, cell division, and embryogenesis [13]. In addition to B. napus and A. thaliana, TSA treatment has been efficient at improving the rate of microspore embryogenesis in several other plants, including pakchoi [14], wheat [15,16], and barley [17], indicating that the HDAC-dependent mechanism in microspore embryogenesis appears to be conserved in different plants. Besides suppressing HDAC activities, the inhibition of DNA methyltransferases catalytic activities by the inhibitors 5-azacytidine (AC) and 2′-deoxy-5-azacytidine (DAC) can remarkably increase the frequency of microspore embryogenesis [18], suggesting that decreased DNA methylation levels are also responsible for microspore embryogenesis, which is likely achieved through increased chromatin accessibility for transcription activation. Similar results in promoting microspore embryogenesis were also observed when the microspores were treated with BIX-01294, which can efficiently inhibit the activities of H3K9me2 methyltransferase [19]. In summary, a low DNA methylation level, low H3K9me2 level, and high acetylation level are critical for the initiation of microspore embryogenesis, whereas, after reprogramming, microspore embryogenesis is accompanied by increased DNA methylation levels and H3K9me2 levels [19,20] at later developmental stages, which may promote haploid embryo differentiation (Figure 1).
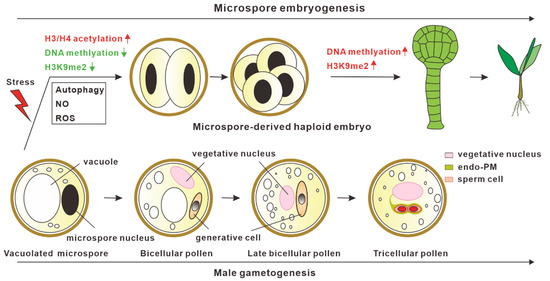
Figure 1.
Model for in vitro microspore embryogenesis. After stress treatment, vacuolated microspores switch from normal male gametogenesis into the embryogenic pathway. Epigenetic modifications, autophagy, reactive oxygen species (ROS), and nitric oxide (NO) are involved in promoting the initiation of microspore embryogenesis.
Besides epigenetic modifications, autophagy, reactive oxygen species (ROS), and nitric oxide (NO) have been shown to be involved in the initiation process of microspore embryogenesis (Figure 1) [12,21,22]. Recent studies have revealed that autophagy-mediated cytoplasm clearance is not only critical for promoting pollen germination [23] and pollen tube growth [24], but it is also important for microspore embryogenesis [25,26,27]. Two independent studies have demonstrated that the cell death of microspores after stress treatment is accompanied by the activation of autophagy, and blocking autophagy prevented the cell death of microspores and increased the frequency of microspore embryogenesis in B. napus, indicating that enhanced autophagic activities play a role in preventing microspore embryogenesis [27]. A similar role of autophagy in Hordeum vulgare microspore embryogenesis was also found. The suppression of autophagosome formation by 3-methyladenine or the inhibition of autophagic body degradation in the vacuoles by E-64 can promote microspore embryogenesis [26]. In addition, ROS and NO were also shown to play important roles in response to stress-induced microspore cell death and microspore reprogramming in barley microspore embryogenesis [28]. Stress-treated microspore embryonic suspension exhibited high ROS levels, high NO signals, and enhanced microspore cell deaths [28,29]. Treating microspores with MnCl2 (O2− scavenger), ascorbate (H2O2 scavenger), and cPTIO (NO scavenger) led to reduced cell death and increased embryogenesis initiation efficiency. Hence, investigating these mechanisms will not only help us understand the mechanism for microspore embryogenesis initiation, but also provide an opportunity to improve haploid induction.
2. CENH3-Mediated Haploid Embryogenesis
CENH3 (CENP-A in humans, Cse4p in Saccharomyces cerevisiae, and HTR12 in Arabidopsis) [30,31] is the centromere-specific histone H3 variant which contains a variable N-terminal domain (NTD) and a conserved histone fold domain (HFD). The core functions of CENH3 primarily include two aspects: recruiting CENH3/H4 reloading factor to nucleosomes and providing a platform for kinetochore binding and assembling. The NTD of CENH3 is essential for CENH3 loading onto the centromeres of meiotic chromosomes, rather than its deposition and function in mitotic nuclei [32,33]. Recent studies have revealed that both the quality (stability of CENH3 on nucleosomes and recognizability of CENH3 by CENH3/H4 reloading factors or kinetochore complex proteins) and quantity (loading amount of CENH3 nucleosomes on centromeres) of CENH3 in centromeres are important for CENH3′s function [34,35,36,37,38,39], and defects in CENH3 will lead to chromosome elimination.
Based on the characteristics of CENH3, GFP-CENH3 and GFP-tailswap (GFP fused with the N-terminal tail of a CENH3 variant, with NTD replaced by conserved Histone3.3 NTD) were designed to rescue the developmental defects in a cenh3-1 mutant. Expressing GFP-tailswap in a cenh3-1 mutant can partially recuse the embryonic lethal phenotype, but it is accompanied by severe male sterility [33,40]. More importantly, expressing GFP-tailswap or GFP-CENH3 in a cenh3-1 mutant can produce aneuploid and haploid plants due to chromosome elimination. When GFP-tailswap/cenh3-1 pistils were pollinated by wild-type (WT) pollen grains, over 30% of the progenies were haploids, though with paternal genomes. Haploid-induced rates (HIR) slightly decreased when GFP-tailswap expressing pollen grains were used for the cross with the WT plants. Expressing CENH3 from B. rapa, Lepidium oleraceum, or Z. mays in the Arabidopsis cehn3-1 mutant can also lead to haploid formation, which is similar to the expression GFP-CENH3 or GFP-tailswap in cenh3-1 [41]. In addition, point mutations in CENH3 can also induce haploids during the outcrossing with WT plants [39,42,43]. Under this scenario, the CENH3-mediated haploid induction system appears to be conserved in different plants.
