Continuous Cropping Inhibits Photosynthesis of Polygonatum odoratum
Abstract
:1. Introduction
2. Results
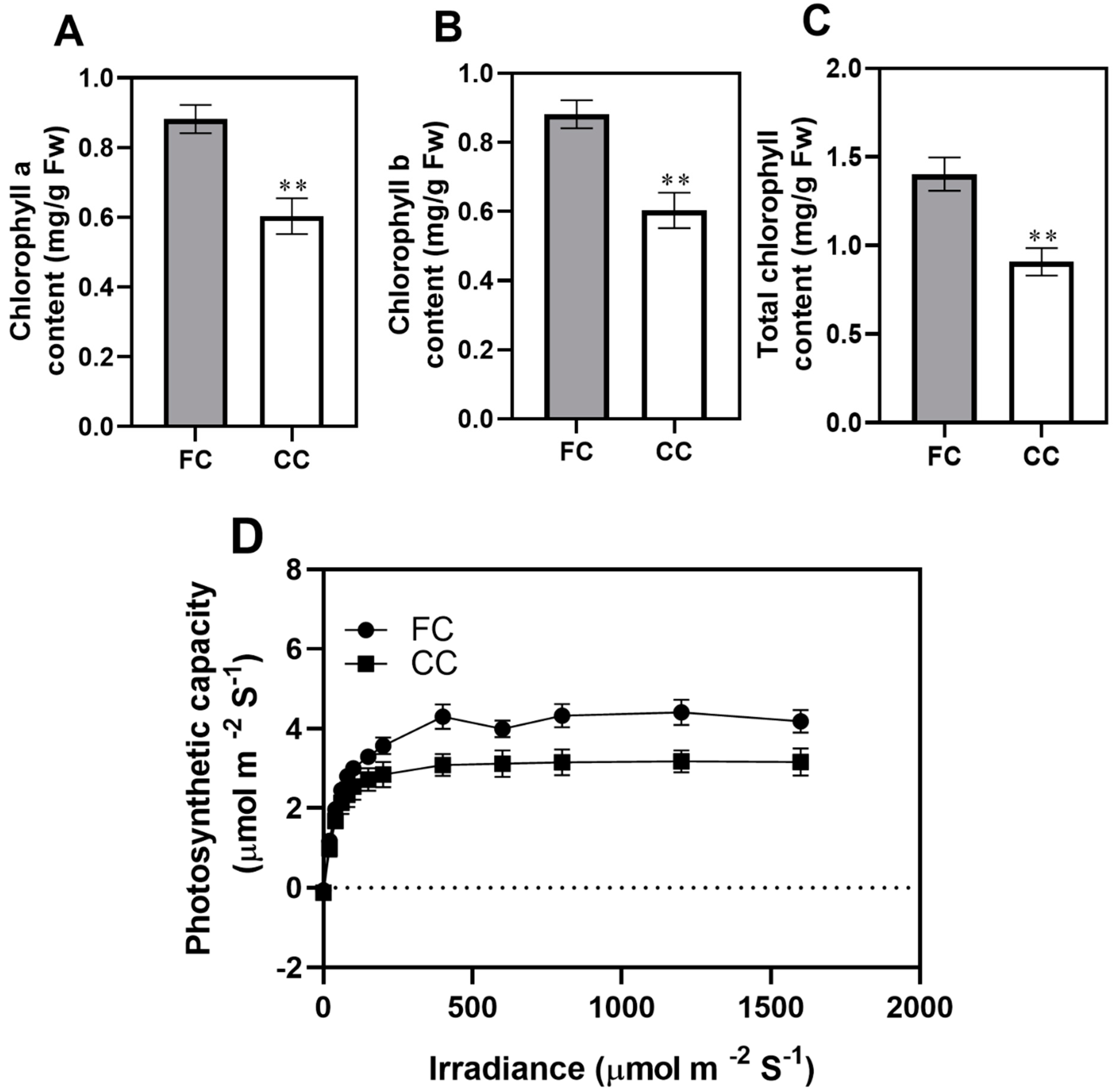
2.1. Effect of Continuous Cropping on the Photosynthetic Physiological Characteristics in P. odoratum
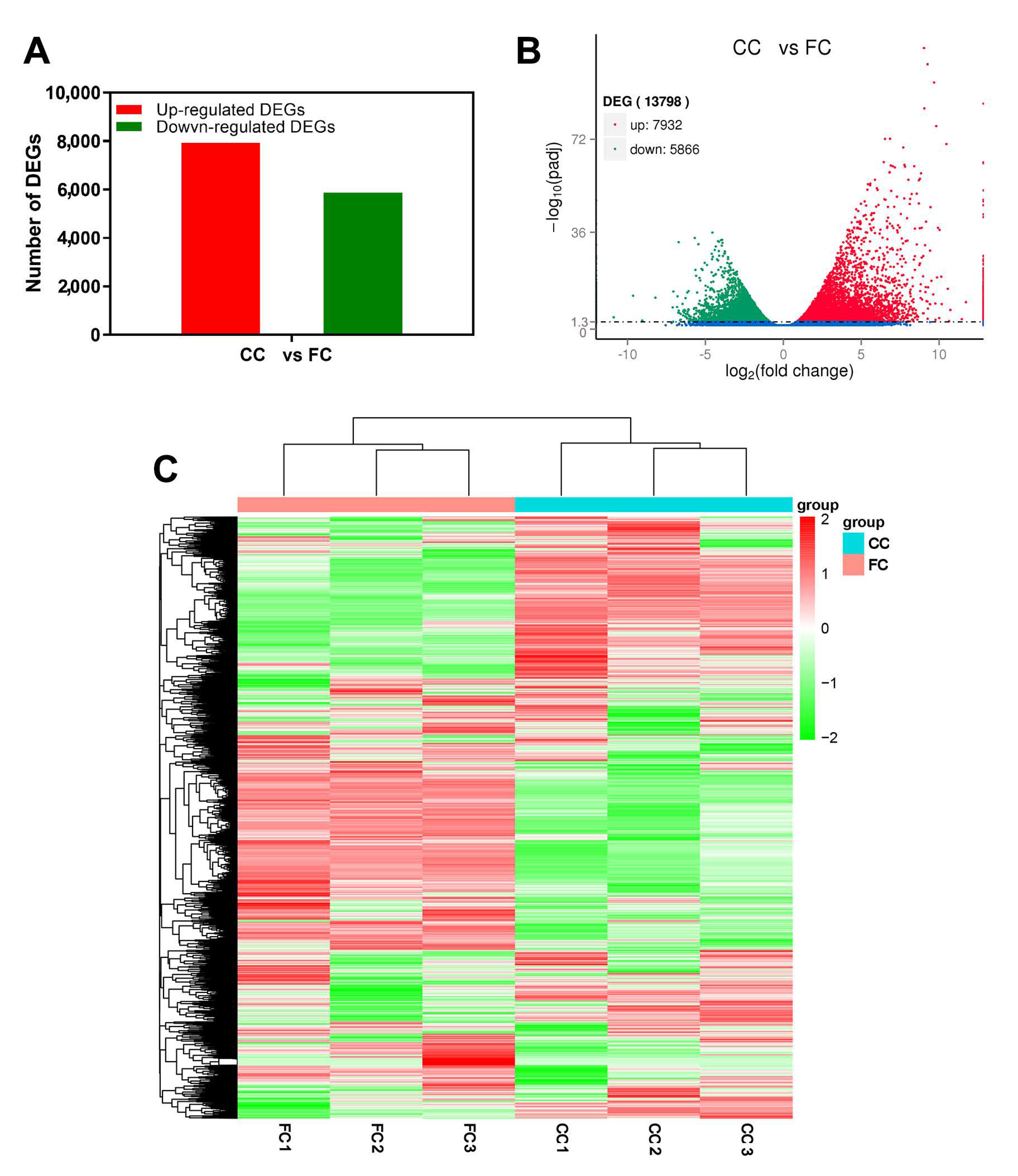
2.2. Gene Differential Expressed in Response to Continuous Cropping in P. odoratum
2.3. Transcripts with Increased Levels of Encoded Proteins Involved in Photosynthesis and Carbon Assimilation of P. odoratum
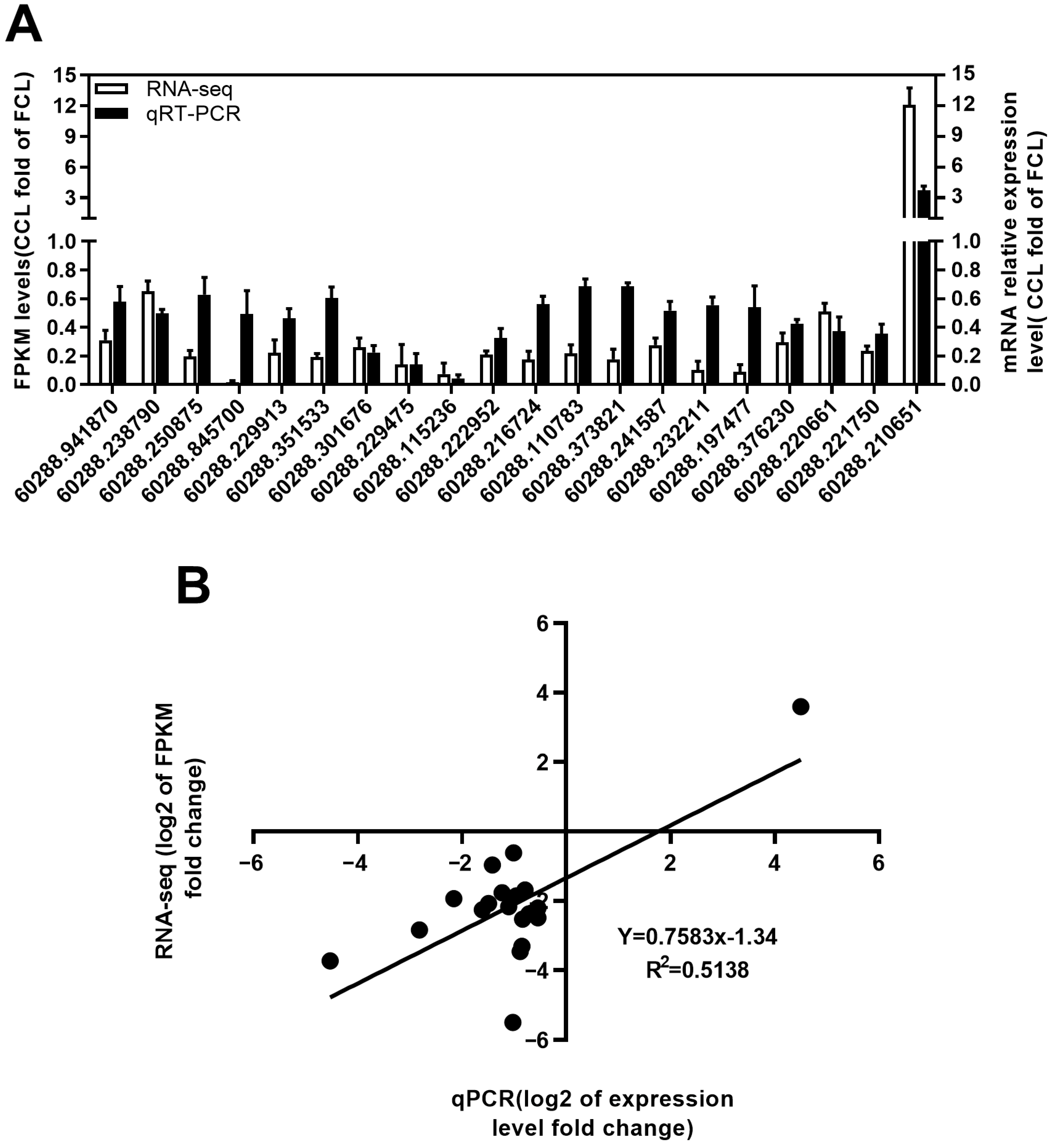
2.4. qRT-PCR Validation of RNA-Seq Expression Changes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Chlorophyll Content Measurements
4.3. Chlorophyll Fluorescence Parameters Measurements
4.4. RNA Isolation, Quantification, and Qualification
4.5. RNA-Sequencing and Analysis
4.6. Quantitative RT-PCR Validation
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zeeshan Ul Haq, M.; Yu, J.; Yao, G.; Yang, H.; Iqbal, H.A.; Tahir, H.; Cui, H.; Liu, Y.; Wu, Y. A systematic review on the continuous cropping obstacles and control strategies in medicinal plants. Int. J. Mol. Sci. 2023, 24, 12470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Lang, D.Y.; Zhang, E.H.; Bai, C.C.; Wang, H.Z. Diurnal changes in photosynthesis and antioxidants of Angelica sinensis as influenced by cropping systems. Photosynthetica 2013, 51, 252–258. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Wu, H.; Wang, J.; Wu, Y.; Lin, S.; Khan, M.U.; Zhang, Z.; Lin, W. Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 2016, 6, 26601. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fei, Y.; Liu, L.; Xiao, Y.; Pang, Y.; Kang, J.; Wang, Z. Polygonatum odoratum polysaccharides modulate gut microbiota and mitigate experimentally induced obesity in rats. Int. J. Mol. Sci. 2018, 19, 3587. [Google Scholar] [CrossRef]
