The Glycine-Rich RNA-Binding Protein Is a Vital Post-Transcriptional Regulator in Crops
Abstract
1. Introduction
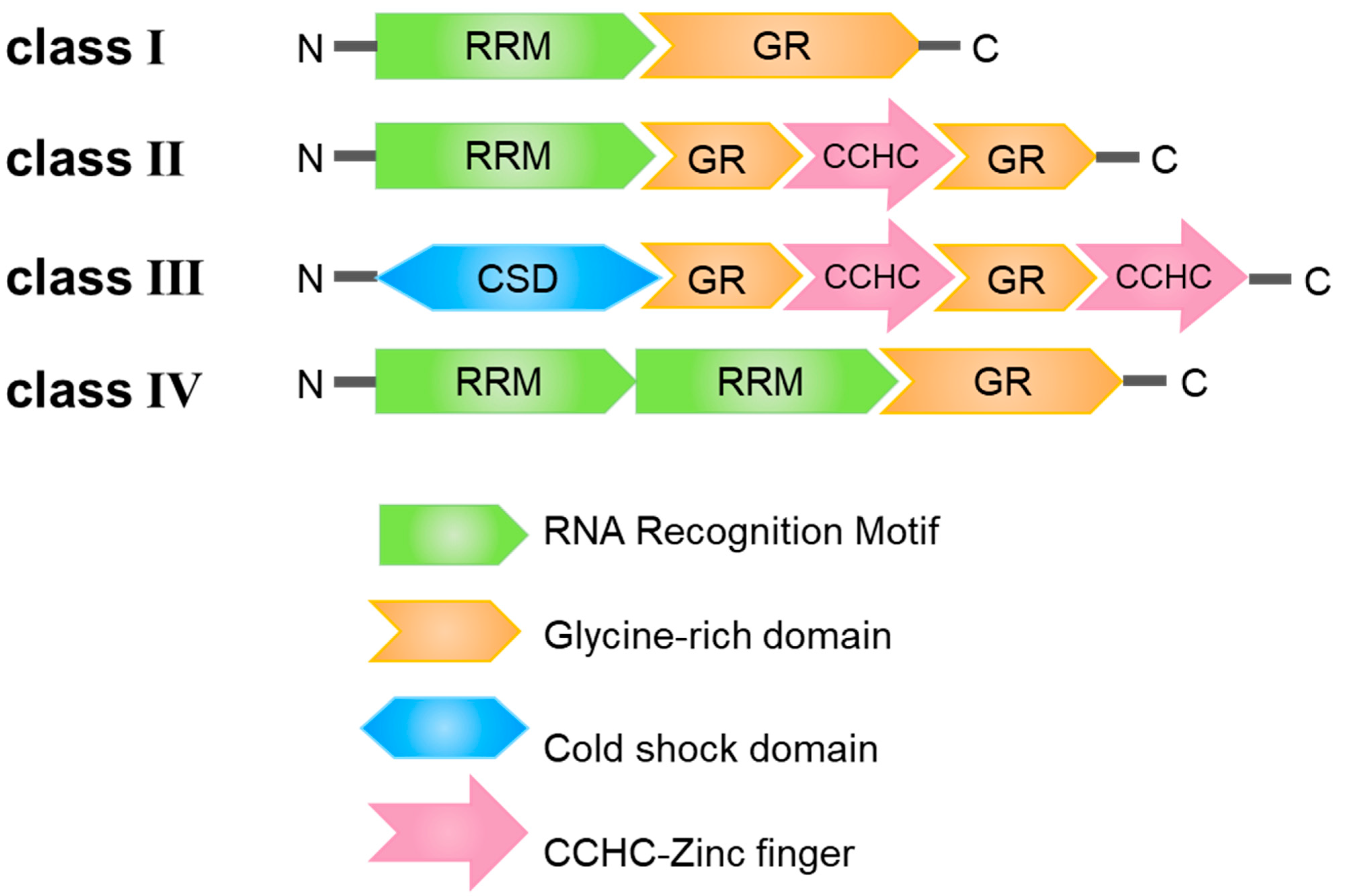
2. Structural Features of the GR-RBPs
2.1. Glycine-Rich Domain
2.2. RNA Recognition Motif
2.3. Cold-Shock Domain
2.4. CCHC Motif
3. Roles of GR-RBPs in RNA Metabolism
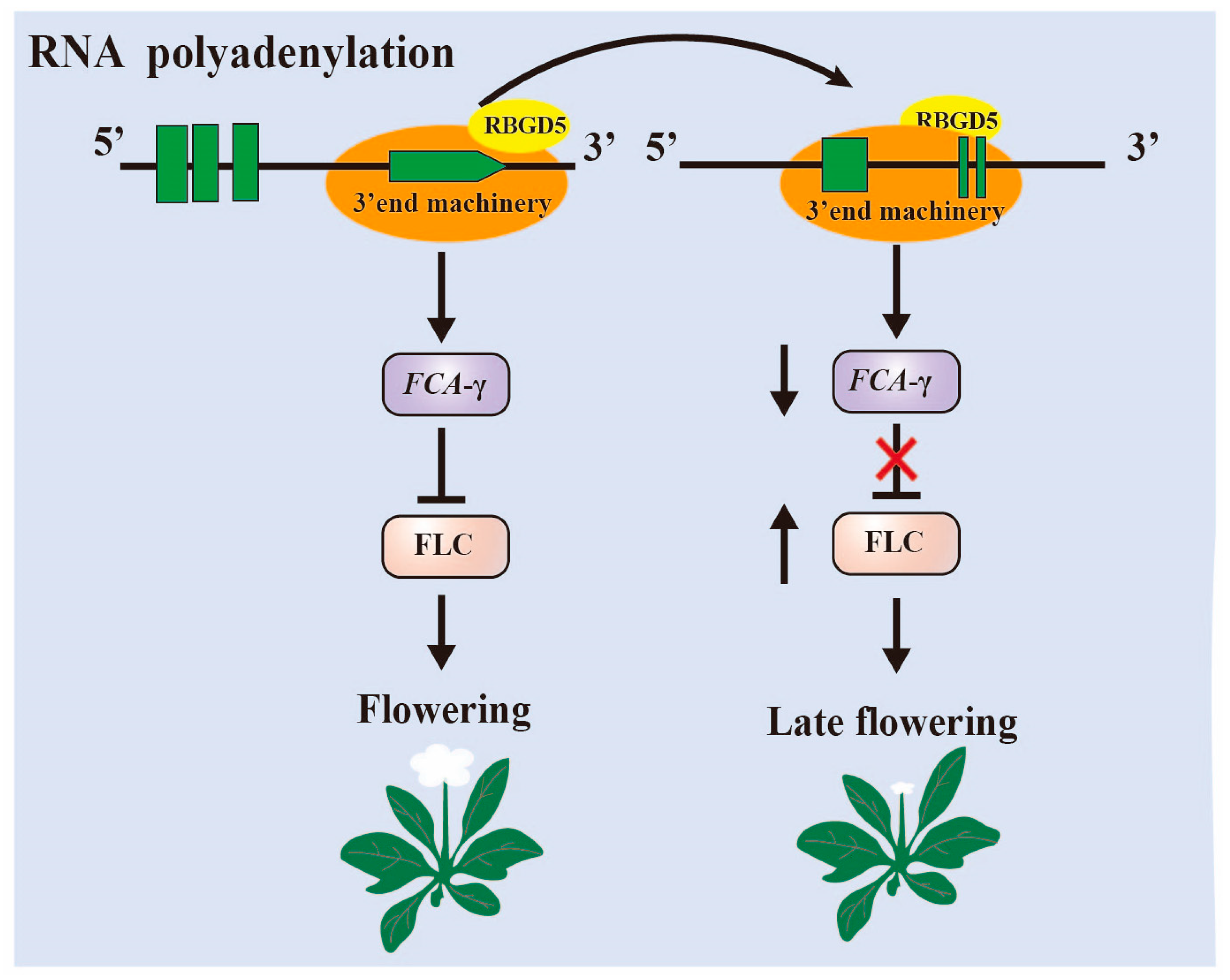
3.1. RNA Alternative Splice and Polyadenylation
3.2. miRNA Biogenesis
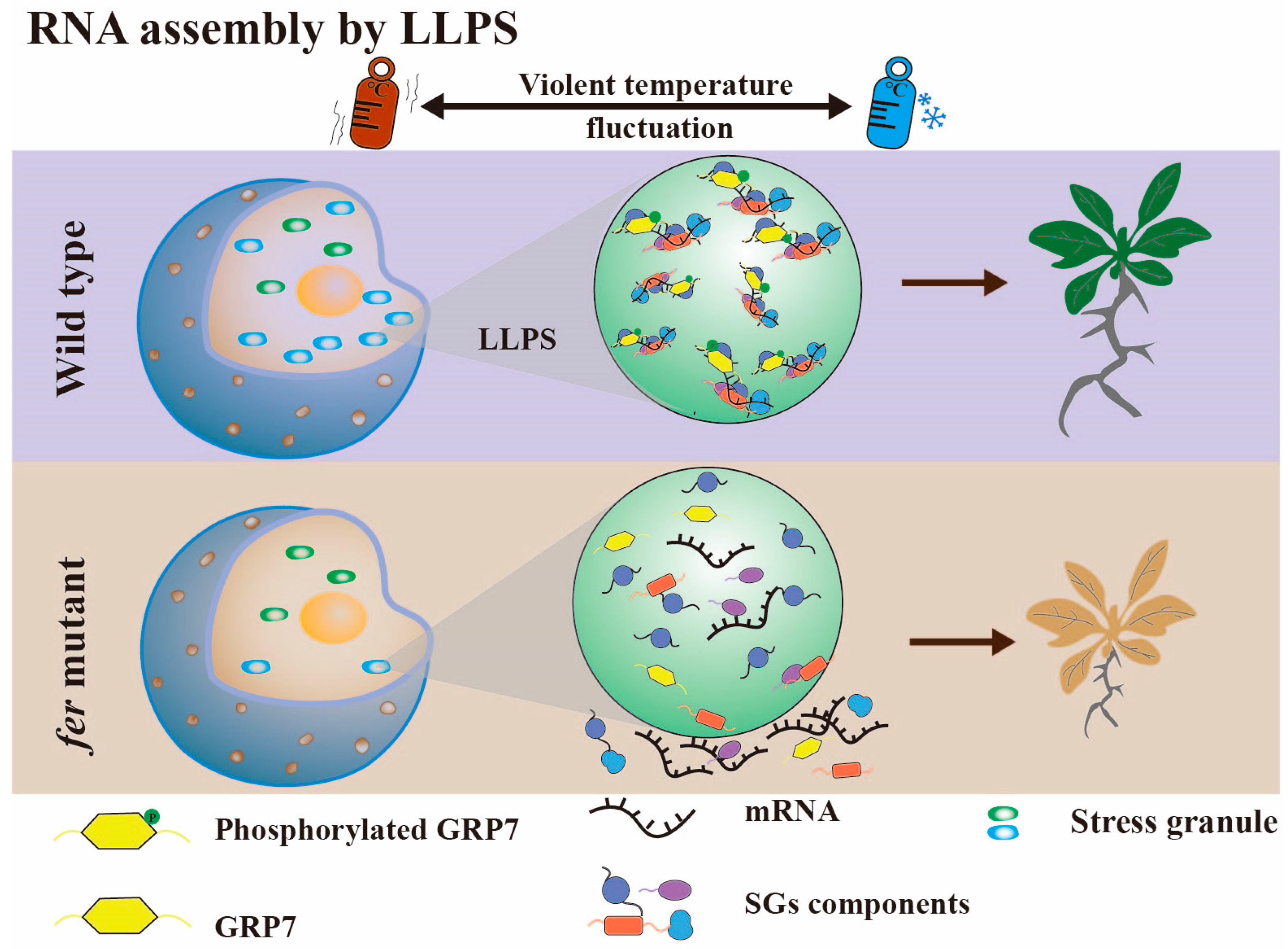
3.3. RNA Assembly by Liquid–Liquid Phase Separation
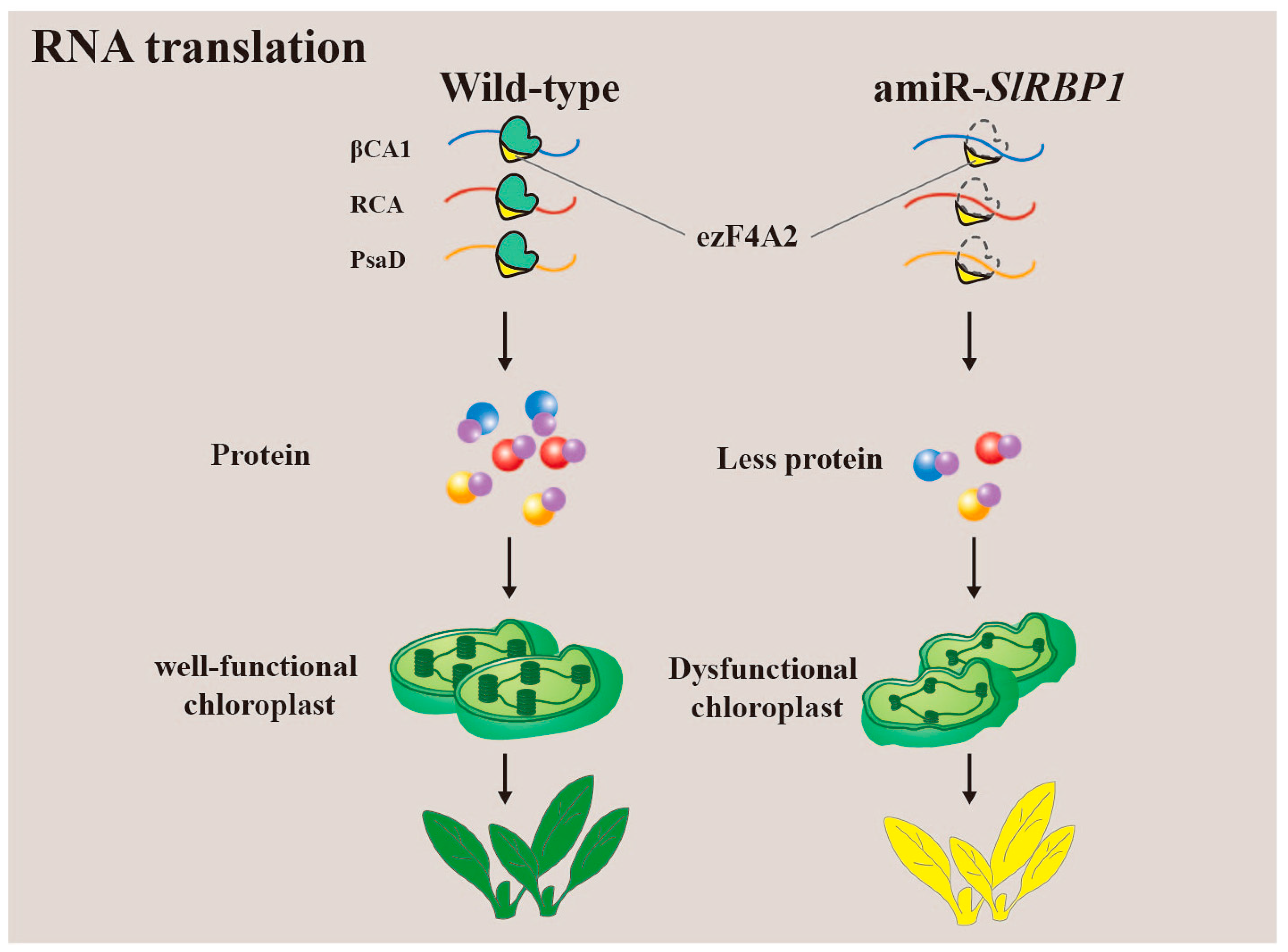
3.4. Translation of RNA
4. Roles of GR-RBPs in Crops
4.1. GR-RBPs in Grain Crops
4.1.1. GR-RBPs in Rice
4.1.2. GR-RBPs in Wheat
4.1.3. GR-RBPs in Maize
4.1.4. GR-RBPs in Other Crops
4.2. GR-RBPs in Horticultural Crops
4.2.1. GR-RBPs in Fruits
4.2.2. GR-RBPs in Vegetables
4.2.3. GR-RBPs in Ornamental Plant
4.3. GR-RBPs in Other Crops
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muthusamy, M.; Kim, J.H.; Kim, J.A.; Lee, S.I. Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview. Int. J. Mol. Sci. 2021, 22, 6731. [Google Scholar] [CrossRef] [PubMed]
- Joshna, C.R.; Saha, P.; Atugala, D.; Chua, G.; Muench, D.G. Plant PUF RNA-binding proteins: A wealth of diversity for post-transcriptional gene regulation. Plant Sci. 2020, 297, 110505. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C. The increasing diversity and complexity of the RNA-binding protein repertoire in plants. Proc. Biol. Sci. 2020, 287, 20201397. [Google Scholar] [CrossRef]
- Lorković, Z.J. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009, 14, 229–236. [Google Scholar] [CrossRef]
- Marondedze, C.; Thomas, L.; Serrano, N.L.; Lilley, K.S.; Gehring, C. The RNA-binding protein repertoire of Arabidopsis thaliana. Sci. Rep. 2016, 6, 29766. [Google Scholar] [CrossRef]
- Bach-Pages, M.; Castello, A.; Preston, G.M. Plant RNA Interactome Capture: Revealing the Plant RBPome. Trends Plant Sci. 2017, 22, 449–451. [Google Scholar] [CrossRef]
- Castro-Bustos, S.; Maruri-Lopez, I.; Ortega-Amaro, M.A.; Serrano, M.; Ovando-Vazquez, C.; Jimenez-Bremont, J.F. An interactome analysis reveals that Arabidopsis thaliana GRDP2 interacts with proteins involved in post-transcriptional processes. Cell Stress Chaperones 2022, 27, 165–176. [Google Scholar] [CrossRef]
- Amir, M.; Kumar, V.; Dohare, R.; Islam, A.; Ahmad, F.; Hassan, M.I. Sequence, structure and evolutionary analysis of cold shock domain proteins, a member of OB fold family. J. Evol. Biol. 2018, 31, 1903–1917. [Google Scholar] [CrossRef]
- Ma, L.; Cheng, K.; Li, J.; Deng, Z.; Zhang, C.; Zhu, H. Roles of Plant Glycine-Rich RNA-Binding Proteins in Development and Stress Responses. Int. J. Mol. Sci. 2021, 22, 5849. [Google Scholar] [CrossRef]
- Sachetto-Martins, G.; Franco, L.O.; de Oliveira, D.E. Plant glycine-rich proteins: A family or just proteins with a common motif? Biochim. Et Biophys. Acta (BBA) Gene Struct. Expr. 2000, 1492, 1–14. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 2010, 61, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Zhao, Y.X.; Xiao, H.L.; Zheng, Y.L.; Yue, B. Genome-wide identification, evolution, and expression analysis of RNA-binding glycine-rich protein family in maize. J. Integr. Plant Biol. 2014, 56, 1020–1031. [Google Scholar] [CrossRef]
- Kwak, K.J.; Kang, H.; Han, K.H.; Ahn, S.J. Molecular cloning, characterization, and stress-responsive expression of genes encoding glycine-rich RNA-binding proteins in Camelina sativa L. Plant Physiol. Bioch 2013, 68, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zeng, Q.-C.; Lu, X.-P.; Yu, D.-Q.; Li, W.-Z. Characterization and Expression Analysis of Four Glycine-Rich RNA-Binding Proteins Involved in Osmotic Response in Tobacco (Nicotiana tabacum cv. Xanthi). Agric Sci. China 2010, 9, 1577–1587. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, J.; Yang, Z.; Zhao, C.; Zhu, M.; Ma, D.; Dong, T.; Zhou, Z.; Liu, M.; Yang, D.; et al. Genome-wide identification and expression analysis of glycine-rich RNA-binding protein family in sweet potato wild relative Ipomoea trifida. Gene 2019, 686, 177–186. [Google Scholar] [CrossRef]