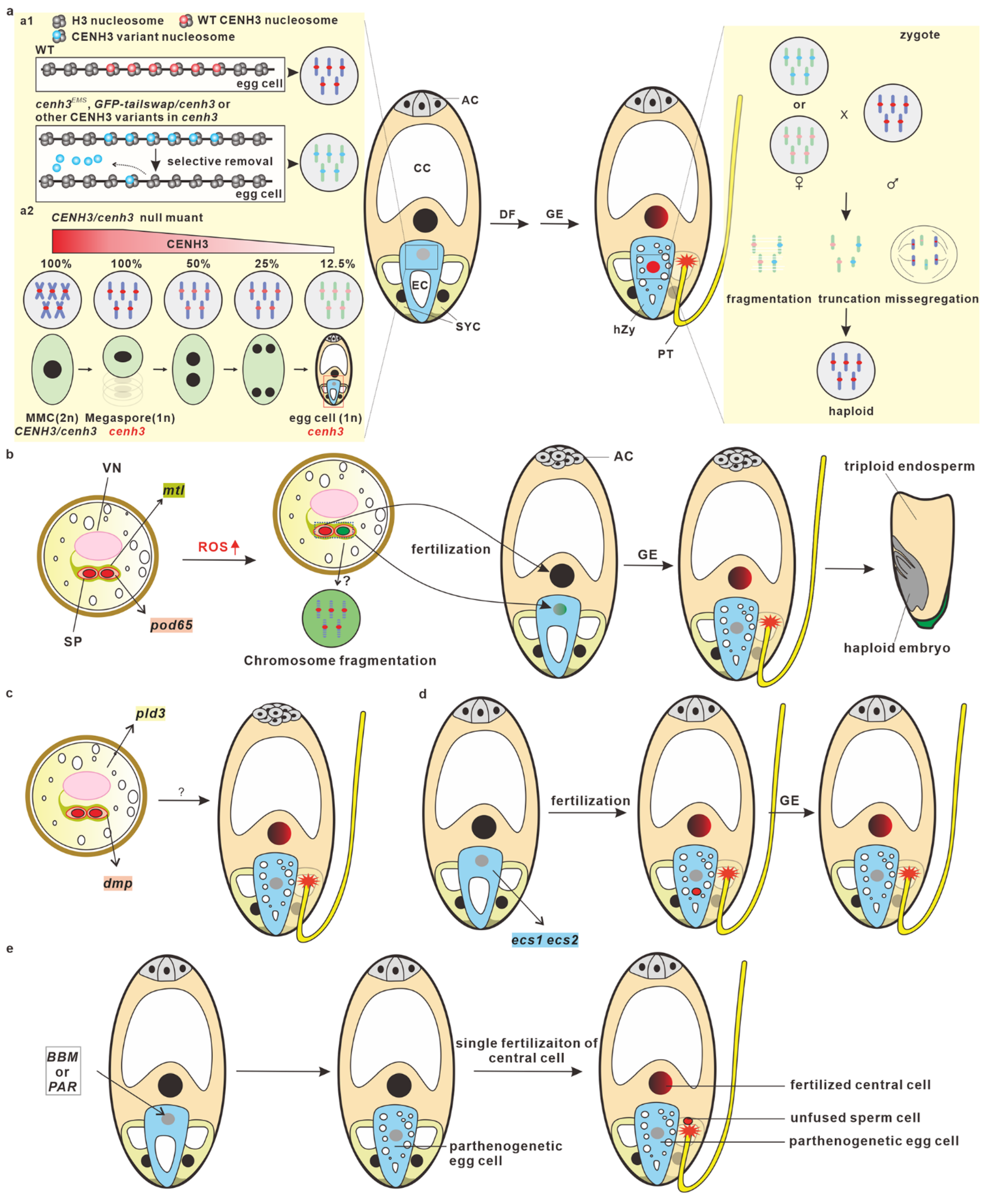
The potential mechanisms underlying CENH3-mediated haploid induction have been summarized and discussed, and several hypotheses and theories have been proposed in recent reviews [36,38,44,45,46,47,48,49,50,51,52]. CENH3-induced haploids are the result of post-fertilization genome elimination [53]. Based recent published results, fertilization is normally completed to form zygotes, but uniparental genomes will be eliminated at early embryonic developmental stages, and eventually they will form the haploid mature embryos. Mutations or modifications in CENH3 may impair the recruitment of CENH3 to centromeres or reduce their stability, resulting in the selective dispossession of CENH3 variants from centromeres in egg cells and zygotes [54]. In a heterozygous cenh3 null mutant, the CENH3 amount on the centromeres of the cenh3 embryo sac are significantly diluted during post-meiotic cell division, prior to gamete formation [55]. The inconsistency of the centromere strength caused by CENH3 among the two parental genomes in zygotes thereby leads to the delay of CENH3 reloading and/or kinetochore assembly in uniparental genomes, resulting in chromosome missegregation, truncation, or fragmentation. Chromosomal irregularities can produce micronuclei or lead to the missegregation of chromosomes [56]. The chromosomal truncation and fragmentation may form micronuclei, which may re-emerge with the nucleus to produce aneuploids or eventually degrade to produce haploid embryos (Figure 2a).
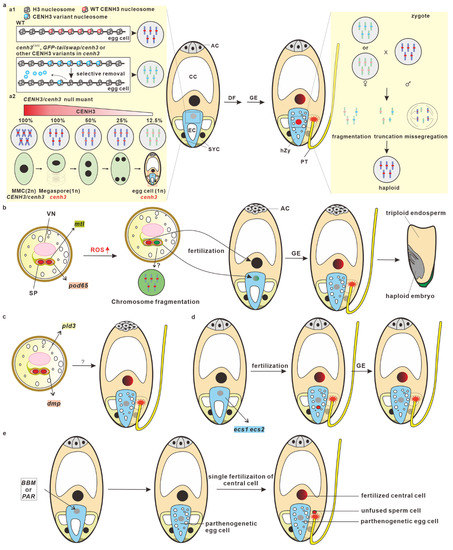
Figure 2.
Model of in vivo haploid embryogenesis. (a) CENH3-mediated haploid embryogenesis is accomplished through post-fertilization genome elimination. This model shows CENH3-mediated maternal genome elimination after fertilization. In a cenh3 EMS mutant, the GFP-tailswap/cenh3 mutant or cenh3 mutants which express the CENH3 variant from other species, selectively eviction of CENH3 variants in mature egg cells and the zygotes produce “weak” centromeres (a1). In a heterozygous model of a cenh3 null mutant, the quantity of CENH3 on the centromeres is significantly diluted after megasporogenesis, which also generates “weak” centromeres in cenh3 egg cells (a2). After being pollinated by wild-type pollen grains, the maternal genome (gray), which contains “weak” centromeres, rather than the normal paternal genome (red), is selectively eliminated in zygotes and early embryos. (b) Mutations in MTL and POD induces haploid embryogenesis through ROS-triggered spermatid chromosome fragmentation. MTL is localized at the endo-PM of vegetative cells while POD65 is localized in sperm cells. An ROS level increase leads to spermatid chromosome fragmentation. Sperm cells (green) with fragmented chromosomes can fertilize egg cells, but the paternal genomes will be eliminated after fertilization (gray). (c) Vegetative cell-expressed PLD3 and sperm cell-expressed DMP may induce maternal haploid embryogenesis. (d) Mutation in egg cell-specifically expressed ECS1/2 induces haploid embryogenesis, which may be due to the karyogamy defect and genome elimination after fertilization. (e) The ectopic expression of BBM and PAR in egg cells induces haploid embryogenesis through parthenogenesis. WT, wild-type; MMC, megaspore mother cell; AC, antipodal cell; CC, central cell; EC, egg cell; SYC, synergid cell; hZy, haploid zygote; DF, double fertilization; GE, genome elimination; PT, pollen tube.
3. Parental Factor-Mediated Haploid Embryogenesis
Although the paternal inducer line of maize (Stock6), which can be used to induce maternal haploids, was discovered in 1959 [57], the specific gene responsible for haploid generation was not identified until recently. In this section, we will summarize the advances in identifying both maternal and parental factors that could induce haploid embryos and discuss the mechanism underlying these parental factors in haploid embryogenesis.
