A Structure Variation in qPH8.2 Detrimentally Affects Plant Architecture and Yield in Rice
Abstract
1. Introduction
2. Results
2.1. QTLs Detected for Plant Height in the CSSL Population
2.2. Genetic Validation of qPH8.2
2.3. Fine Mapping of qPH8.2
2.4. Frequency of the 42 kb Insertion in Rice Germplasm
2.5. Pleiotropic Effects of qPH8.2 on Photosynthetic Characteristics and Plant Yield
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Trait Measurement
4.3. DNA Extraction and Genotype Analysis
4.4. QTL Mapping
4.5. The Insertion Analysis in Rice Germplasm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wing, R.A.; Purugganan, M.D.; Zhang, Q. The rice genome revolution: From an ancient grain to Green Super Rice. Nat. Rev. Genet. 2018, 19, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, P.; Zhang, X.; Li, X.; Yan, X.; Fu, D.; Wu, G. The genetic and molecular basis of crop height based on a rice model. Planta 2018, 247, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Su, S.; Wang, L.; Bai, S.; Xu, J.; Li, Z.; Betts, N.; Liang, W.; Wang, W.; Shi, J.; et al. Combined genome-wide association study and epistasis analysis reveal multifaceted genetic architectures of plant height in Asian cultivated rice. Plant Cell Environ. 2023, 46, 1295–1311. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi-Tanaka, M.; Fujisawa, Y.; Kobayashi, M.; Ashikari, M.; Iwasaki, Y.; Kitano, H.; Matsuoka, M. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 2000, 97, 11638–11643. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. Green revolution: Preparing for the 21st century. Genome 1999, 42, 646–655. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Asano, K.; Takashi, T.; Miura, K.; Qian, Q.; Kitano, H.; Matsuoka, M.; Ashikari, M. Genetic and molecular analysis of utility of sd1 alleles in rice breeding. Breeding Sci. 2007, 57, 53–58. [Google Scholar] [CrossRef][Green Version]
- Oikawa, T.; Koshioka, M.; Kojima, K.; Yoshida, H.; Kawata, M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 2004, 55, 687–700. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Mi, X.; Shan, J.; Li, X.; Xu, J.; Lin, H. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016, 12, e1006386. [Google Scholar] [CrossRef]
- Qi, J.; Qian, Q.; Bu, Q.; Li, S.; Chen, Q.; Sun, J.; Liang, W.; Zhou, Y.; Chu, C.; Li, X.; et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008, 147, 1947–1959. [Google Scholar] [CrossRef]
- Jiang, D.; Fang, J.; Lou, L.; Zhao, J.; Yuan, S.; Yin, L.; Sun, W.; Peng, L.; Guo, B.; Li, X. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE 2015, 10, e0118169. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Yan, A.; Shao, K.; Wang, S.; Wang, Y.; Chen, Z.; Xu, J. Large Vascular Bundle Phloem Area 4 enhances grain yield and quality in rice via source-sink-flow. Plant Physiol. 2023, 191, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yan, J.; Zhang, Y.; Zhang, F.; Guan, Z.; Yao, Y.; Chang, Y.; Tu, H.; Li, X.; Wang, H.; et al. Serine protease NAL1 exerts pleiotropic functions through degradation of TOPLESS-related corepressor in rice. Nat. Plants 2023, 9, 1130–1142. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Yan, W.; Wang, P.; Chen, H.; Zhou, H.; Li, Q.; Wang, C.; Ding, Z.; Zhang, Y.; Yu, S.; Xing, Y.; et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Ali, M.L.; Sanchez, P.L.; Yu, S.; Lorieux, M.; Eizenga, G.C. Chromosome segment substitution lines: A powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 2010, 3, 218–234. [Google Scholar] [CrossRef]
- Wang, D.; Sun, W.; Yuan, Z.; Sun, Q.; Fan, K.; Zhang, C.; Yu, S. Identification of a novel QTL and candidate gene associated with grain size using chromosome segment substitution lines in rice. Sci. Rep. 2021, 11, 189. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.; Zhang, F.; et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Qin, P.; Lu, H.; Du, H.; Wang, H.; Chen, W.; Chen, Z.; He, Q.; Ou, S.; Zhang, H.; Li, X.; et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 2021, 184, 3542–3558. [Google Scholar] [CrossRef]
- Zhang, F.; Xue, H.; Dong, X.; Li, M.; Zheng, X.; Li, Z.; Xu, J.; Wang, W.; Wei, C. Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes. Genome Res. 2022, 32, 853–863. [Google Scholar] [CrossRef]
- Shang, L.; Li, X.; He, H.; Yuan, Q.; Song, Y.; Wei, Z.; Lin, H.; Hu, M.; Zhao, F.; Zhang, C.; et al. A super pan-genomic landscape of rice. Cell Res. 2022, 32, 878–896. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, Z.; Wang, Y.; Sun, W.; Tang, X.; Sun, Y.; Yu, S. Genetic dissection of seed dormancy in rice (Oryza sativa L.) by using two mapping populations derived from common parents. Rice 2020, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sun, Y.; Wang, D.; Sun, W.; Yu, Y.; Hu, Z.; Yu, S. Dissection of heterotic loci for grain yield using interconnected chromosome segment substitution lines in rice. The Crop J. 2022, 10, 323–331. [Google Scholar] [CrossRef]