- Czolpinska, M.; Rurek, M. Plant Glycine-Rich Proteins in Stress Response: An Emerging, Still Prospective Story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef]
- Mateos, J.L.; Staiger, D. Toward a systems view on RNA-binding proteins and associated RNAs in plants: Guilt by association. Plant Cell 2023, 35, 1708–1726. [Google Scholar] [CrossRef]
- Condit, C.M.; Meagher, R.B. Expression of a gene encoding a glycine-rich protein in petunia. Mol. Cell Biol. 1987, 7, 4273–4279. [Google Scholar] [CrossRef]
- Mangeon, A.; Junqueira, R.M.; Sachetto-Martins, G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 2010, 5, 99–104. [Google Scholar] [CrossRef]
- Kar, B.; Nayak, S.; Joshi, R.K. Classification and comparative analysis of Curcuma longa L. expressed sequences tags (ESTs) encoding glycine-rich proteins (GRPs). Bioinformation 2012, 8, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Lummis, S.C.R. The roles of aromatic residues in the glycine receptor transmembrane domain. BMC Neurosci. 2018, 19, 53. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Chou, H.L.; Zhang, L.; Hwang, S.K.; Starkenburg, S.R.; Doroshenk, K.A.; Kumamaru, T.; Okita, T.W. RNA-Binding Protein RBP-P Is Required for Glutelin and Prolamine mRNA Localization in Rice Endosperm Cells. Plant Cell 2018, 30, 2529–2552. [Google Scholar] [CrossRef]
- Lancaster, A.K.; Nutter-Upham, A.; Lindquist, S.; King, O.D. PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 2014, 30, 2501–2502. [Google Scholar] [CrossRef]
- Zhu, S.; Gu, J.; Yao, J.; Li, Y.; Zhang, Z.; Xia, W.; Wang, Z.; Gui, X.; Li, L.; Li, D.; et al. Liquid-liquid phase separation of RBGD2/4 is required for heat stress resistance in Arabidopsis. Dev. Cell 2022, 57, 583–597. [Google Scholar] [CrossRef]
- O’Shea, E.K.; Lumb, K.J.; Kim, P.S. Peptide ‘Velcro’: Design of a heterodimeric coiled coil. Curr. Biol. 1993, 3, 658–667. [Google Scholar] [CrossRef]
- Steinert, P.M.; Mack, J.W.; Korge, B.P.; Gan, S.Q.; Haynes, S.R.; Steven, A.C. Glycine loops in proteins: Their occurrence in certain intermediate filament chains, loricrins and single-stranded RNA binding proteins. Int. J. Biol. Macromol. 1991, 13, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Hanano, S.; Sugita, M.; Sugiura, M. Isolation of a novel RNA-binding protein and its association with a large ribonucleoprotein particle present in the nucleoplasm of tobacco cells. Plant Mol. Biol. 1996, 31, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Ringli, C.; Keller, B.; Ryser, U. Glycine-rich proteins as structural components of plant cell walls. Cell Mol. Life Sci. 2001, 58, 1430–1441. [Google Scholar] [CrossRef]
- Xu, D.; Lei, M.; Wu, R. Expression of the rice Osgrpl promoter-Gus reporter gene is specifically associated with cell elongation/expansion and differentiation. Plant Mol. Biol. 1995, 28, 455–471. [Google Scholar] [CrossRef]
- Clery, A.; Blatter, M.; Allain, F.H. RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol. 2008, 18, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Dreyfuss, G.; Swanson, M.S.; Piñol-Roma, S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem. Sci. 1988, 13, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Maris, C.; Dominguez, C.; Allain, F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Hudson, B.P.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004, 11, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Sudol, M.; Sliwa, K.; Russo, T. Functions of WW domains in the nucleus. FEBS Lett. 2001, 490, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, L.; Hou, Y.; Wang, L.; Deng, X.; Hang, R.; Chen, D.; Zhang, X.; Zhang, Y.; Liu, C.; et al. Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Res. 2015, 25, 864–876. [Google Scholar] [CrossRef]
- Allain, F.H.; Gilbert, D.E.; Bouvet, P.; Feigon, J. Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J. Mol. Biol. 2000, 303, 227–241. [Google Scholar] [CrossRef]