3.1. Paternal Players in Haploid Embryogenesis
Four paternal factors (MTL/PLA1/NLD, DMP, PLD3, and POD65) have been identified from maize (Figure 2b,c). RMZM2G47124, the first-identified maternal haploid-inducing gene cloned from qhir1, was named MATRILINEAL (MTL) [58], PHOSPHOLIPASE A1 (PLA1) [59], and NOT LIKE DAD (NLD) [60] by three different groups. MTL/PLA1/NLD (hereafter referred to as MTL) belongs to the phospholipase family and is an enzyme that hydrolyzes the phospholipids that function in membrane remodeling [61,62,63]. MTL is localized at the pollen endo-plasma membrane, a special plasma membrane that originates in the plasma membrane of vegetative cells and closely surrounds two sperm cells [64]. A 4 bp insertion leads to a frame-shift mutation in the MTL, which results in seed abortion and haploid induction. The second-identified maternal haploid inducer gene is ZmDMP, which encodes a DUF679 domain membrane protein and is specifically expressed in sperm cells. ZmDMP mutation induced an HIR of 0.1~0.3%, but it significantly increased HIR at a five-to-six-fold in combination of MTL mutation [65]. In addition to maize, recent studies have demonstrated that the role of DMP in haploid induction is conserved between monocots and eudicots, which has been achieved in A. thaliana, B. napus [66,67], B. oleracea [68], Medicago truncatula [69], Nicotiana tabacum [67,70], S. lycopersicum [71], and S. tuberosum [72]. In addition to MTL and DMP, vegetative cell-expressed ZmPLD3 and sperm cell-expressed ZmPOD65 were also shown to be able to induce haploids in maize [73,74]. ZmPLD3 belongs to phospholipase D (PLD) family, and it is localized in the ER, plastids, the Golgi apparatus, and the cytosol of vegetative cells [73], whereas ZmPOD65 encodes a peroxidase (POD) protein and is highly expressed in pollen at the tricellular stage [74].
3.2. Maternal Factor in Haploid Embryogenesis
In addition to the paternal factors in haploid embryogenesis, recent reports have demonstrated that egg cell-expressed maternal factors can also be used to induce haploid embryos. EGG CELL-SPECIFIC1/2 (ECS1/2) encodes egg cell-specifically expressed aspartic proteases, which are secreted to the synergid cell region upon fertilization to avoid polytubey by degrading the pollen tube attractant LURE1 [75]. Recently, two independent studies demonstrated that the mutation of ECS1 and ECS2 can also induce haploid embryogenesis [76,77]. Unfused sperm nuclei were observed in zygotes and early embryos, suggesting that karyogamy defects occurred in the sperm–egg fertilization, and the haploids from the ecs1 ecs2 mutant progenies may have resulted from the post-fertilization genomic elimination [77]. In summary, ECS-mediated haploid induction is likely caused by pseudogamy and potential post-fertilization genomic elimination (Figure 2d).
3.3. Synergistic Effects on Haploid Embryogenesis
Since a low efficiency of haploid embryogenesis is observed in most inducer lines, efficiency has become a major barrier for its application in crop breeding. To improve the efficiency of haploid embryogenesis, the combination of MTL and DMP [65] or MTL and PLD3 [73] was performed to test whether they could improve haploid production. The mutation of PLD3 or DMP in mtl mutant background could significantly increase its haploid induction rate, but the HIR was still lower than the expected. A dmp–mtl–pld3 triple mutant was also created to test whether it could increase the HIR. However, dmp–mtl–pld3 triple-homozygous plants could not be obtained, which was likely due to the pollen developmental defect or the fertilization defect in triple mutants. Hence, it is still worth testing other combinations of haploid inducer genes to improve the efficiency of haploid induction in further studies.
3.4. Mechanism for Haploid Embryogenesis
Two mechanisms have been proposed to explain the haploid formation in maize. The first is a single fertilization-induced haploid and the second is post-fertilization genome elimination. The former may produce haploid embryos through the parthenogenesis of the egg cell while the central cell fertilizes normally to form the endosperm. In the latter, double fertilization occurs normally, but the zygote undergoes uniparental genome elimination that results in haploid embryo formation.
The mechanisms for haploid embryogenesis are primarily focused on MTL-induced haploid embryogenesis, and whether it is conserved among different inducers, such as DMP, remains largely unknown. Several recent studies have demonstrated that MTL-induced haploids may form through post-fertilization genome elimination, rather than single fertilization-mediated parthenogenesis. The markers, including B chromosomes and CENH3-YFP derived from the paternal genome, were detected in haploid progenies [78], suggesting that the egg cells were fertilized successfully, and that uniparental genome elimination occurred during haploid induction. In addition, when WT pistils were pollinated by mtl mutant pollen grains (which were carrying the Cas9 and gRNA expression cassette) [79], genome-edited haploids without the Cas9 expression cassette were detected in the progenies. Since the CRISPR/Cas9 system only exists in the paternal genome, this result strongly suggests that the paternal genome is transmitted to the egg cell upon fertilization and is eventually eliminated after fertilization. A multi-omics analysis of mtl pollen grains revealed that ROS signals were involved in post-fertilization genome elimination [74]. Elevated ROS levels may cause DNA damage in pollen from the mtl mutant [80], which may lead to chromosome fragmentation in sperm cells and then induce genome elimination after fertilization. Spermatid chromosome fragmentation in the CAU5 haploid inducer line was detected through single nucleus sequencing [81]. In addition, the in vitro treatment of pollen grains with ROS-inducing reagents can also result in sperm DNA fragmentation and lead to the formation of haploids when pollinated to the WT plants. Studies on the sperm cell-expressed peroxidase gene ZmPOD65 further confirmed the role of ROS in haploid embryogenesis [74]. Peroxidases wildly exist in the plant kingdom [82], and they convert hydrogen peroxide (H2O2) into H2O during the POD catalytic reaction [83]. Therefore, POD65 may be functional in the removal of H2O2 in sperm cells, and POD65 mutation can cause H2O2 to burst in sperm cells, which leads to sperm DNA fragmentation and, eventually, haploid production.
In summary, vegetative cell-expressed MTL and sperm cell-expressed POD65 are involved in the regulation of ROS levels, as well as the expression of ROS-related genes in pollen grains. Mutations in MTL and POD65 do not appear to impair fertilization, but the zygotes generated from the cross between mtl or pod65 mutants and WT plants will undergo genome elimination to form haploid embryos (Figure 2b). An MTL mutation may cause an imbalance in the hydrolyzed phospholipids (such as PC) between sperm cells and vegetative cells, resulting in the over-accumulation of PCs in sperm cells, which disrupts mitochondrial homeostasis and leads to increased ROS levels [84]. Elevated ROS levels in sperm cells will induce DNA damage and impair the expression of ROS-related genes, leading to the chromatin fragmentation. The fragmented chromatins of sperm cells are then eliminated after fertilization, leading to the formation of haploid embryos. This genome elimination mechanism is different from that in CENH3-mediated haploid embryogenesis. In the MTL- or POD65-induced haploid embryos, chromosome fragmentation occurs before fertilization and the fragmented paternal genome cannot function properly after fertilization and is eventually eliminated, whereas the CENH3-induced post-fertilization genome elimination is largely due to the incompatibility of the parental genome.