- Takehara, S.; Sakuraba, S.; Mikami, B.; Yoshida, H.; Yoshimura, H.; Itoh, A.; Endo, M.; Watanabe, N.; Nagae, T.; Matsuoka, M.; et al. A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin. Nat. Commun. 2020, 11, 2143. [Google Scholar] [CrossRef]
- Asano, K.; Miyao, A.; Hirochika, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. SSD1, which encodes a plant-specific novel protein, controls plant elongation by regulating cell division in rice. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, D.; Surapaneni, M.; Mesapogu, S.; Neelamraju, S. Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor. Appl. Genet. 2019, 132, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.; Kim, T.; Trinugroho, J.; Cordón-Preciado, V.; Wijayatilake, N.; Bhatia, A.; Rutherford, A.; Cardona, T. The evolution and evolvability of photosystem II. Annu Rev. Plant Biol. 2023, 74, 225–257. [Google Scholar] [CrossRef]
- Ahmad, N.; Zaidi, S.; Mansoor, S. Alternative routes to improving photosynthesis in field crops. Trends Plant Sci. 2020, 25, 958–960. [Google Scholar] [CrossRef]
- Yuan, Y.; Bayer, P.; Batley, J.; Edwards, D. Current status of structural variation studies in plants. Plant Biotechnol. J. 2021, 19, 2153–2163. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1230. [Google Scholar] [CrossRef]