- Varani, L.; Gunderson, S.I.; Mattaj, I.W.; Kay, L.E.; Neuhaus, D.; Varani, G. The NMR structure of the 38 kDa U1A protein—PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat. Struct. Biol. 2000, 7, 329–335. [Google Scholar] [CrossRef]
- ElAntak, L.; Tzakos, A.G.; Locker, N.; Lukavsky, P.J. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: Structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J. Biol. Chem. 2007, 282, 8165–8174. [Google Scholar] [CrossRef]
- Chaikam, V.; Karlson, D.T. Comparison of structure, function and regulation of plant cold shock domain proteins to bacterial and animal cold shock domain proteins. BMB Rep. 2010, 43, 1–8. [Google Scholar] [CrossRef]
- Sasaki, K.; Imai, R. Pleiotropic roles of cold shock domain proteins in plants. Front. Plant Sci. 2011, 2, 116. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Heinemann, U.; Roske, Y. Cold-Shock Domains-Abundance, Structure, Properties, and Nucleic-Acid Binding. Cancers 2021, 13, 190. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Park, S.J.; Kwak, K.J.; Kim, Y.O.; Kim, J.Y.; Song, J.; Jang, B.; Jung, C.H.; Kang, H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 2007, 35, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Radkova, M.; Revalska, M.; Zhiponova, M.; Iantcheva, A. Evaluation of the role of Medicago truncatula Zn finger CCHC type protein after heterologous expression in Arabidopsis thaliana. Biotechnol. Biotechnol. Equip. 2021, 35, 1686–1695. [Google Scholar] [CrossRef]
- Summers, M.F. Zinc finger motif for single-stranded nucleic acids? Investigations by nuclear magnetic resonance. J. Cell Biochem. 1991, 45, 41–48. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Pang, Y.; Yu, H.; Zhang, W.; Zhao, X.; Yu, J. The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism. RNA Biol. 2021, 18, 2107–2126. [Google Scholar] [CrossRef]
- Han, G.; Qiao, Z.; Li, Y.; Wang, C.; Wang, B. The Roles of CCCH Zinc-Finger Proteins in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8327. [Google Scholar] [CrossRef]
- Sun, A.; Li, Y.; He, Y.; Zou, X.; Chen, F.; Ji, R.; You, C.; Yu, K.; Li, Y.; Xiao, W.; et al. Comprehensive Genome-Wide Identification, Characterization, and Expression Analysis of CCHC-Type Zinc Finger Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 892105. [Google Scholar] [CrossRef]
- Song, H.; Kim, H.; Hwang, B.H.; Yi, H.; Hur, Y. Natural variation in glycine-rich region of Brassica oleracea cold shock domain protein 5 (BoCSDP5) is associated with low temperature tolerance. Genes Genom. 2020, 42, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Kim, J.S.; Kang, H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 2005, 42, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Gu, L.; Choi, M.J.; Kim, R.J.; Suh, M.C.; Kang, H. Comparative Functional Analysis of Wheat (Triticum aestivum) Zinc Finger-Containing Glycine-Rich RNA-Binding Proteins in Response to Abiotic Stresses. PLoS ONE 2014, 9, e96877. [Google Scholar] [CrossRef] [PubMed]
- Ambrosone, A.; Costa, A.; Leone, A.; Grillo, S. Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci. 2012, 182, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Lutz, C.S.; Moreira, A. Alternative mRNA polyadenylation in eukaryotes: An effective regulator of gene expression. Wiley Interdiscip. Rev. RNA 2011, 2, 22–31. [Google Scholar] [CrossRef]
- Xing, D.; Li, Q.Q. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip. Rev. RNA 2011, 2, 445–458. [Google Scholar] [CrossRef]
- Xiao, J.; Li, C.; Xu, S.; Xing, L.; Xu, Y.; Chong, K. JACALIN-LECTIN LIKE1 Regulates the Nuclear Accumulation of GLYCINE-RICH RNA-BINDING PROTEIN7, Influencing the RNA Processing of FLOWERING LOCUS C Antisense Transcripts and Flowering Time in Arabidopsis. Plant Physiol. 2015, 169, 2102–2117. [Google Scholar] [CrossRef]
- Streitner, C.; Köster, T.; Simpson, C.G.; Shaw, P.; Danisman, S.; Brown, J.W.; Staiger, D. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res. 2012, 40, 11240–11255. [Google Scholar] [CrossRef]