4. Transcription Factors Triggered Haploid Embryogenesis
Normal zygotic embryogenesis is triggered by the fusion of a sperm cell and an egg cell. However, recent reports have demonstrated that egg cells can bypass fertilization to trigger embryogenesis by expressing the transcriptional factors BABY BOOM (BBM) or PARTHENOGENESIS (PAR). BBM, an AINTEGUMENTA-like (AIL) transcription factor, belongs to the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) family, which was previously shown to play a critical role in inducing or enhancing somatic embryogenesis [85,86,87,88,89,90,91,92]. In addition to triggering somatic embryogenesis, recent studies have demonstrated that the ectopic expression of BBM in egg cells can trigger egg cells to initiate embryogenesis in multiple sexual plants [93,94,95,96,97,98,99], indicating the conserved role of BBM in promoting embryogenesis. For example, in rice, the ectopic expression of BBM1 in egg cells can produce embryo-like structures without fertilization [96]. An expression pattern analysis revealed that OsBBM1 is expressed in sperm cells, and only the paternal allele of OsBBM1 is expressed in 2.5 HAP (hours after pollination) zygotes. Both the maternal and paternal transcripts could be detected in 6.5 HAP zygotes, possibly due to the auto-activation ability of BBM. Recently, the ectopic expression of BnBBM in the egg cells of A. thaliana, B. napus, and S. lycopersicon also bypassed fertilization for embryogenesis [99], although how BnBBM triggered the egg cells to initiate the embryo program remains unknown. Besides BBM, PAR has also been reported to be able to induce parthenogenesis in sexual plants (Figure 2e). PAR encodes a zinc finger domain protein with an EAR motif, which may function as a transcription factor in dandelion (Taraxacum officinale). The ectopic expression of ToPAR in an egg cell can also promote that egg cell to produce embryo-like structures without fertilization in sexually reproductive lettuce [100]. There are two PAR homologs, named DAZ3 and TREE1, in the Arabidopsis genome. It is worth investigating whether the homologs of PAR can also trigger egg cells to initiate embryogenesis in other plants, especially in crops.
5. Application of Haploid Embryogenesis
5.1. Haploid Breeding
The main application of haploid embryogenesis is to generate haploid plants which can accelerate the gain of homozygous plants and efficiently shorten breeding times. Most haploid induction systems have been tested in multiple crops (Table 1). The CENH3-mediated haploid induction system can be used to generate both maternal and paternal haploids, whereas the other haploid induction systems discussed above can only produce maternal haploids. The CENH3-mediated paternal haploid induction method can also be used to introduce a nuclear genome of interest into the targeted cytoplasm, such as the cytoplasmic male sterile (CMS) line. Therefore, the CENH3-mediated haploid induction system is also a useful tool for the establishment or improvement of the CMS line for hybrid seed production. However, CENH3-mediated haploid induction systems have not been successfully used in crop breeding [49], primarily due to the low HIR in crops and the complex steps required to construct CENH3 induction lines. In addition, the MTL-mediated haploid induction system has only been validated in monocots [58,59,60,79,80,101,102,103,104], while the DMP-mediated haploid induction system has been confirmed in both monocots and eudicots [65,66,67,68,69,70,71,72,105]. Hence, it is worth optimizing the haploid induction systems to promote their application in crop breeding in future studies.

Table 1.
List of in vivo haploid systems tested in various species.
5.2. Genome Editing
In addition to haploid breeding, a haploid induction system combined with CRISPR/Cas9 technology could be used for genome editing in crops that are resistant to genetic transformation. Gene-editing strategies, named haploid induction editing technology (HI-EDIT) and haploid inducer mediated genome editing (IMGE), have been developed based on in vivo haploid induction systems, respectively [79,108]. In the HI-EDIT system, CRISPR-Cas9 technology was combined with the MTL-mediated or CENH3-mediated haploid induction system to create a one-step genome editing method. Targeted genome loci could be efficiently edited in the haploid progenies and steadily inherited in the next generation, and at the same time, the Cas9 construct could be completely removed through genome elimination. This method is based on the post-fertilization genome elimination of the haploid induction systems. In maize, when pollinated by mtl pollen grains that carry the Cas9 construct, five of six maize germplasms received a more than 3% editing efficiency increase in the haploids via the HI-EDIT method. In Arabidopsis, a CENH3 haploid induction system, in combination with CRISPR-Cas9 technology, was developed for the HI-EDIT method. The results demonstrated that when Arabidopsis Landsberg erecta pollen grains pollinated to the Arabidopsis cenh3-1 mutant (Columbia ecotype) expressing CENH3 from maize and the Cas9 construct [109], 16.9% of the targeted genes in the haploid progenies were edited [79]. Thus, the HI-Edit method works effectively in both monocots and eudicots, which can provide a power tool for wide applications in genome-editing technology of the commercial variety. Similarly, Baobao Wang et al. [108] developed an approach, named IMGE, for genome editing. They introduced a CRISPR/Cas9 cassette into the CAU5 inducer line and tested its ability for genome editing in haploids, and they reported an approximately 4.1% increase in editing efficiency in the haploids. Hence, HI-EDIT and IMGE can be efficiently used for genome editing in crops that are resistant to genetic transformation, and their use avoids the interference of transgenes on crop traits through uniparental chromosome elimination.
5.3. Heterosis Fixation
Another application of haploid embryogenesis is for heterosis fixation, when used in combination with the MiMe (Mitosis instead of Meiosis) system [110,111,112]. In MiMe system, the combination of the mutations in the genes which are responsible for abolishing meiotic recombination, separating the sister chromatids and skipping the second division during meiosis, respectively, will shift meiosis into a mitosis-like division [113,114]. Since MiMe gametes are diploid, the introduction of a haploid induction system into the MiMe background can produce clonal propagation seeds with hybrid genotypes [96,102,115]. For example, osd1–pair1–rec8–mtl quadruple mutants (named Fix, for Fixation of hybrids) were used for heterosis fixation in rice [102]. Approximately 6.2% of the Fix diploid progenies with the same genotype as the mother were obtained, indicating that the Fix system can produce clonal seeds and fix heterozygosity in the F1 generation of hybrid rice. Recent results have demonstrated that the heterotic phenotypes and synthetic apomixis traits of these clonal seeds could be stably transmitted to the next generation [116]. Similarly, the MiMe system, in combination with the ectopic expression of BBM1 in egg cells, was also successfully used for heterosis fixation in rice, and clonal seeds were obtained from 11% and 29% of the progeny of the two transgenic lines of the MiMe plus BBM1-ee (ectopic expression of OsBBM1 in egg cells) constructs [96], respectively. In further studies, more attention should be paid to increasing the efficiency of clonal seeds.
6. Conclusions and Perspectives
There are two alternative roads for generating haploid embryos, as described above. Microspore embryogenesis can not only be applied for haploid breeding, but it also is an ideal in vitro system for investigating cell fate determination and plant embryogenesis. Epigenetic reprogramming is critical for the initiation of microspore embryogenesis, which appears to be similar to somatic embryogenesis. It worth comparing the mechanisms behinds the different roads to embryogenesis, including zygotic embryogenesis, microspore embryogenesis, and somatic embryogenesis, in further studies. In addition, as the single-cell epigenome sequencing technologies develop, profiling DNA modifications and histone modifications during the initial stages of microspore embryogenesis will greatly aid us in understanding the mechanisms behind microspore reprogramming and haploid embryogenesis initiation.