- Gonzalez, G.M.; Cellini, F.; Fotopoulos, V.; Balestrini, R.; Arbona, V. New approaches to improve crop tolerance to biotic and abiotic stresses. Physiol. Plant. 2022, 174, e13547. [Google Scholar] [CrossRef]
- Mohnike, L.; Rekhter, D.; Huang, W.; Feussner, K.; Tian, H.; Herrfurth, C.; Zhang, Y.; Feussner, I. The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 2021, 33, 735–749. [Google Scholar] [CrossRef]
- Pandey, J.K.; Gopal, R. Laser-induced chlorophyll fluorescence: A technique for detection of dimethoate effect on chlorophyll content and photosynthetic activity of wheat plant. J. Fluoresc. 2011, 21, 785–791. [Google Scholar] [CrossRef]
- Kayoumu, M.; Iqbal, A.; Muhammad, N.; Li, X.; Li, L.; Wang, X.; Gui, H.; Qi, Q.; Ruan, S.; Guo, R.; et al. Phosphorus availability affects the photosynthesis and antioxidant system of contrasting low-p-tolerant cotton genotypes. Antioxidants 2023, 12, 466. [Google Scholar] [CrossRef]
- Subramani, M.; Urrea, C.A.; Habib, R.; Bhide, K.; Thimmapuram, J.; Kalavacharla, V. Comparative transcriptome analysis of tolerant and sensitive genotypes of common bean (Phaseolus vulgaris L.) in response to terminal drought stress. Plants 2023, 12, 210. [Google Scholar] [CrossRef]
- Pan, X.; Guan, L.; Lei, K.; Li, J.; Zhang, X. Transcriptional and physiological data revealed cold tolerance in a photo-thermo sensitive genic male sterile line Yu17S. BMC Plant Biol. 2022, 22, 44. [Google Scholar] [CrossRef]
- Zhang, X.; Ye, N.; Liang, C.; Mou, S.; Fan, X.; Xu, J.; Xu, D.; Zhuang, Z. De novo sequencing and analysis of the Ulva linza transcriptome to discover putative mechanisms associated with its successful colonization of coastal ecosystems. BMC Genom. 2012, 13, 565. [Google Scholar] [CrossRef] [PubMed]