- Wu, Z.; Zhu, D.; Lin, X.; Miao, J.; Gu, L.; Deng, X.; Yang, Q.; Sun, K.; Zhu, D.; Cao, X.; et al. RNA Binding Proteins RZ-1B and RZ-1C Play Critical Roles in Regulating Pre-mRNA Splicing and Gene Expression during Development in Arabidopsis. Plant Cell 2016, 28, 55–73. [Google Scholar] [CrossRef]
- Terzi, L.C.; Simpson, G.G. Regulation of flowering time by RNA processing. Curr. Top. Microbiol. Immunol. 2008, 326, 201–218. [Google Scholar] [CrossRef]
- Koster, T.; Meyer, K.; Weinholdt, C.; Smith, L.M.; Lummer, M.; Speth, C.; Grosse, I.; Weigel, D.; Staiger, D. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 2014, 42, 9925–9936. [Google Scholar] [CrossRef] [PubMed]
- Luders, J.; Winkel, A.R.; Reichel, M.; Bitterer, V.W.; Scheibe, M.; Widmann, C.; Butter, F.; Koster, T. Identification of Pri-miRNA Stem-Loop Interacting Proteins in Plants Using a Modified Version of the Csy4 CRISPR Endonuclease. Int. J. Mol. Sci. 2022, 23, 8961. [Google Scholar] [CrossRef] [PubMed]
- Wiedner, H.J.; Giudice, J. It’s not just a phase: Function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biol. 2021, 28, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zheng, C.; Lu, D.; Song, C.P.; Zhang, L. Phase separation in plants: New insights into cellular compartmentalization. J. Integr. Plant Biol. 2021, 63, 1835–1855. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhu, S.; Kumar, P.; MacMicking, J.D. A phase-separated nuclear GBPL circuit controls immunity in plants. Nature 2021, 594, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, T.; Wang, B.; Lin, Q.; Zhu, S.; Li, C.; Ma, Y.; Tang, J.; Xing, J.; Li, X.; et al. RALF1-FERONIA complex affects splicing dynamics to modulate stress responses and growth in plants. Sci. Adv. 2020, 6, eaaz1622. [Google Scholar] [CrossRef]
- Xu, F.; Wang, L.; Li, Y.; Shi, J.; Staiger, D.; Chen, W.; Wang, L.; Yu, F. The Receptor Kinase FER Mediates Phase Separation of Glycine-Rich RNA-Binding Protein 7 to Confer Temperature Resilience in Arabidopsis. bioRxiv 2022, 2022, 483201. [Google Scholar] [CrossRef]
- Ma, L.; Yang, Y.; Wang, Y.; Cheng, K.; Zhou, X.; Li, J.; Zhang, J.; Li, R.; Zhang, L.; Wang, K.; et al. SlRBP1 promotes translational efficiency via SleIF4A2 to maintain chloroplast function in tomato. Plant Cell 2022, 34, 2747–2764. [Google Scholar] [CrossRef]
- Habibpourmehraban, F.; Atwell, B.J.; Haynes, P.A. Unique and Shared Proteome Responses of Rice Plants (Oryza sativa) to Individual Abiotic Stresses. Int. J. Mol. Sci. 2022, 23, 15552. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 2010, 33, 759–768. [Google Scholar] [CrossRef]
- Xu, W.; Dou, Y.; Geng, H.; Fu, J.; Dan, Z.; Liang, T.; Cheng, M.; Zhao, W.; Zeng, Y.; Hu, Z.; et al. OsGRP3 Enhances Drought Resistance by Altering Phenylpropanoid Biosynthesis Pathway in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 7045. [Google Scholar] [CrossRef] [PubMed]
- Sahi, C.; Agarwal, M.; Singh, A.; Grover, A. Molecular characterization of a novel isoform of rice (Oryza sativa L.) glycine rich-RNA binding protein and evidence for its involvement in high temperature stress response. Plant Sci. 2007, 173, 144–155. [Google Scholar] [CrossRef]
- Wang, F.; Bai, M.Y.; Deng, Z.; Oses-Prieto, J.A.; Burlingame, A.L.; Lu, T.; Chong, K.; Wang, Z.Y. Proteomic study identifies proteins involved in brassinosteroid regulation of rice growth. J. Integr. Plant Biol. 2010, 52, 1075–1085. [Google Scholar] [CrossRef]
- Takebe, N.; Nakamura, A.; Watanabe, T.; Miyashita, A.; Satoh, S.; Iwai, H. Cell wall Glycine-rich Protein2 is involved in tapetal differentiation and pollen maturation. J. Plant Res. 2020, 133, 883–895. [Google Scholar] [CrossRef]
- Xue, G.P.; Rae, A.L.; White, R.G.; Drenth, J.; Richardson, T.; McIntyre, C.L. A strong root-specific expression system for stable transgene expression in bread wheat. Plant Cell Rep. 2016, 35, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Chou, H.L.; Zhang, L.; Okita, T.W. Targeted Endoplasmic Reticulum Localization of Storage Protein mRNAs Requires the RNA-Binding Protein RBP-L. Plant Physiol. 2019, 179, 1111–1131. [Google Scholar] [CrossRef]