The studies of in vivo haploid embryogenesis in flowering plants greatly promote the use of haploid technology in plant breeding, especially for maize. Based on the current in vivo haploid induction system discussed above, haploid embryogenesis may be induced by uniparental centromere defects, sperm chromosome fragmentation, and the ectopic expression of transcriptional factors related to embryo initiation in egg cells. These genes responsible for haploid induction are closely related to plant reproduction and, in particular, gamete development and fertilization. For example, MTL, POD65, DMP, and PLD3 are involved male gametophyte development, ECS is involved in egg cell development, and BBM and PAR are involved in embryogenesis initiation. In addition to haploid embryogenesis, sperm-central cell fertilization to form endosperm is necessary for haploid generation. Thus, basic studies related to the molecular and cellular mechanisms for gamete development, fertilization, and embryogenesis initiation will greatly aid us in establishing or optimizing haploid indcution systems.
At present, there are still several questions that remain to be answered about these haploid induction systems. First, a high haploid induction rate is critical for their applications in plant breeding. Presently, in vivo haploid induction systems are typically accompanied by a high frequency of seed abortion. Reducing seed abortion and increasing haploid induction efficiency are key points for the improvement of in vivo haploid induction systems. Second, haploid induction systems have been established for only a few species, primarily for grass and Arabidopsis. Expanding these haploid induction systems to other important economic crops must be investigated with great care. Thus, more attention should be paid to the mechanisms of haploid embryogenesis, which will not only aid us in understanding haploid embryo formation, but it will also help us to optimize these haploid induction systems and expand the applications of haploid technology in crop breeding.
Author Contributions
Writing—original draft preparation, K.S., M.Q. and P.Z.; writing—review and editing, K.S. and P.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Major Project of Hubei Hongshan Laboratory, grant number 2022hszd017; the Science and Technology Department of Hubei Province, grant number 2022CFA071 and the National Natural Science Foundation of China, grant number 31970340.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Meng, D.; Liu, C.; Chen, S.; Jin, W. Haploid induction and its application in maize breeding. Mol. Breed. 2021, 41, 20. [Google Scholar] [CrossRef]
- Zhang, F.L.; Takahata, Y. Inheritance of microspore embryogenic ability in Brassica crops. Theor. Appl. Genet. 2001, 103, 254–258. [Google Scholar] [CrossRef]
- Satpute, G.K.; Long, H.; Seguí-Simarro, J.M.; Risueño, M.C.; Testillano, P.S. Cell architecture during gametophytic and embryogenic microspore development in Brassica napus L. Acta Physiol. Plant. 2005, 27, 665–674. [Google Scholar] [CrossRef]
- Shariatpanahi, M.E.; Bal, U.; Heberle-Bors, E.; Touraev, A. Stresses applied for the re-programming of plant microspores towards in vitro embryogenesis. Physiol. Plant. 2006, 127, 519–534. [Google Scholar] [CrossRef]
- Tang, X.C.; He, Y.Q.; Wang, Y.; Sun, M.X. The role of arabinogalactan proteins binding to Yariv reagents in the initiation, cell developmental fate, and maintenance of microspore embryogenesis in Brassica napus L. cv. Topas. J. Exp. Bot. 2006, 57, 2639–2650. [Google Scholar] [CrossRef]
- Germanà, M.A. Anther culture for haploid and doubled haploid production. Plant Cell Tissue Organ Cult. 2010, 104, 283–300. [Google Scholar] [CrossRef]
- Prem, D.; Solis, M.T.; Barany, I.; Rodriguez-Sanz, H.; Risueno, M.C.; Testillano, P.S. A new microspore embryogenesis system under low temperature which mimics zygotic embryogenesis initials, expresses auxin and efficiently regenerates doubled-haploid plants in Brassica napus. BMC Plant Biol. 2012, 12, 127. [Google Scholar] [CrossRef]
- Rivas-Sendra, A.; Campos-Vega, M.; Calabuig-Serna, A.; Seguí-Simarro, J.M. Development and characterization of an eggplant (Solanum melongena) doubled haploid population and a doubled haploid line with high androgenic response. Euphytica 2017, 213, 89. [Google Scholar] [CrossRef]
- Testillano, P.; Georgiev, S.; Mogensen, H.L.; Coronado, M.J.; Dumas, C.; Risueno, M.C.; Matthys-Rochon, E. Spontaneous chromosome doubling results from nuclear fusion during in vitro maize induced microspore embryogenesis. Chromosoma 2004, 112, 342–349. [Google Scholar] [CrossRef][Green Version]
- Pintos, B.; Manzanera, J.A.; Bueno, M.A. Antimitotic agents increase the production of doubled-haploid embryos from cork oak anther culture. J. Plant Physiol. 2007, 164, 1595–1604. [Google Scholar] [CrossRef]
- Tang, X.; Liu, Y.; He, Y.; Ma, L.; Sun, M.X. Exine dehiscing induces rape microspore polarity, which results in different daughter cell fate and fixes the apical-basal axis of the embryo. J. Exp. Bot. 2013, 64, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Testillano, P.S. Microspore embryogenesis: Targeting the determinant factors of stress-induced cell reprogramming for crop improvement. J. Exp. Bot. 2019, 70, 2965–2978. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Soriano, M.; Cordewener, J.; Muino, J.M.; Riksen, T.; Fukuoka, H.; Angenent, G.C.; Boutilier, K. The histone deacetylase inhibitor trichostatin a promotes totipotency in the male gametophyte. Plant Cell 2014, 26, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Y.; Gao, Y.; Jiang, X.; Zhang, M.; Wu, H.; Liu, Z.; Feng, H. Effects of histone deacetylase inhibitors on microspore embryogenesis and plant regeneration in Pakchoi (Brassica rapa ssp. chinensis L.). Sci. Hortic. 2016, 209, 61–66. [Google Scholar] [CrossRef]
- Jiang, F.; Ryabova, D.; Diedhiou, J.; Hucl, P.; Randhawa, H.; Marillia, E.F.; Foroud, N.A.; Eudes, F.; Kathiria, P. Trichostatin A increases embryo and green plant regeneration in wheat. Plant Cell Rep. 2017, 36, 1701–1706. [Google Scholar] [CrossRef]
- Castillo, A.M.; Valero-Rubira, I.; Burrell, M.A.; Allue, S.; Costar, M.A.; Valles, M.P. Trichostatin A Affects Developmental Reprogramming of Bread Wheat Microspores towards an Embryogenic Route. Plants 2020, 9, 1442. [Google Scholar] [CrossRef]
