Integrative Analysis of Metabolome and Transcriptome Revealed Lutein Metabolism Contributed to Yellow Flower Formation in Prunus mume
Abstract
1. Introduction
2. Results
2.1. Description of Flower Color Parameters for the Yellow Variety
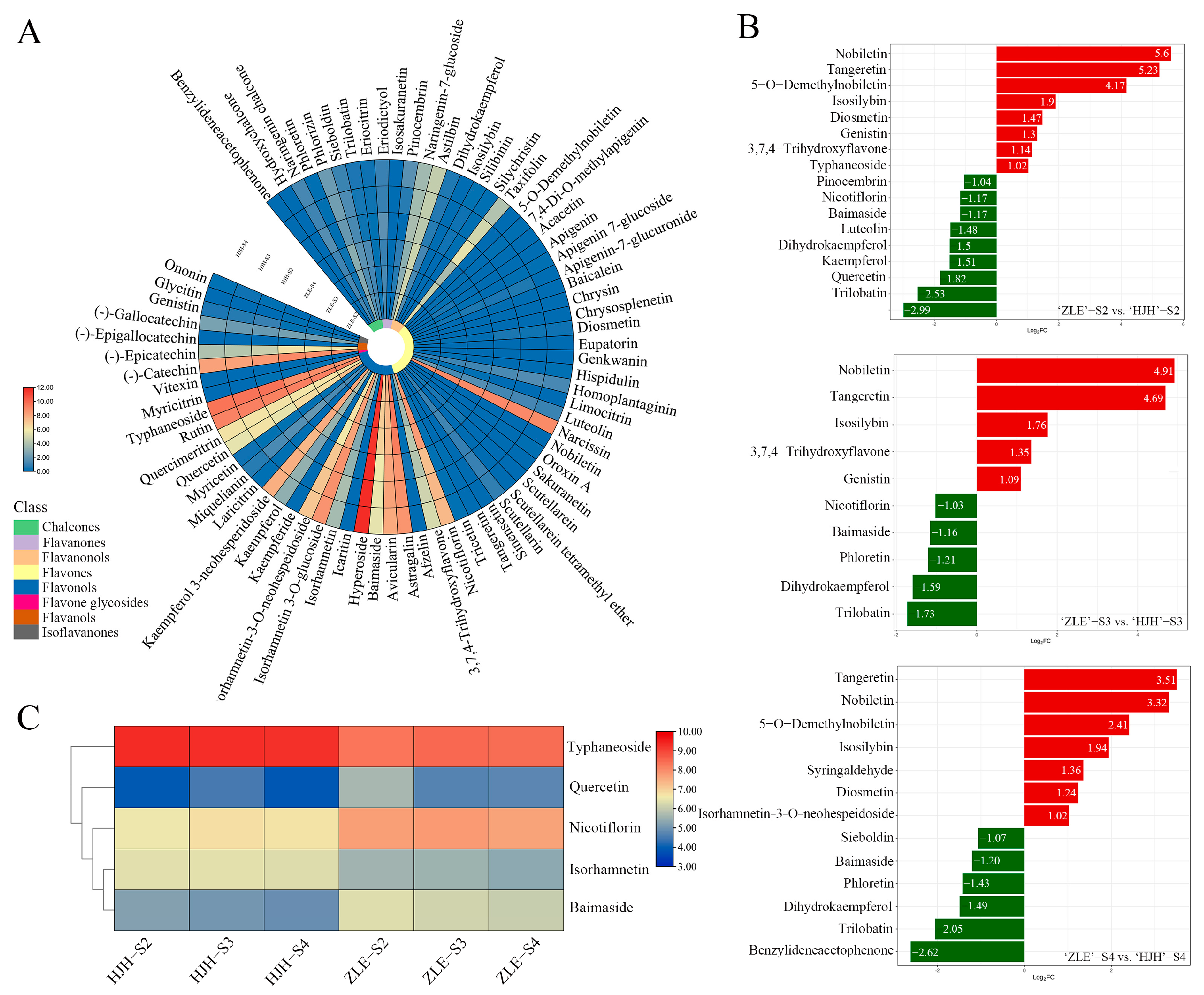
2.2. Identification of Flavonoids and Carotenoids in Yellow Flowers
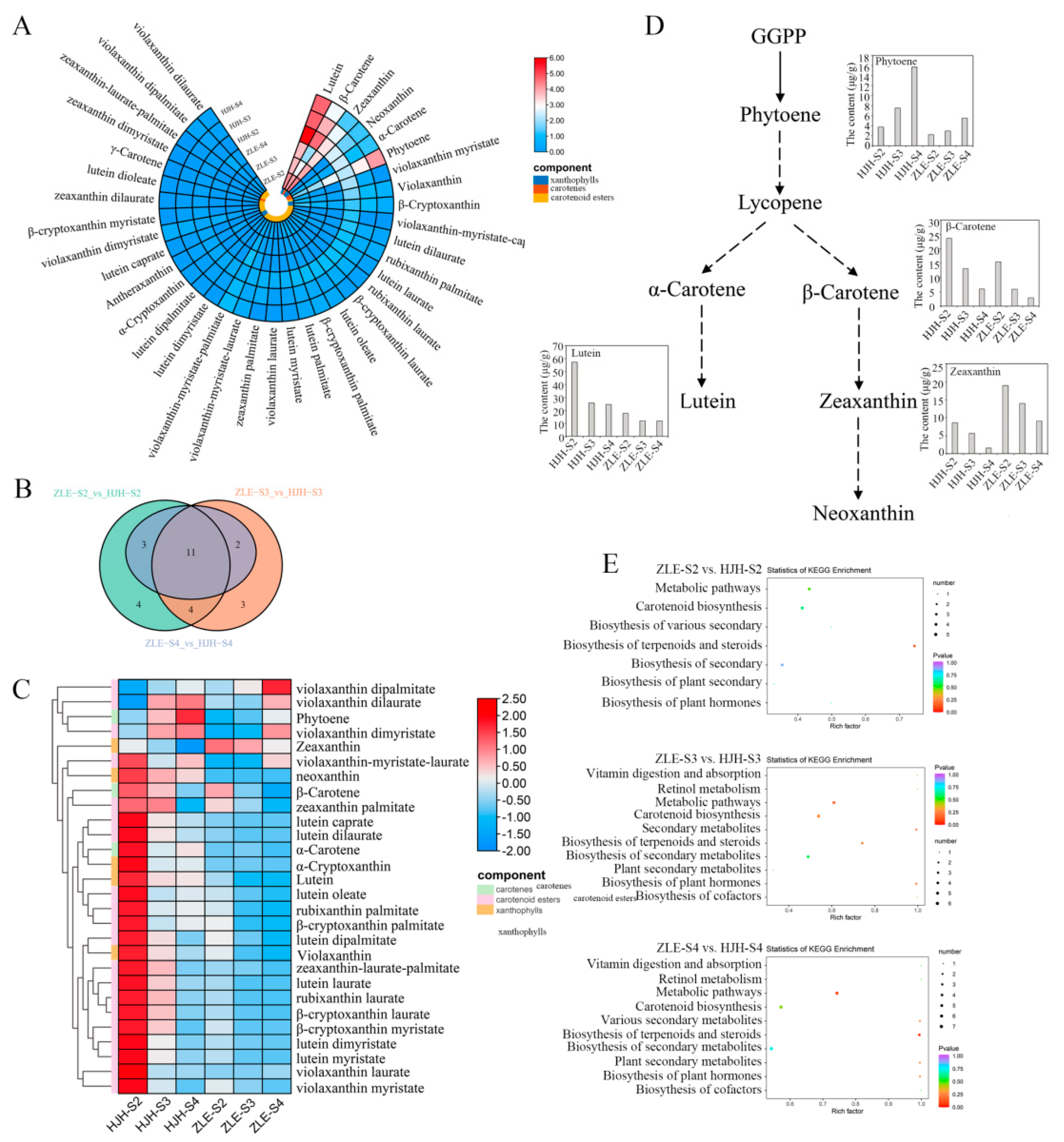