- Gamuyao, R.; Chin, J.; Pariasca-Tanaka, J.; Pesaresi, P.; Catausan, S.; Dalid, C.; Slamet-Loedin, I.; Tecson-Mendoza, E.; Wissuwa, M.; Heuer, S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 2012, 488, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.; Zhang, Y.; Jamil, M.; Hepworth, J.; Charnikhova, T.; Dimkpa, S.; Meharg, C.; Wright, M.; Liu, J.; Meng, X.; et al. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proc. Natl. Acad. Sci. USA 2014, 111, 2379–2384. [Google Scholar] [CrossRef]
- Yu, H.; Xie, W.; Li, J.; Zhou, F.; Zhang, Q. A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol. J. 2014, 12, 28–37. [Google Scholar] [CrossRef]
- Wang, P.; Zhou, G.; Cui, K.; Li, Z.; Yu, S. Clustered QTL for source leaf size and yield traits in rice (Oryza sativa L.). Mol. Breed. 2012, 29, 99–113. [Google Scholar] [CrossRef]
- Dong, H.; Zhao, H.; Xie, W.; Han, Z.; Li, G.; Yao, W.; Bai, X.; Hu, Y.; Guo, Z.; Lu, K.; et al. A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 2016, 12, e1006412. [Google Scholar] [CrossRef] [PubMed]
- Takai, T.; Adachi, S.; Taguchi-Shiobara, F.; Sanoh-Arai, Y.; Iwasawa, N.; Yoshinaga, S.; Hirose, S.; Taniguchi, Y.; Yamanouchi, U.; Wu, J. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci. Rep. 2013, 3, 2149. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Zhao, H.; Yao, W.; Ouyang, Y.; Yang, W.; Wang, G.; Lian, X.; Xing, Y.; Chen, L.; Xie, W. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015, 43, 1018–1022. [Google Scholar] [CrossRef]
- Panaud, O.; Chen, X.; McCouch, S. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen Genet. 1996, 252, 597–607. [Google Scholar] [CrossRef]