- Beauvais-Fluck, R.; Slaveykova, V.I.; Cosio, C. Comparative study of Cu uptake and early transcriptome responses in the green microalga Chlamydomonas reinhardtii and the macrophyte Elodea nuttallii. Environ. Pollut. 2019, 250, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Olsson, S.; Puente-Sanchez, F.; Gomez, M.J.; Aguilera, A. Transcriptional response to copper excess and identification of genes involved in heavy metal tolerance in the extremophilic microalga Chlamydomonas acidophila. Extremophiles 2015, 19, 657–672. [Google Scholar] [CrossRef]
- Ni, X.; Jin, C.; Liu, A.; Chen, Y.; Hu, Y. Physiological and transcriptomic analyses to reveal underlying phenolic acid action in consecutive monoculture problem of Polygonatum odoratum. BMC Plant Biol. 2021, 21, 362. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, K.; Zhou, Y.; Chen, Y.; Jin, C.; Hu, Y. Integrated analysis of microRNA and RNA-Seq reveals phenolic acid secretion metabolism in continuous cropping of Polygonatum odoratum. Plants 2023, 12, 943. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, T.; Fu, F.F.; El-Kassaby, Y.A.; Wang, G. Metabolome and transcriptome analyses reveal the regulatory mechanisms of photosynthesis in developing ginkgo biloba leaves. Int. J. Mol. Sci. 2021, 22, 2601. [Google Scholar] [CrossRef]
- Wong, A.L.; Totah, N.N.; Iavarone, A.T.; Pesavento, J.J. Quick and facile preparation of histone proteins from the green microalga Chlamydomonas reinhardtii and other photosynthetic organisms. Methods 2020, 184, 102–111. [Google Scholar] [CrossRef]
- Tian, T.; Qiao, G.; Wen, Z.; Deng, B.; Qiu, Z.; Hong, Y.; Wen, X. Comparative transcriptome analysis reveals the molecular regulation underlying the adaptive mechanism of cherry (Cerasus pseudocerasus Lind L.) to shelter covering. BMC Plant Biol. 2020, 20, 27. [Google Scholar] [CrossRef]