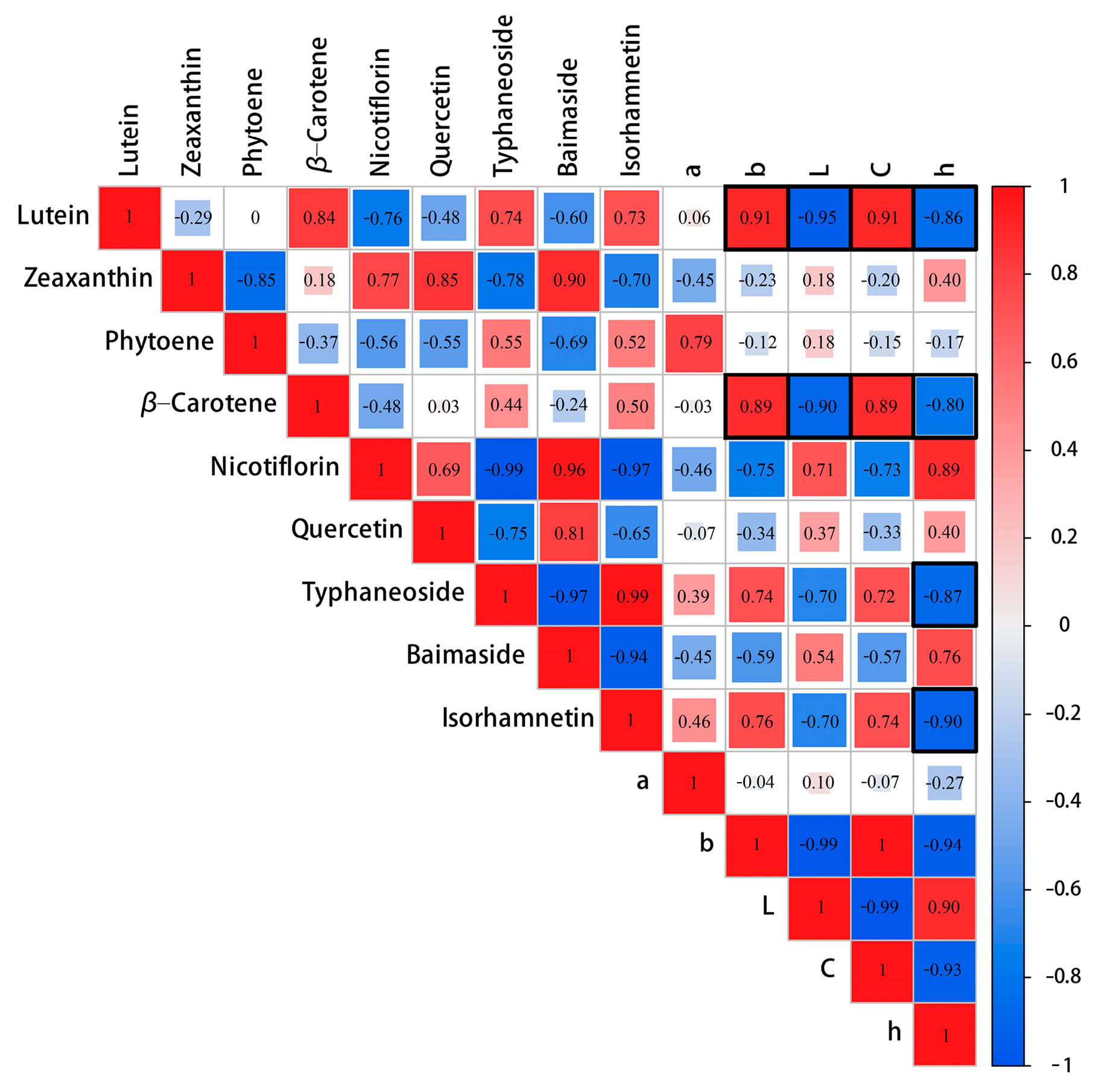
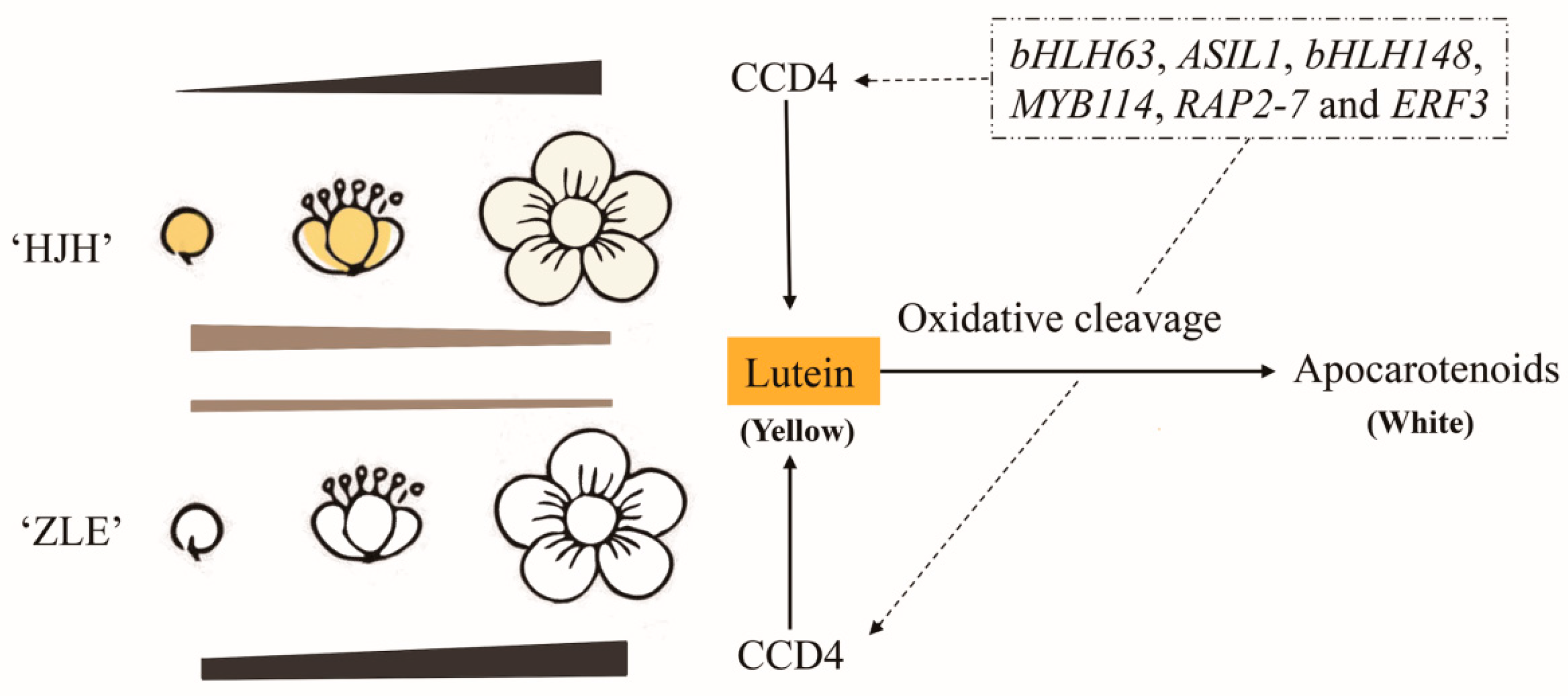
2.3. Lutein Determined the Yellow Flower