- Pandey, P.; Daghma, D.S.; Houben, A.; Kumlehn, J.; Melzer, M.; Rutten, T. Dynamics of post-translationally modified histones during barley pollen embryogenesis in the presence or absence of the epi-drug trichostatin A. Plant Reprod. 2017, 30, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Nowicka, A.; Juzon, K.; Krzewska, M.; Dziurka, M.; Dubas, E.; Kopec, P.; Zielinski, K.; Zur, I. Chemically-induced DNA de-methylation alters the effectiveness of microspore embryogenesis in triticale. Plant Sci. 2019, 287, 110189. [Google Scholar] [CrossRef]
- Berenguer, E.; Barany, I.; Solis, M.T.; Perez-Perez, Y.; Risueno, M.C.; Testillano, P.S. Inhibition of Histone H3K9 Methylation by BIX-01294 Promotes Stress-Induced Microspore Totipotency and Enhances Embryogenesis Initiation. Front. Plant Sci. 2017, 8, 1161. [Google Scholar] [CrossRef]
- Solis, M.T.; Rodriguez-Serrano, M.; Meijon, M.; Canal, M.J.; Cifuentes, A.; Risueno, M.C.; Testillano, P.S. DNA methylation dynamics and MET1a-like gene expression changes during stress-induced pollen reprogramming to embryogenesis. J. Exp. Bot. 2012, 63, 6431–6444. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Britt, A.B.; Tripathi, L.; Sharma, S.; Upadhyaya, H.D.; Ortiz, R. Haploids: Constraints and opportunities in plant breeding. Biotechnol. Adv. 2015, 33, 812–829. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.; Ferrie, A.M.R.; Chellamma, S.; Samuel, J.P.; Phillips, G.C. Androgenesis-Based Doubled Haploidy: Past, Present, and Future Perspectives. Front. Plant Sci. 2021, 12, 751230. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhou, X.M.; Zhao, L.L.; Cheung, A.Y.; Sun, M.X. Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy 2020, 16, 2180–2192. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhuang, M.; Xu, X.; Li, S.; Yang, M.; Li, N.; Du, X.; Hu, K.; Peng, X.; Huang, W.; et al. Autophagy and its mediated mitochondrial quality control maintain pollen tube growth and male fertility in Arabidopsis. Autophagy 2022, 1–16. [Google Scholar] [CrossRef]
- Corral-Martínez, P.; Parra-Vega, V.; Seguí-Simarro, J.M. Novel features of Brassica napus embryogenic microspores revealed by high pressure freezing and freeze substitution: Evidence for massive autophagy and excretion-based cytoplasmic cleaning. J. Exp. Bot. 2013, 64, 3061–3075. [Google Scholar] [CrossRef]
- Barany, I.; Berenguer, E.; Solis, M.T.; Perez-Perez, Y.; Santamaria, M.E.; Crespo, J.L.; Risueno, M.C.; Diaz, I.; Testillano, P.S. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 2018, 69, 1387–1402. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, E.; Minina, E.A.; Carneros, E.; Bárány, I.; Bozhkov, P.V.; Testillano, P.S. Suppression of Metacaspase- and Autophagy-Dependent Cell Death Improves Stress-Induced Microspore Embryogenesis in Brassica napus. Plant Cell Physiol. 2021, 61, 2097–2110. [Google Scholar] [CrossRef]
- Rodriguez-Serrano, M.; Barany, I.; Prem, D.; Coronado, M.J.; Risueno, M.C.; Testillano, P.S. NO, ROS, and cell death associated with caspase-like activity increase in stress-induced microspore embryogenesis of barley. J. Exp. Bot. 2012, 63, 2007–2024. [Google Scholar] [CrossRef]
- Zur, I.; Dubas, E.; Golemiec, E.; Szechynska-Hebda, M.; Golebiowska, G.; Wedzony, M. Stress-related variation in antioxidative enzymes activity and cell metabolism efficiency associated with embryogenesis induction in isolated microspore culture of triticale (x Triticosecale Wittm.). Plant Cell Rep. 2009, 28, 1279–1287. [Google Scholar] [CrossRef]
- Wieland, G.; Orthaus, S.; Ohndorf, S.; Diekmann, S.; Hemmerich, P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell. Biol. 2004, 24, 6620–6630. [Google Scholar] [CrossRef]
- Talbert, P.B.; Masuelli, R.; Tyagi, A.P.; Comai, L.; Henikoff, S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 2002, 14, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Lermontova, I.; Koroleva, O.; Rutten, T.; Fuchs, J.; Schubert, V.; Moraes, I.; Koszegi, D.; Schubert, I. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 2011, 68, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Shibata, F.; Ramahi, J.S.; Nagaki, K.; Chen, C.; Murata, M.; Chan, S.W. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 2011, 7, e1002121. [Google Scholar] [CrossRef] [PubMed]
- Sanei, M.; Pickering, R.; Kumke, K.; Nasuda, S.; Houben, A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. USA 2011, 108, E498–E505. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, T.; Earnshaw, W.C. The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 2014, 30, 496–508. [Google Scholar] [CrossRef]
- Ishii, T.; Karimi-Ashtiyani, R.; Houben, A. Haploidization via Chromosome Elimination: Means and Mechanisms. Annu. Rev. Plant Biol. 2016, 67, 421–438. [Google Scholar] [CrossRef]
- Comai, L.; Maheshwari, S.; Marimuthu, M.P.A. Plant centromeres. Curr. Opin. Plant Biol. 2017, 36, 158–167. [Google Scholar] [CrossRef]
- Wang, N.; Dawe, R.K. Centromere Size and Its Relationship to Haploid Formation in Plants. Mol. Plant 2018, 11, 398–406. [Google Scholar] [CrossRef]
- Kuppu, S.; Ron, M.; Marimuthu, M.P.A.; Li, G.; Huddleson, A.; Siddeek, M.H.; Terry, J.; Buchner, R.; Shabek, N.; Comai, L.; et al. A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol. J. 2020, 18, 2068–2080. [Google Scholar] [CrossRef]
- Ravi, M.; Kwong, P.N.; Menorca, R.M.; Valencia, J.T.; Ramahi, J.S.; Stewart, J.L.; Tran, R.K.; Sundaresan, V.; Comai, L.; Chan, S.W. The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics 2010, 186, 461–471. [Google Scholar] [CrossRef]
- Maheshwari, S.; Tan, E.H.; West, A.; Franklin, F.C.; Comai, L.; Chan, S.W. Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 2015, 11, e1004970. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Ashtiyani, R.; Ishii, T.; Niessen, M.; Stein, N.; Heckmann, S.; Gurushidze, M.; Banaei-Moghaddam, A.M.; Fuchs, J.; Schubert, V.; Koch, K.; et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. USA 2015, 112, 11211–11216. [Google Scholar] [CrossRef] [PubMed]
- Kuppu, S.; Tan, E.H.; Nguyen, H.; Rodgers, A.; Comai, L.; Chan, S.W.; Britt, A.B. Point Mutations in Centromeric Histone Induce Post-zygotic Incompatibility and Uniparental Inheritance. PLoS Genet. 2015, 11, e1005494. [Google Scholar] [CrossRef]
- Britt, A.B.; Kuppu, S. Cenh3: An Emerging Player in Haploid Induction Technology. Front. Plant Sci. 2016, 7, 357. [Google Scholar] [CrossRef]
- Watts, A.; Kumar, V.; Bhat, S.R. Centromeric histone H3 protein: From basic study to plant breeding applications. J. Plant Biochem. Biotechnol. 2016, 25, 339–348. [Google Scholar] [CrossRef]