- Sun, W.; Zhou, Q.; Yao, Y.; Qiu, X.; Xie, K.; Yu, S. Identification of genomic regions and the isoamylase gene for reduced grain chalkiness in rice. PLoS ONE 2015, 10, e0122013. [Google Scholar] [CrossRef]
- Grömping, U. Relative importance for linear regression in R: The package relaimpo. J. Stat. Softw. 2006, 17, 1–27. [Google Scholar] [CrossRef]
- McCouch, S.R. Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. The Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
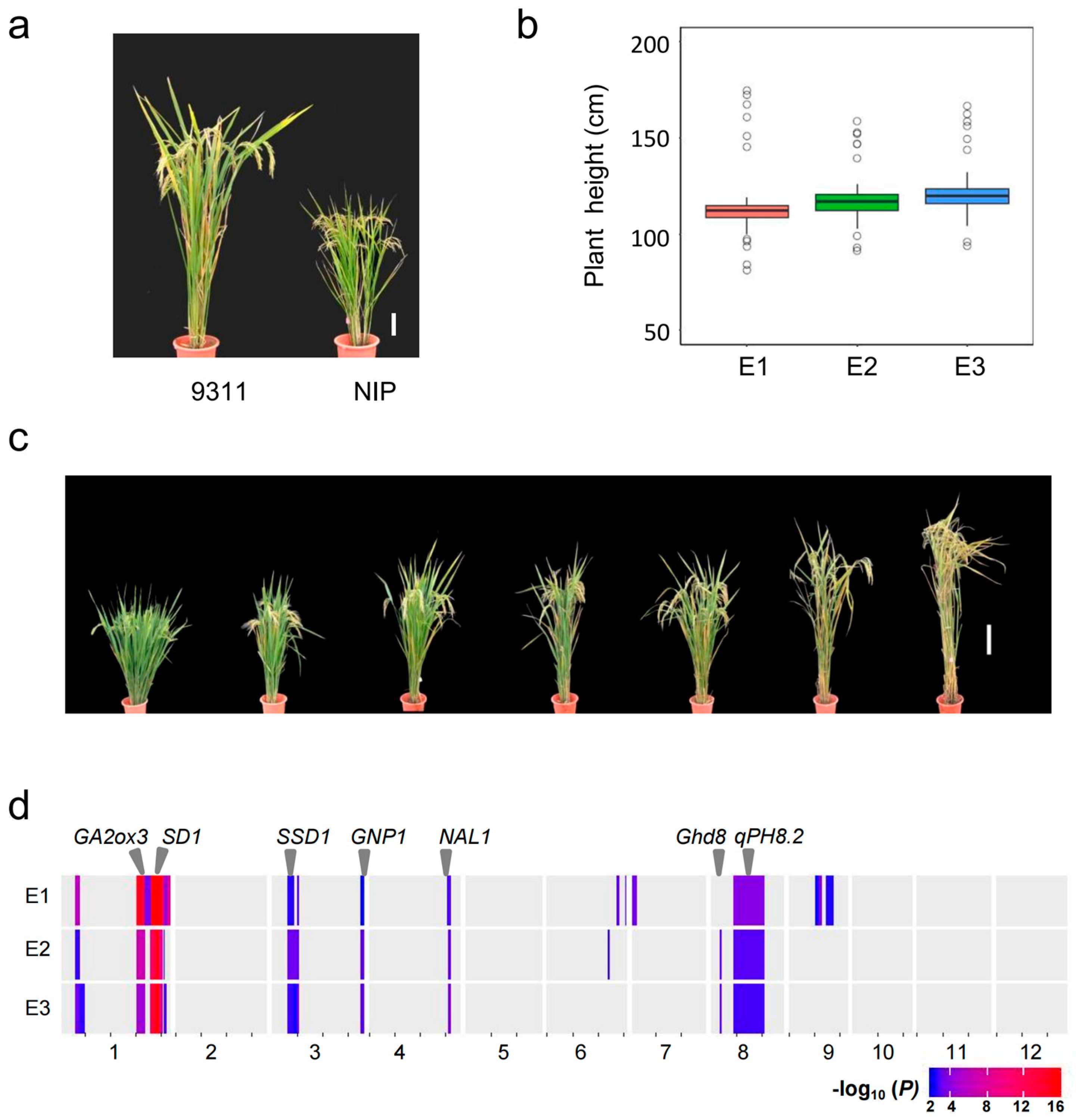
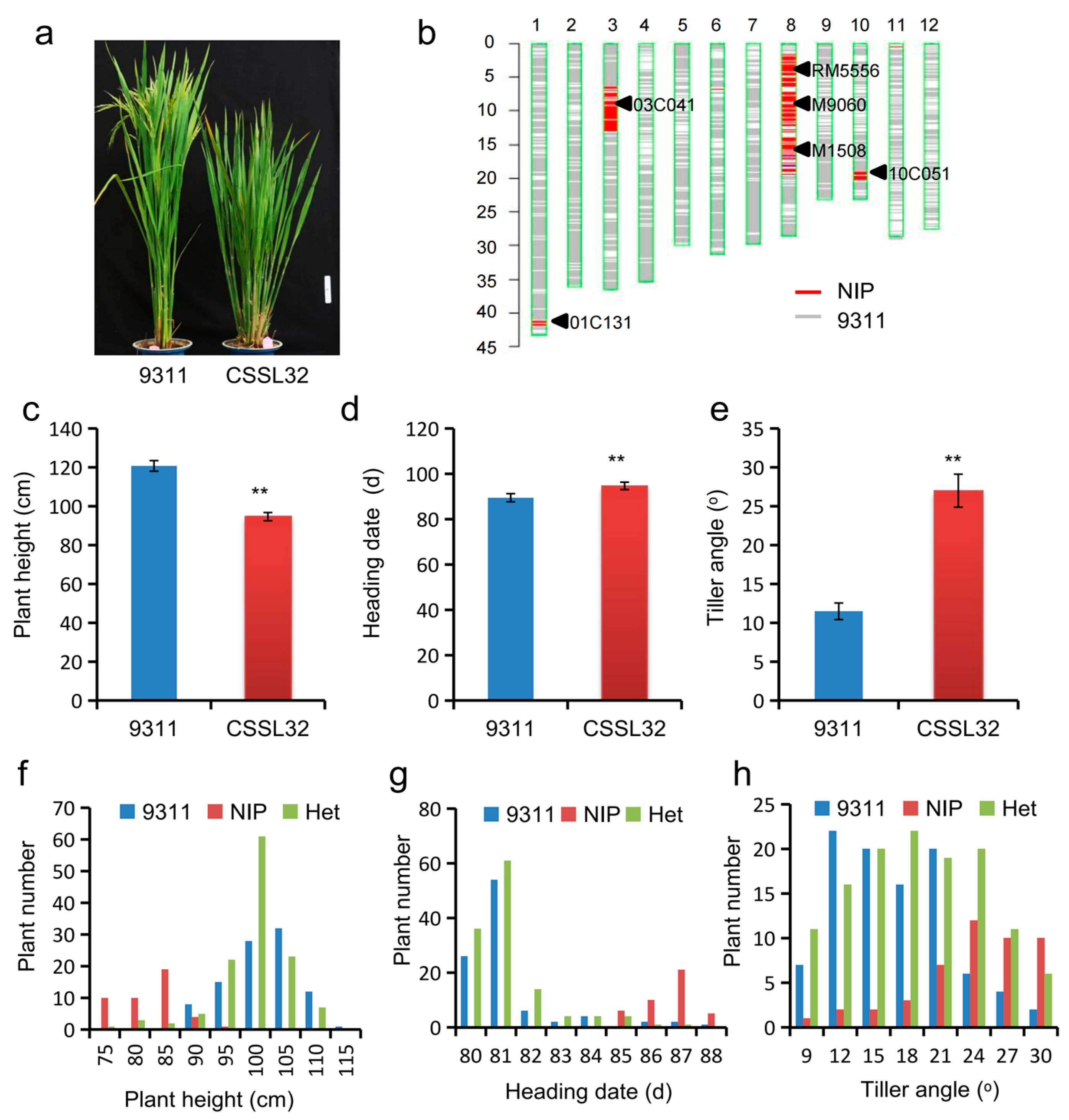
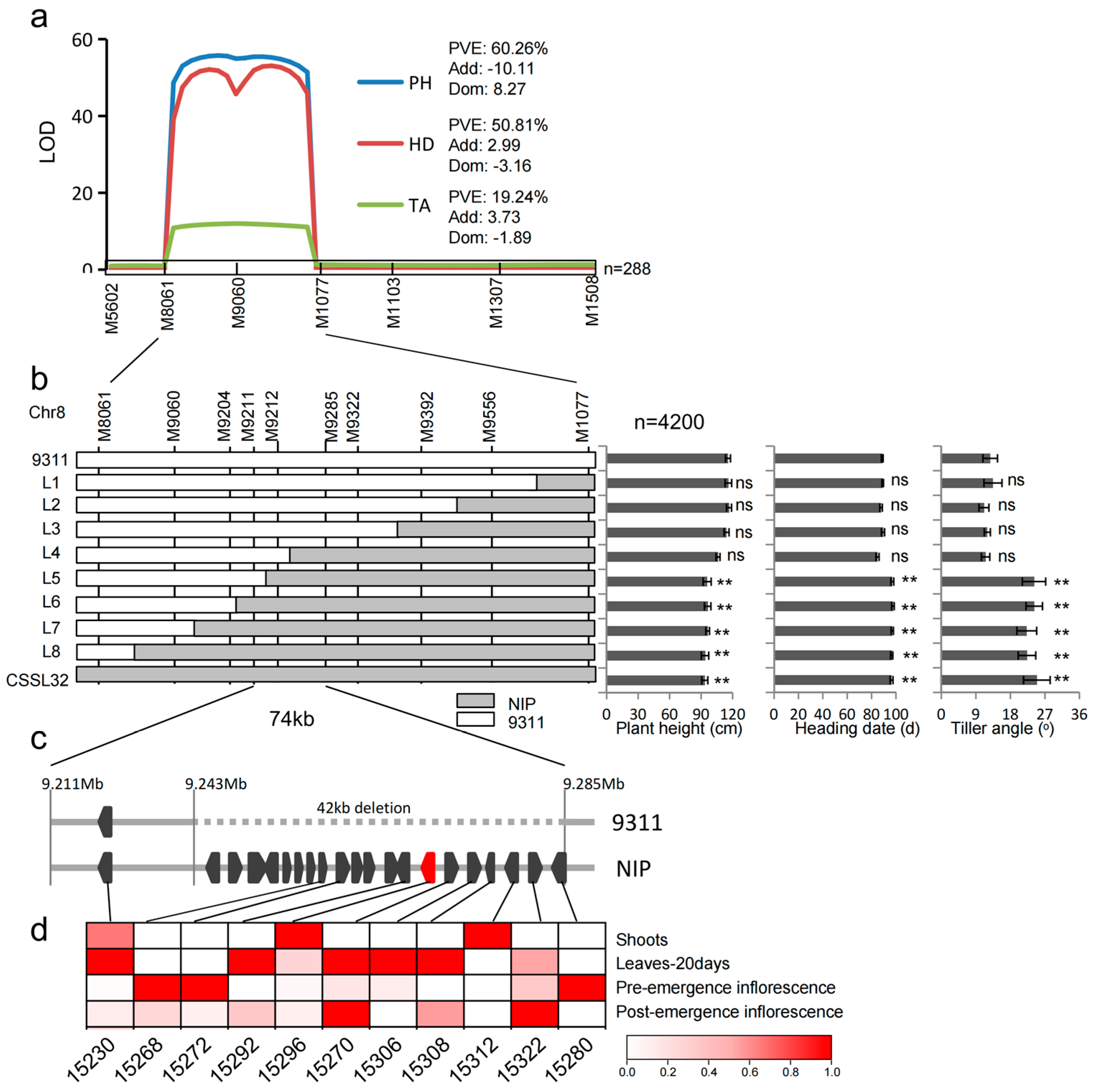
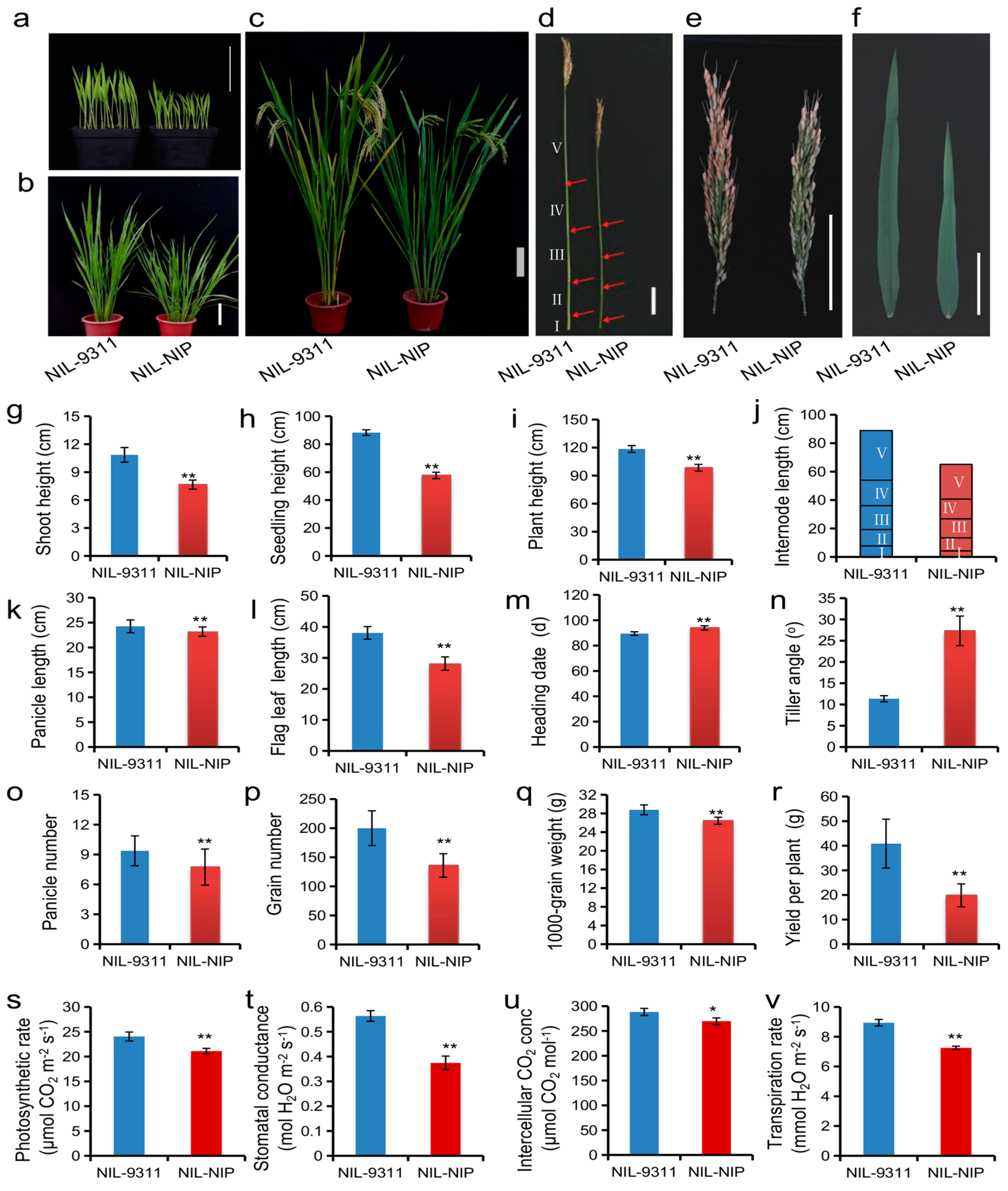
| Environment a | QTL | Chr | Interval (Mb) | Effect | p Value | PVE (%) | Gene |
|---|---|---|---|---|---|---|---|
| E1 | qPH1.1 | 1 | 6.1–7.6 | 0.10 | 1.15 × 10−6 | 6.3 | |
| qPH1.2 | 1 | 30.3–33.3 | 0.15 | 2.00 × 10−15 | 16.6 | GA2ox3 | |
| qPH1.3 | 1 | 38.1–39 | 0.20 | 2.00 × 10−16 | 41.1 | SD1 | |
| qPH3.1 | 3 | 10.3–10.5 | −0.06 | 2.37 × 10−3 | 3.2 | SSD1 | |
| qPH3.2 | 3 | 34.9–end | 0.06 | 9.06 × 10−3 | 2.1 | GNP1 | |
| qPH4 | 4 | 31.4–31.6 | −0.06 | 4.01 × 10−4 | 3.6 | NAL1 | |
| qPH8.1 | 8 | 3.5–4 | −0.03 | 5.11 × 10−2 | 0.7 | Ghd8 | |
| qPH8.2 | 8 | 9.1–20.6 | −0.07 | 1.46 × 10−4 | 5.8 | ||
| E2 | qPH1.1 | 1 | 6.1–7.6 | 0.59 | 4.44 × 10−3 | 5.6 | |
| qPH1.2 | 1 | 30.3–33.3 | 0.56 | 1.65 × 10−7 | 11.2 | GA2ox3 | |
| qPH1.3 | 1 | 38.1–39 | 1.23 | 2.00 × 10−16 | 35.2 | SD1 | |
| qPH3.1 | 3 | 10.3–10.5 | −0.37 | 6.75 × 10−4 | 5.7 | SSD1 | |
| qPH3.2 | 3 | 34.9–end | 0.65 | 1.25 × 10−3 | 1.7 | GNP1 | |