- Zhang, L.; Zou, D.; Zeng, N.; Li, L.; Xiao, Z. Slaked lime improves growth, antioxidant capacity and reduces Cd accumulation of peanut (Arachis hypogaea L.) under Cd stress. Sci. Rep. 2022, 12, 4388. [Google Scholar] [CrossRef]
- Morey, J.S.; Monroe, E.A.; Kinney, A.L.; Beal, M.; Johnson, J.G.; Hitchcock, G.L.; Van Dolah, F.M. Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genom. 2011, 12, 346. [Google Scholar] [CrossRef]
- Mu, X.H.; Chen, Q.W.; Chen, F.J.; Yuan, L.X.; Mi, G.H. A RNA-Seq Analysis of the response of photosynthetic system to low nitrogen supply in maize leaf. Int. J. Mol. Sci. 2017, 18, 2624. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.G.; Zhang, M.Y.; Zhou, J.J.; Gao, X.; Zhu, S.D.; Yuan, L.Y.; Hou, X.L.; Liu, T.K.; Chen, G.H.; Tang, X.Y.; et al. Transcriptome analysis and differential gene expression profiling of wucai (Brassica campestris L.) in response to cold stress. BMC Genom. 2022, 23, 137. [Google Scholar] [CrossRef] [PubMed]
- Song, X.L.; Zhou, G.S.; Xu, Z.Z.; Lv, X.M.; Wang, Y.H. Detection of photosynthetic performance of stipa bungeana seedlings under climatic change using chlorophyll fluorescence imaging. Front. Plant Sci. 2016, 6, 1254. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.S.; Karami, A.; Maggi, F. Photosynthesis and chlorophyll fluorescence of Iranian licorice (Glycyrrhiza glabra L.) accessions under salinity stress. Front. Plant Sci. 2022, 13, 984944. [Google Scholar] [CrossRef]
- Ghani, M.I.; Ali, A.; Atif, M.J.; Ali, M.; Ahanger, M.A.; Chen, X.Y.L.; Cheng, Z.H. Different leafy vegetable cropping systems regulate growth, photosynthesis, and PSII functioning in mono-cropped eggplant by altering chemical properties and upregulating the antioxidant system. Front. Plant Sci. 2023, 14, 1132861. [Google Scholar] [CrossRef]