- Ren, J.; Wu, P.; Trampe, B.; Tian, X.; Lubberstedt, T.; Chen, S. Novel technologies in doubled haploid line development. Plant Biotechnol. J. 2017, 15, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Kursel, L.E.; Malik, H.S. The cellular mechanisms and consequences of centromere drive. Curr. Opin. Cell Biol. 2018, 52, 58–65. [Google Scholar] [CrossRef]
- Comai, L.; Tan, E.H. Haploid Induction and Genome Instability. Trends Genet. 2019, 35, 791–803. [Google Scholar] [CrossRef]
- Kalinowska, K.; Chamas, S.; Unkel, K.; Demidov, D.; Lermontova, I.; Dresselhaus, T.; Kumlehn, J.; Dunemann, F.; Houben, A. State-of-the-art and novel developments of in vivo haploid technologies. Theor. Appl. Genet. 2019, 132, 593–605. [Google Scholar] [CrossRef]
- Wang, S.; Jin, W.; Wang, K. Centromere histone H3- and phospholipase-mediated haploid induction in plants. Plant Methods 2019, 15, 42. [Google Scholar] [CrossRef]
- Karimi-Ashtiyani, R. Centromere engineering as an emerging tool for haploid plant production: Advances and challenges. Doubled Haploid Technol. 2021, 2289, 3–22. [Google Scholar]
- Zhou, J.; Liu, Y.; Guo, X.; Birchler, J.A.; Han, F.; Su, H. Centromeres: From chromosome biology to biotechnology applications and synthetic genomes in plants. Plant Biotechnol. J. 2022, 20, 2051–2063. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Chan, S.W. Haploid plants produced by centromere-mediated genome elimination. Nature 2010, 464, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, M.P.A.; Maruthachalam, R.; Bondada, R.; Kuppu, S.; Tan, E.H.; Britt, A.; Chan, S.W.L.; Comai, L. Epigenetically mismatched parental centromeres trigger genome elimination in hybrids. Sci. Adv. 2021, 7, eabk1151. [Google Scholar] [CrossRef]
- Wang, N.; Gent, J.I.; Dawe, R.K. Haploid induction by a maize cenh3 null mutant. Sci. Adv. 2021, 7, eabe2299. [Google Scholar] [CrossRef]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Coe, E.H. A Line of Maize with High Haploid Frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Meng, D.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L.; et al. A 4-bp Insertion at ZmPLA1 Encoding a Putative Phospholipase A Generates Haploid Induction in Maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef]
- Gilles, L.M.; Khaled, A.; Laffaire, J.B.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Berges, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef]
- Wang, G.; Ryu, S.; Wang, X. Plant Phospholipases: An Overview. Methods Mol. Biol. 2012, 861, 123–137. [Google Scholar] [PubMed]
- Aloulou, A.; Rahier, R.; Arhab, Y.; Noiriel, A.; Abousalham, A. Phospholipases: An overview. Lipases Phospholipases 2018, 1835, 69–105. [Google Scholar]
- Ali, U.; Lu, S.; Fadlalla, T.; Iqbal, S.; Yue, H.; Yang, B.; Hong, Y.; Wang, X.; Guo, L. The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses. Prog. Lipid Res. 2022, 86, 101158. [Google Scholar] [CrossRef] [PubMed]
- Gilles, L.M.; Calhau, A.R.M.; La Padula, V.; Jacquier, N.M.A.; Lionnet, C.; Martinant, J.P.; Rogowsky, P.M.; Widiez, T. Lipid anchoring and electrostatic interactions target NOT-LIKE-DAD to pollen endo-plasma membrane. J. Cell Biol. 2021, 220, e202010077. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Xiao, Q.; Wang, H.; Wen, J.; Tu, J.; Shen, J.; Fu, T.; Yi, B. An in planta haploid induction system in Brassica napus. J. Integr. Plant Biol. 2022, 64, 1140–1144. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Y.; Chen, B.; Liu, J.; Wang, D.; Li, M.; Qi, X.; Liu, C.; Boutilier, K.; Chen, S. Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum. J. Integr. Plant Biol. 2022, 64, 1281–1294. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, K.; Liu, Y.; Zhang, N.; Yang, L.; Zhang, Y.; Wang, Y.; Ji, J.; Fang, Z.; Han, F.; et al. In vivo maternal haploid induction based on genome editing of DMP in Brassica oleracea. Plant Biotechnol. J. 2022, 20, 2242–2244. [Google Scholar] [CrossRef]
- Wang, N.; Xia, X.; Jiang, T.; Li, L.; Zhang, P.; Niu, L.; Cheng, H.; Wang, K.; Lin, H. In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol. J. 2022, 20, 22–24. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Zhang, J.; Jia, M.; Cao, L.; Yu, J.; Zhao, D. Haploid induction in allotetraploid tobacco using DMPs mutation. Planta 2022, 255, 98. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, B.; Wang, D.; Zhu, X.; Li, M.; Zhang, J.; Chen, M.; Wang, M.; Riksen, T.; Liu, J.; et al. In vivo maternal haploid induction in tomato. Plant Biotechnol. J. 2022, 20, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yin, J.; Luo, J.; Tang, D.; Zhu, X.; Wang, J.; Liu, Z.; Wang, P.; Zhong, Y.; Liu, C.; et al. Construction of homozygous diploid potato through maternal haploid induction. aBIOTECH 2022, 3, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, Z.; Yue, Y.; Zhao, H.; Fei, X.; Li, E.; Liu, C.; Chen, S.; Lai, J.; Song, W. Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants 2021, 7, 1579–1588. [Google Scholar] [CrossRef]
- Jiang, C.; Sun, J.; Li, R.; Yan, S.; Chen, W.; Guo, L.; Qin, G.; Wang, P.; Luo, C.; Huang, W.; et al. A reactive oxygen species burst causes haploid induction in maize. Mol. Plant 2022, 15, 943–955. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, X.; Zhao, P.; Peng, X.; Chen, H.; Bleckmann, A.; Bazhenova, A.; Shi, C.; Dresselhaus, T.; Sun, M.X. Fertilized egg cells secrete endopeptidases to avoid polytubey. Nature 2021, 592, 433–437. [Google Scholar] [CrossRef]
- Mao, Y.; Nakel, T.; Serbes, I.E.; Tekleyohans, D.G.; Joshi, S.; Baum, T.; Groß-Hardt, R. ECS1 and ECS2 regulate polyspermy and suppress the formation of haploid plants by promoting double fertilization. BioRxiv 2022. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, C.; Li, s.; Zhang, B.; Dresselhaus, T.; Sun, M.-x. A female in vivo haploid-induction system via mutagenesis of egg cell-specific peptidases. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, X.; Xie, H.; Chen, S.; Jin, W. Fertilization and uniparental chromosome elimination during crosses with maize haploid inducers. Plant Physiol. 2013, 163, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Geng, S.; Zhang, H.; Jia, M.; Wang, Z.; Deng, Z.; Tao, S.; Liao, R.; Wang, F.; Kong, X.; et al. Matrilineal empowers wheat pollen with haploid induction potency by triggering postmitosis reactive oxygen species activity. New Phytol. 2022, 233, 2405–2414. [Google Scholar] [CrossRef]