- Guiltinan, M.J.; Niu, X.P. cDNA encoding a wheat (Triticum aestivum cv Chinese Spring) glycine-rich RNA-binding protein. Plant Mol. Biol. 1996, 30, 1301–1306. [Google Scholar] [CrossRef]
- Xiao, J.; Xu, S.; Li, C.; Xu, Y.; Xing, L.; Niu, Y.; Huan, Q.; Tang, Y.; Zhao, C.; Wagner, D.; et al. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 2014, 5, 4572. [Google Scholar] [CrossRef]
- Gomez, J.; Sanchezmartinez, D.; Stiefel, V.; Rigau, J.; Puigdomenech, P.; Pages, M. A Gene Induced by the Plant Hormone Abscisic-Acid in Response to Water-Stress Encodes a Glycine-Rich Protein. Nature 1988, 334, 262–264. [Google Scholar] [CrossRef]
- Freire, M.A. The Zea mays glycine-rich RNA-binding protein MA16 is bound to a ribonucleotide(s) by a stable linkage. J. Plant Res. 2012, 125, 653–660. [Google Scholar] [CrossRef]
- Pages, M.A.F.a.M. Functional characteristics of the maize RNA-binding protein MA16. Plant Mol. Biol. 1995, 29, 797–807. [Google Scholar] [CrossRef]
- Alba, M.M.; Culianez-Macia, F.A.; Goday, A.; Freire, M.A.; Nadal, B.; Pages, M. The maize RNA-binding protein, MA16, is a nucleolar protein located in the dense fibrillar component. Plant J. 1994, 6, 825–834. [Google Scholar] [CrossRef]
- Gendra, E.; Moreno, A.; Alba, M.M.; Pages, M. Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. Plant J. 2004, 38, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Przybylska, A.; Spychalski, M. Changes in the expression level of genes encoding transcription factors and cell wall-related proteins during Meloidogyne arenaria infection of maize (Zea mays). Mol. Biol. Rep. 2021, 48, 6779–6786. [Google Scholar] [CrossRef] [PubMed]
- Kelley, R.Y.; Williams, W.P.; Mylroie, J.E.; Boykin, D.L.; Harper, J.W.; Windham, G.L.; Ankala, A.; Shan, X. Identification of maize genes associated with host plant resistance or susceptibility to Aspergillus flavus infection and aflatoxin accumulation. PLoS ONE 2012, 7, e36892. [Google Scholar] [CrossRef]
- Li, Y.Z.; Fan, X.W.; Chen, Q.; Zhong, H. A photoperiod-responsive protein compendium and conceptual proteome roadmap outline in maize grown in growth chambers with controlled conditions. PLoS ONE 2017, 12, e0174003. [Google Scholar] [CrossRef]
- Vermel, M.; Guermann, B.; Delage, L.; Grienenberger, J.M.; Maréchal-Drouard, L.; Gualberto, J.M. A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. USA 2002, 99, 5866–5871. [Google Scholar] [CrossRef]
- Di Silvestre, D.; Passignani, G.; Rossi, R.; Ciuffo, M.; Turina, M.; Vigani, G.; Mauri, P.L. Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein-Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium quinoa Plants. Biology 2022, 11, 95. [Google Scholar] [CrossRef]
- Tripet, B.P.; Mason, K.E.; Eilers, B.J.; Burns, J.; Powell, P.; Fischer, A.M.; Copie, V. Structural and biochemical analysis of the Hordeum vulgare L. HvGR-RBP1 protein, a glycine-rich RNA-binding protein involved in the regulation of barley plant development and stress response. Biochemistry 2014, 53, 7945–7960. [Google Scholar] [CrossRef]
- Molina, A.; Mena, M.; Carbonero, P.; GarciaOlmedo, F. Differential expression of pathogen-responsive genes encoding two types of glycine-rich proteins in barley. Plant Mol. Biol. 1997, 33, 803–810. [Google Scholar] [CrossRef]
- Sanan-Mishra, N.; Tuteja, N.; Kumar Sopory, S. Salinity- and ABA-induced up-regulation and light-mediated modulation of mRNA encoding glycine-rich RNA-binding protein from Sorghum bicolor. Biochem. Biophys. Res. Commun. 2002, 296, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Chapman, N.; David, K.; Angenent, G.C.; Seymour, G.B.; de Maagd, R.A. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014, 65, 4527–4541. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, X.; Wang, K.; Li, J.; Zhu, G.; Ren, S.; Deng, Z.; Zhu, B.; Fu, D.; Qu, G.; et al. Molecular and functional diversity of organelle RNA editing mediated by RNA recognition motif-containing protein ORRM4 in tomato. New Phytol. 