| qPH4 | 4 | 31.4–31.6 | −0.26 | 8.31 × 10−4 | 3.0 | NAL1 | |
| qPH8.1 | 8 | 3.5–4 | −0.27 | 1.91 × 10−3 | 2.3 | Ghd8 | |
| qPH8.2 | 8 | 9.1–20.6 | −0.42 | 1.78 × 10−3 | 6.2 | ||
| E3 | qPH1.1 | 1 | 6.1–7.6 | 0.70 | 5.16 × 10−4 | 6.1 | |
| qPH1.2 | 1 | 30.3–33.3 | 0.44 | 2.32 × 10−5 | 10.2 | GA2ox3 | |
| qPH1.3 | 1 | 38.1–39 | 1.17 | 2.00 × 10−16 | 36.9 | SD1 | |
| qPH3.1 | 3 | 10.3–10.5 | −0.42 | 9.28 × 10−5 | 5.2 | SSD1 | |
| qPH3.2 | 3 | 34.9–end | 0.62 | 1.40 × 10−3 | 1.8 | GNP1 | |
| qPH4 | 4 | 31.4–31.6 | −0.28 | 1.75 × 10−4 | 5.2 | NAL1 | |
| qPH8.1 | 8 | 3.5–4 | −0.27 | 1.36 × 10−3 | 1.8 | Ghd8 | |
| qPH8.2 | 8 | 9.1–20.6 | −0.38 | 3.34 × 10−3 | 5.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Sun, Q.; Tian, L.; Sun, Y.; Yu, S. A Structure Variation in qPH8.2 Detrimentally Affects Plant Architecture and Yield in Rice. Plants 2023, 12, 3336. https://doi.org/10.3390/plants12183336
Sun W, Sun Q, Tian L, Sun Y, Yu S. A Structure Variation in qPH8.2 Detrimentally Affects Plant Architecture and Yield in Rice. Plants. 2023; 12(18):3336. https://doi.org/10.3390/plants12183336
Chicago/Turabian StyleSun, Wenqiang, Qiang Sun, Li Tian, Yongjian Sun, and Sibin Yu. 2023. "A Structure Variation in qPH8.2 Detrimentally Affects Plant Architecture and Yield in Rice" Plants 12, no. 18: 3336. https://doi.org/10.3390/plants12183336
APA StyleSun, W., Sun, Q., Tian, L., Sun, Y., & Yu, S. (2023). A Structure Variation in qPH8.2 Detrimentally Affects Plant Architecture and Yield in Rice. Plants, 12(18), 3336. https://doi.org/10.3390/plants12183336