- Li, X.; Meng, D.; Chen, S.; Luo, H.; Zhang, Q.; Jin, W.; Yan, J. Single nucleus sequencing reveals spermatid chromosome fragmentation as a possible cause of maize haploid induction. Nat. Commun. 2017, 8, 991. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Dunand, C. Specific functions of individual class III peroxidase genes. J. Exp. Bot. 2009, 60, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Kaothien, P.; Matsui, T.; Kawaoka, A.; Shinmyo, A. Molecular biology and application of plant peroxidase genes. Appl. Microbiol. Biotechnol. 2003, 60, 665–670. [Google Scholar] [CrossRef]
- Ruban, A.; Houben, A. Highly reactive chemicals meet haploidization. Mol. Plant 2022, 15, 937–939. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbe, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Irikova, T.; Grozeva, S.; Denev, I. Identification of BABY BOOM and LEAFY COTYLEDON genes in sweet pepper (Capsicum annuum L.) genome by their partial gene sequences. Plant Growth Regul. 2012, 67, 191–198. [Google Scholar] [CrossRef]
- Salvo, S.A.; Hirsch, C.N.; Buell, C.R.; Kaeppler, S.M.; Kaeppler, H.F. Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS ONE 2014, 9, e111407. [Google Scholar] [CrossRef]
- Florez, S.L.; Erwin, R.L.; Maximova, S.N.; Guiltinan, M.J.; Curtis, W.R. Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol. 2015, 15, 121. [Google Scholar] [CrossRef]
- Lutz, K.A.; Martin, C.; Khairzada, S.; Maliga, P. Steroid-inducible BABY BOOM system for development of fertile Arabidopsis thaliana plants after prolonged tissue culture. Plant Cell Rep. 2015, 34, 1849–1856. [Google Scholar] [CrossRef]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM Transcription Factor Activates the LEC1-ABI3-FUS3-LEC2 Network to Induce Somatic Embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Kumar, V. BABY BOOM (BBM): A candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 2018, 40, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Podio, M.; Ozias-Akins, P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod. 2017, 30, 41–52. [Google Scholar] [CrossRef]
- Bui, L.T.; Pandzic, D.; Youngstrom, C.E.; Wallace, S.; Irish, E.E.; Szovenyi, P.; Cheng, C.L. A fern AINTEGUMENTA gene mirrors BABY BOOM in promoting apogamy in Ceratopteris richardii. Plant J. 2017, 90, 122–132. [Google Scholar] [CrossRef]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef]
- Zhang, Z.; Conner, J.; Guo, Y.; Ozias-Akins, P. Haploidy in Tobacco Induced by PsASGR-BBML Transgenes via Parthenogenesis. Genes 2020, 11, 1072. [Google Scholar] [CrossRef]
- Chahal, L.S.; Conner, J.A.; Ozias-Akins, P. Phylogenetically Distant BABY BOOM Genes from Setaria italica Induce Parthenogenesis in Rice. Front. Plant Sci. 2022, 13, 863908. [Google Scholar] [CrossRef]
- Chen, B.; Maas, L.; Figueiredo, D.; Zhong, Y.; Reis, R.; Li, M.; Horstman, A.; Riksen, T.; Weemen, M.; Liu, H.; et al. BABY BOOM regulates early embryo and endosperm development. Proc. Natl. Acad. Sci. USA 2022, 119, e2201761119. [Google Scholar] [CrossRef]
- Underwood, C.J.; Vijverberg, K.; Rigola, D.; Okamoto, S.; Oplaat, C.; Camp, R.; Radoeva, T.; Schauer, S.E.; Fierens, J.; Jansen, K.; et al. A PARTHENOGENESIS allele from apomictic dandelion can induce egg cell division without fertilization in lettuce. Nat. Genet. 2022, 54, 84–93. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R.; et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 2019, 37, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhong, Y.; Qi, X.; Chen, M.; Liu, Z.; Chen, C.; Tian, X.; Li, J.; Jiao, Y.; Wang, D.; et al. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol. J. 2020, 18, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Sun, Y.; Yang, S.; Zhi, H.; Yin, T.; Ma, X.; Zhang, H.; Diao, X.; Guo, Y.; Li, X.; et al. Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol. J. 2021, 19, 1089–1091. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, B.; Li, M.; Wang, D.; Jiao, Y.; Qi, X.; Wang, M.; Liu, Z.; Chen, C.; Wang, Y.; et al. A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat. Plants 2020, 6, 466–472. [Google Scholar] [CrossRef]
- Ravi, M.; Marimuthu, M.P.; Tan, E.H.; Maheshwari, S.; Henry, I.M.; Marin-Rodriguez, B.; Urtecho, G.; Tan, J.; Thornhill, K.; Zhu, F.; et al. A haploid genetics toolbox for Arabidopsis thaliana. Nat. Commun. 2014, 5, 5334. [Google Scholar] [CrossRef] [PubMed]
- Kelliher, T.; Starr, D.; Wang, W.; McCuiston, J.; Zhong, H.; Nuccio, M.L.; Martin, B. Maternal Haploids Are Preferentially Induced by CENH3-tailswap Transgenic Complementation in Maize. Front. Plant Sci. 2016, 7, 414. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhu, L.; Zhao, B.; Zhao, Y.; Xie, Y.; Zheng, Z.; Li, Y.; Sun, J.; Wang, H. Development of a Haploid-Inducer Mediated Genome Editing System for Accelerating Maize Breeding. Mol. Plant 2019, 12, 597–602. [Google Scholar] [CrossRef]
- Feng, C.; Yuan, J.; Bai, H.; Liu, Y.; Su, H.; Liu, Y.; Shi, L.; Gao, Z.; Birchler, J.A.; Han, F. The deposition of CENH3 in maize is stringently regulated. Plant J. 2020, 102, 6–17. [Google Scholar] [CrossRef]
- Jacquier, N.M.A.; Gilles, L.M.; Pyott, D.E.; Martinant, J.P.; Rogowsky, P.M.; Widiez, T. Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 2020, 6, 610–619. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; Conner, J.A. Clonal Reproduction through Seeds in Sight for Crops. Trends Genet. 2020, 36, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Underwood, C.J.; Mercier, R. Engineering Apomixis: Clonal Seeds Approaching the Fields. Annu. Rev. Plant. Biol. 2022, 73, 201–225. [Google Scholar] [CrossRef] [PubMed]
- d’Erfurth, I.; Jolivet, S.; Froger, N.; Catrice, O.; Novatchkova, M.; Mercier, R. Turning meiosis into mitosis. PLoS Biol. 2009, 7, e1000124. [Google Scholar] [CrossRef] [PubMed]
- Mieulet, D.; Jolivet, S.; Rivard, M.; Cromer, L.; Vernet, A.; Mayonove, P.; Pereira, L.; Droc, G.; Courtois, B.; Guiderdoni, E.; et al. Turning rice meiosis into mitosis. Cell Res. 2016, 26, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, M.P.; Jolivet, S.; Ravi, M.; Pereira, L.; Davda, J.N.; Cromer, L.; Wang, L.; Nogue, F.; Chan, S.W.; Siddiqi, I.; et al. Synthetic clonal reproduction through seeds. Science 2011, 331, 876. [Google Scholar] [CrossRef]
- Liu, C.; He, Z.; Zhang, Y.; Hu, F.; Li, M.; Liu, Q.; Huang, Y.; Wang, J.; Zhang, W.; Wang, C.; et al. Synthetic apomixis enables stable transgenerational transmission of heterotic phenotypes in hybrid rice. Plant Commun. 2022, 4, 100470. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).