2020, 228, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, G.; Li, R.; Yan, S.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. The RNA Editing Factor SlORRM4 Is Required for Normal Fruit Ripening in Tomato. Plant Physiol. 2017, 175, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Zeng, N.; Qu, G.; Fu, D.; Zhu, B.; Luo, Y.; Ostersetzer-Biran, O.; Zhu, H. Glycine-rich RNA-binding cofactor RZ1AL is associated with tomato ripening and development. Hortic. Res. 2022, 9, uhac134. [Google Scholar] [CrossRef]
- Ruggieri, G.M.; Triassi, A.; Alvarez, C.E.; Gola, A.; Wiggenhauser, J.; Budde, C.O.; Lara, M.V.; Drincovich, M.F.; Müller, G.L. Overexpression of glycine-rich RNA-binding protein in tomato renders fruits with higher protein content after cold storage. Biol. Plant. 2018, 62, 501–510. [Google Scholar] [CrossRef]
- Tang, Y.; Huang, C.; Li, Y.; Wang, Y.; Zhang, C. Genome-wide identification, phylogenetic analysis, and expression profiling of glycine-rich RNA-binding protein (GRPs) genes in seeded and seedless grapes (Vitis vinifera). Physiol. Mol. Biol. Plants 2021, 27, 2231–2243. [Google Scholar] [CrossRef]
- Wang, B.; Wang, G.; Shen, F.; Zhu, S. A Glycine-Rich RNA-Binding Protein, CsGR-RBP3, Is Involved in Defense Responses Against Cold Stress in Harvested Cucumber (Cucumis sativus L.) Fruit. Front. Plant Sci. 2018, 9, 540. [Google Scholar] [CrossRef]
- Kim, M.K.; Jung, H.J.; Kim, D.H.; Kang, H. Characterization of glycine-rich RNA-binding proteins in Brassica napus under stress conditions. Physiol. Plant 2012, 146, 297–307. [Google Scholar] [CrossRef]
- Lu, X.; Cheng, Y.; Gao, M.; Li, M.; Xu, X. Molecular Characterization, Expression Pattern and Function Analysis of Glycine-Rich Protein Genes Under Stresses in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Front. Genet. 2020, 11, 774. [Google Scholar] [CrossRef]
- Tan, Y.; Qin, Y.; Li, Y.; Li, M.; Ma, F. Overexpression of MpGR-RBP1, a glycine-rich RNA-binding protein gene from Malus prunifolia (Willd.) Borkh., confers salt stress tolerance and protects against oxidative stress in Arabidopsis. Plant Cell Tissue Organ. Cult. (PCTOC) 2014, 119, 635–646. [Google Scholar] [CrossRef]
- Wang, S.; Wang, R.; Liang, D.; Ma, F.; Shu, H. Molecular characterization and expression analysis of a glycine-rich RNA-binding protein gene from Malus hupehensis Rehd. Mol. Biol. Rep. 2012, 39, 4145–4153. [Google Scholar] [CrossRef]
- Sukarta, O.C.A.; Zheng, Q.; Slootweg, E.J.; Mekken, M.; Mendel, M.; Putker, V.; Bertran, A.; Brand, A.; Overmars, H.; Pomp, R.; et al. GLYCINE-RICH RNA-BINDING PROTEIN 7 potentiates effector-triggered immunity through an RNA recognition motif. Plant Physiol. 2022, 189, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Yang, Z.; Song, W.; Miao, J.; Zhao, H.; Ji, P.; Li, T.; Si, J.; Yin, Z.; Jing, M.; et al. The Phytophthora sojae effector PsFYVE1 modulates immunity-related gene expression by targeting host RZ-1A protein. Plant Physiol. 2022, 191, 925–945. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yu, R.; Li, W.; Geng, L.; Jing, X.; Zhu, C.; Liu, H. Identification and characterisation of a glycine-rich RNA-binding protein as an endogenous suppressor of RNA silencing from Nicotiana glutinosa. Planta 2019, 249, 1811–1822. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.J.; Kim, H.-S.; Jang, H.Y.; Kang, H.; Ahn, S.-J. Diverse roles of glycine-rich RNA-binding protein 7 in the response of camelina (Camelina sativa) to abiotic stress. Acta Physiol. Plant. 2016, 38, 129. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, D.W.; Wang, Y.C.; Zheng, L.; Yang, C.P. A glycine-rich RNA-binding protein can mediate physiological responses in transgenic plants under salt stress. Mol. Biol. Rep. 2012, 39, 1047–1053. [Google Scholar] [CrossRef]
- Ayarpadikannan, S.; Chung, E.S.; So, H.A.; Kim, K.M.; Schraufnagle, K.R.; Lee, J.H. Overexpression of SaRBP1 enhances tolerance of Arabidopsis to salt stress. Plant Cell Tissue Organ. Cult. (PCTOC) 2014, 118, 327–338. [Google Scholar] [CrossRef]