- Yue, J.; Shi, D.; Zhang, L.; Zhang, Z.; Fu, Z.; Ren, Q.; Zhang, J. The photo-inhibition of camphor leaves (Cinnamomum camphora L.) by NaCl stress based on physiological, chloroplast structure and comparative proteomic analysis. Peer J. 2020, 8, e9443. [Google Scholar] [CrossRef]
- Yadavalli, V.; Malleda, C.; Subramanyam, R. Protein-protein interactions by molecular modeling and biochemical characterization of PSI-LHCI supercomplexes from Chlamydomonas reinhardtii. Mol. Biosyst. 2011, 7, 3143–3151. [Google Scholar] [CrossRef]
- Ling, N.; Li, W.; Xu, G.; Qi, Z.; Ji, C.; Liu, X.; Cui, D.; Sun, Y. Transcriptomic sequencing reveals the response of Dunaliella salina to copper stress via the increased photosynthesis and carbon mechanism. Mol. Omics. 2021, 17, 769–782. [Google Scholar] [CrossRef]
- Tang, C.; Yang, C.; Yu, H.; Tian, S.; Huang, X.; Wang, W.; Cai, P. Electromagnetic radiation disturbed the photosynthesis of microcystis aeruginosa at the proteomics level. Sci. Rep. 2018, 8, 479. [Google Scholar] [CrossRef]
- Hayano-Kanashiro, C.; Calderon-Vazquez, C.; Ibarra-Laclette, E.; Herrera-Estrella, L.; Simpson, J. Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS ONE 2009, 4, e7531. [Google Scholar] [CrossRef]
- Wada, S.; Takagi, D.; Miyake, C.; Makino, A.; Suzuki, Y. Responses of the photosynthetic electron transport reactions stimulate the oxidation of the reaction center chlorophyll of photosystem I, P700, under drought and high temperatures in rice. Int. J. Mol. Sci. 2019, 20, 2068. [Google Scholar] [CrossRef]
- Salunke, R.; Mourier, T.; Banerjee, M.; Pain, A.; Shanmugam, D. Highly diverged novel subunit composition of apicomplexan F-type ATP synthase identified from Toxoplasma gondii. PLoS Biol. 2018, 16, e2006128. [Google Scholar] [CrossRef]