- Tsuyoshi Nomata, Y.K.a.N.S. Cloning and Characterization of Glycine-Rich RNA-Binding Protein cDNAs in the Moss Physcomitrella patens. Plant Cell Physiol. 2004, 45, 48–56. [Google Scholar] [CrossRef]
- Teng, K.; Tan, P.; Xiao, G.; Han, L.; Chang, Z.; Chao, Y. Heterologous expression of a novel Zoysia japonica salt-induced glycine-rich RNA-binding protein gene, ZjGRP, caused salt sensitivity in Arabidopsis. Plant Cell Rep. 2017, 36, 179–191. [Google Scholar] [CrossRef]
- Gao, M.; Zhang, C.; Angel, W.; Kwak, O.; Allison, J.; Wiratan, L.; Hallworth, A.; Wolf, J.; Lu, H. Circadian Regulation of the GLYCINE-RICH RNA-BINDING PROTEIN Gene by the Master Clock Protein CIRCADIAN CLOCK-ASSOCIATED 1 Is Important for Plant Innate Immunity. J. Exp. Bot. 2022, 74, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Jung, H.J.; Lee, H.J.; Kim, K.A.; Goh, C.H.; Woo, Y.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 2008, 55, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Bartel, D.P. Widespread Influence of 3’-End Structures on Mammalian mRNA Processing and Stability. Cell 2017, 169, 905–917. [Google Scholar] [CrossRef] [PubMed]


| Gene | Plant Species | Roles | Phenotype | Reference |
|---|---|---|---|---|
| CqGR-RBP2 | Chenopodium quinoa | Drought resistance | Not determined | [88] |
| HvGR-RBP1 | Hordeum vulgare | Regulates the timing of anthesis and leaf senescence; cold resistance | Not determined | [89] |
| HvGRP3 | Hordeum vulgare | Fungal pathogens and cold stress induce | Not determined | [90] |
| MA16 | Zea mays | rRNA metabolism | Not determined | [80,81,82,83] |
| OsRBP-P OsRBP-L | Oryza sativa | Correct localization of mRNA | The rbp-l/rbp-p knockdown mutant exhibits plant dwarfism, chlorophyll deficiency, sterility to late flowering, and low spikelet fertility | [23,76] |
| OsGRP1 | Oryza sativa | Enhances cell elongation; cold resistance | Overexpression of OsGRP1 in Arabidopsis suppresses the dwarf phenotype of the mutant bri1-5; recovery cold adaptation to cold-sensitive E. coli mutant strains | [30,69,73] |
| OsGRP2 | Oryza sativa | Cell wall construction; floral organ early development | Not determined | [74] |
| OsGRP3 | Oryza sativa | Drought resistance; mRNA stability | Overexpression of OsGRP3 in rice had better growth and higher rate after drought treatment, while transgenic plants with OsGRP3 knocked out or knocked down had lower survival rate. | [71] |
| OsGRP4 | Oryza sativa | Cold resistance; Heat resistance; mRNA export; | Under low temperatures, promotes seed germination and seedling growth | [72] |
| OsGRP6 | Oryza sativa | Cold resistance; mRNA export | Recovery cold adaptation to cold-sensitive E. coli mutant strains | [69] |
| OsRZ-2 | Oryza sativa | Cold resistance | The overexpression of OsRZ-2 restore the cold-sensitive phenotype of grp7 mutants | [70] |
| SbGR-RNP | Sorghum bicolor | Salinity- and ABA-induced upregulation; light response | Not determined | [91] |
| TaGRP2 | Triticum aestivum | Flowering repressors | The flowering was delayed in overexpression TaGRP2 plants and accelerated in RNAi-TaGRP2 plants | [78] |
| TaRZ1 | Triticum aestivum | Cold resistance; salt resistance | Overexpression TaRZ1 in Arabidopsis delays seed germination and inhibits seedling growth under salt stress, and seedling growth was inhibited at low temperature. | [53] |
| TaRZ2 | Triticum aestivum | Cold resistance; salt resistance | Overexpression TaRZ2 in Arabidopsis delays seed germination under salt and drought stress and enhances the freezing tolerance. | [53] |
| TaRZ3 | Triticum aestivum | Cold resistance | Overexpression TaRZ3 in Arabidopsis delays seed germination under salt and drought stress | [53] |
| whGRP-1 | Triticum aestivum | ABA-inducible gene | Not determined | [77] |
| ZmGRP1 | Zea mays | Photoperiodic mRNA splicing and export | Not determined | [86] |