- Koc, I.; Yuksel, I.; Caetano-Anolles, G. Metabolite-centric reporter pathway and tripartite network analysis of arabidopsis under cold stress. Front. Bioeng. Biotechnol. 2018, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Waititu, J.K.; Zhang, X.; Chen, T.; Zhang, C.; Zhao, Y.; Wang, H. Transcriptome analysis of tolerant and susceptible maize genotypes reveals novel insights about the molecular mechanisms underlying drought responses in leaves. Int. J. Mol. Sci. 2021, 22, 6980. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymology 1987, 148, 350–382. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.; Tu, N.; Hu, Y.; Jin, C.; Luo, Y.; Liu, A.; Zhang, X. Integrated transcriptome and microRNA profiles analysis reveals molecular mechanisms underlying the consecutive monoculture problem of Polygonatum odoratum. Cell. Mol. Biol. 2020, 66, 47–52. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome. Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhou, Y.; Ye, J.; Jin, C.; Hu, Y. Continuous Cropping Inhibits Photosynthesis of Polygonatum odoratum. Plants 2023, 12, 3374. https://doi.org/10.3390/plants12193374
Wang Y, Zhou Y, Ye J, Jin C, Hu Y. Continuous Cropping Inhibits Photosynthesis of Polygonatum odoratum. Plants. 2023; 12(19):3374. https://doi.org/10.3390/plants12193374
Chicago/Turabian StyleWang, Yan, Yunyun Zhou, Jing Ye, Chenzhong Jin, and Yihong Hu. 2023. "Continuous Cropping Inhibits Photosynthesis of Polygonatum odoratum" Plants 12, no. 19: 3374. https://doi.org/10.3390/plants12193374
APA StyleWang, Y., Zhou, Y., Ye, J., Jin, C., & Hu, Y. (2023). Continuous Cropping Inhibits Photosynthesis of Polygonatum odoratum. Plants, 12(19), 3374. https://doi.org/10.3390/plants12193374