| ZmGRP2 | Zea mays | Insect and fungal infection resistance | Upregulation of ZmGRP2 can help maize to better fight against Meloidogyne arenaria infection. | [85] |
| Gene | Plant Species | Roles | Phenotype | Reference |
|---|---|---|---|---|
| BoCSDP5 | Brassica oleracea | Cold resistance | Not determined | [51] |
| BnGRP1 | Brassica napus | Cold resistance | Accelerated germination by overexpression BnGRP1 in Arabidopsis | [99] |
| CsGR-RBP3 | Cucumis sativa | Cold resistance | Arabidopsis plant overexpressing CsGR-RBP3 shows strong cold tolerance at 0 °C and −20 °C | [98] |
| LeRBP1 | Solanum lycopersicum | Cold resistance | Not determined | [96] |
| MpGR-RBP1 | Malus prunifolia | Salt stress tolerance; oxidative stress tolerance | Overexpression MpGR-RBP1 in Arabidopsis accelerates seed germination and seedling growth when plants were exposed to high salt or oxidative stress | [101] |
| SlORRM4 | Solanum lycopersicum | Mitochondrial RNA editing | The slorrm4 mutant shows delayed fruit ripening | [94] |
| SlRZ1A-like | Solanum lycopersicum | Regulates target RNA transcription and translation | The rzla-l mutant fruit became smaller and ultimately less red in color | [95] |
| SlRBP1 | Solanum lycopersicum | Regulates correct translation of target RNA; chloroplast development | The slrbp1 knockdown mutant exhibits dwarf tomato plants with yellow leaves, smaller flowers, and fruit | [68] |
| VviGRP2, VviRZ-1A VviGRP3 VviGRP5 VviGRP7 | Vitis vinifera | Vitis vinifera mesocarp development | Not determined | [97] |
| Gene | Plant Species | Roles | Phenotype | Reference |
|---|---|---|---|---|
| CsGRP2s | Camelina sativa | Cold resistance; RNA chaperone activity | Overexpression CsGRP2s had the ability to complement cold-sensitive Escherichia coli mutants at low temperatures | [14] |
| CsGRP7a | Camelina sativa | Cold resistance; RNA chaperone activity | Overexpression CsGRP7a in camelina grows sluggishly under salt stress, but its root grows better under cold stress than WT | [106] |
| LbGRP1 | Limonium bicolor | Salt resistance | Overexpression LbGRP1 in tobacco significantly improved superoxide dismutase, catalase activities, and proline levels under salt-stress conditions | [107] |
| NgRBP | Nicotiana glutinosa | Inhibition of RNA silencing; viral resistance | Knockdown of NgRBP enhanced resistance to PVX and cucumber mosaic virus | [105] |
| NbRZ-1A | Nicotiana benthamiana | mRNA alternative splicing; pathogen resistance | Not determined | [104] |
| NbGRP7 | Nicotiana benthamiana | Participating in plant effector-triggered immunity | Mutation or ectopic expression of the NbGRP7 compromises Rx1-mediated defense | [103] |
| PpGRPs | Physcomitrella patens | Low temperature response | Not determined | [109] |
| SaRBP1 | Suaeda asparagoides | Salt resistance | Overexpression SaRBP1 Arabidopsis seedlings displays longer primary roots, more fresh weight, higher number of lateral roots, and higher survival rates than WT | [108] |
| ZjGRP | Zoysia japonica | Salt resistance | Overexpression of ZjGRP increases resistance to salt stress in Arabidopsis | [110] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, K.; Zhang, C.; Lu, Y.; Li, J.; Tang, H.; Ma, L.; Zhu, H. The Glycine-Rich RNA-Binding Protein Is a Vital Post-Transcriptional Regulator in Crops. Plants 2023, 12, 3504. https://doi.org/10.3390/plants12193504
Cheng K, Zhang C, Lu Y, Li J, Tang H, Ma L, Zhu H. The Glycine-Rich RNA-Binding Protein Is a Vital Post-Transcriptional Regulator in Crops. Plants. 2023; 12(19):3504. https://doi.org/10.3390/plants12193504
Chicago/Turabian StyleCheng, Ke, Chunjiao Zhang, Yao Lu, Jinyan Li, Hui Tang, Liqun Ma, and Hongliang Zhu. 2023. "The Glycine-Rich RNA-Binding Protein Is a Vital Post-Transcriptional Regulator in Crops" Plants 12, no. 19: 3504. https://doi.org/10.3390/plants12193504
APA StyleCheng, K., Zhang, C., Lu, Y., Li, J., Tang, H., Ma, L., & Zhu, H. (2023). The Glycine-Rich RNA-Binding Protein Is a Vital Post-Transcriptional Regulator in Crops. Plants, 12(19), 3504. https://doi.org/10.3390/plants12193504