Coloration in P. mume
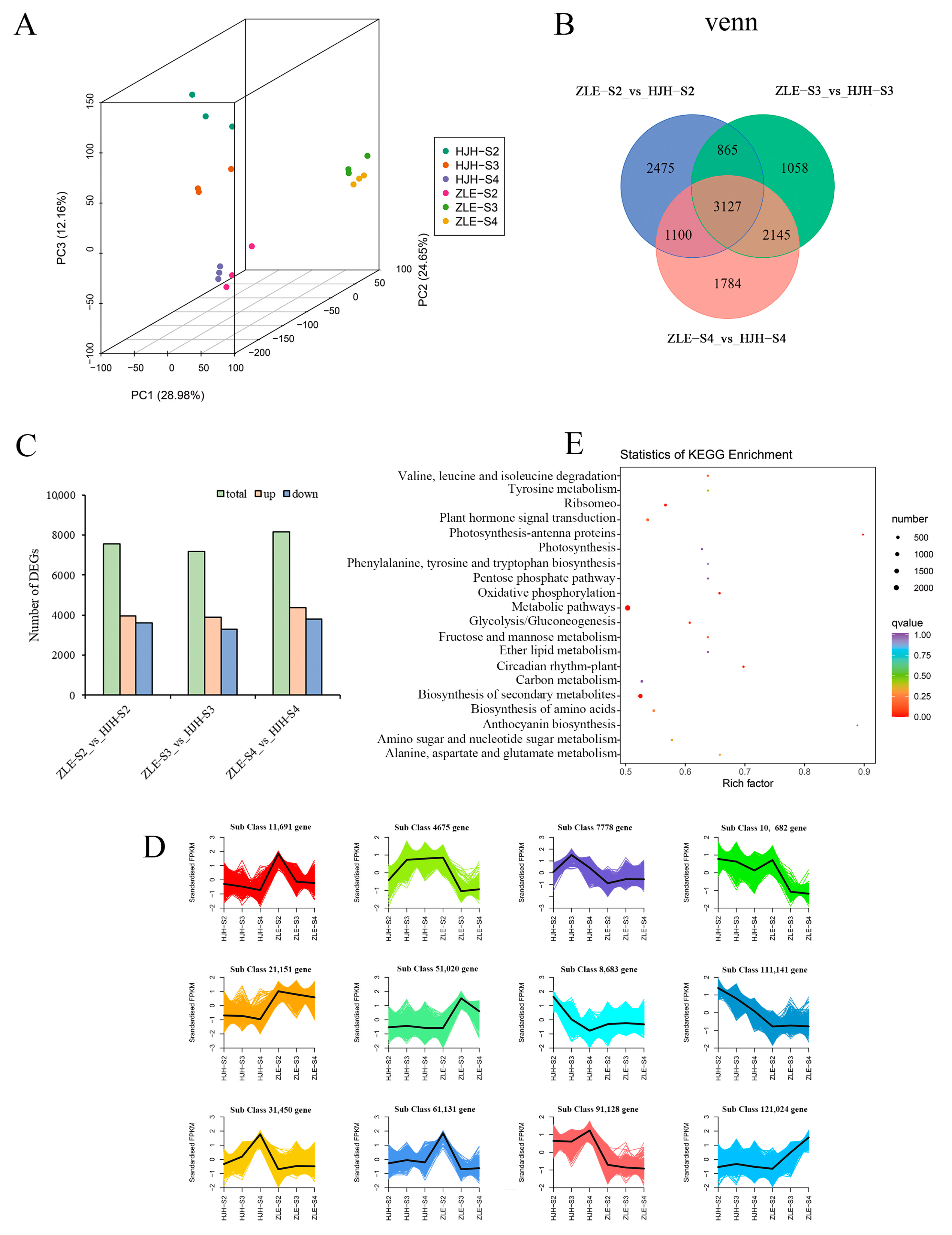
2.4. Transcriptome Sequencing, Annotation and Analysis of DEGs
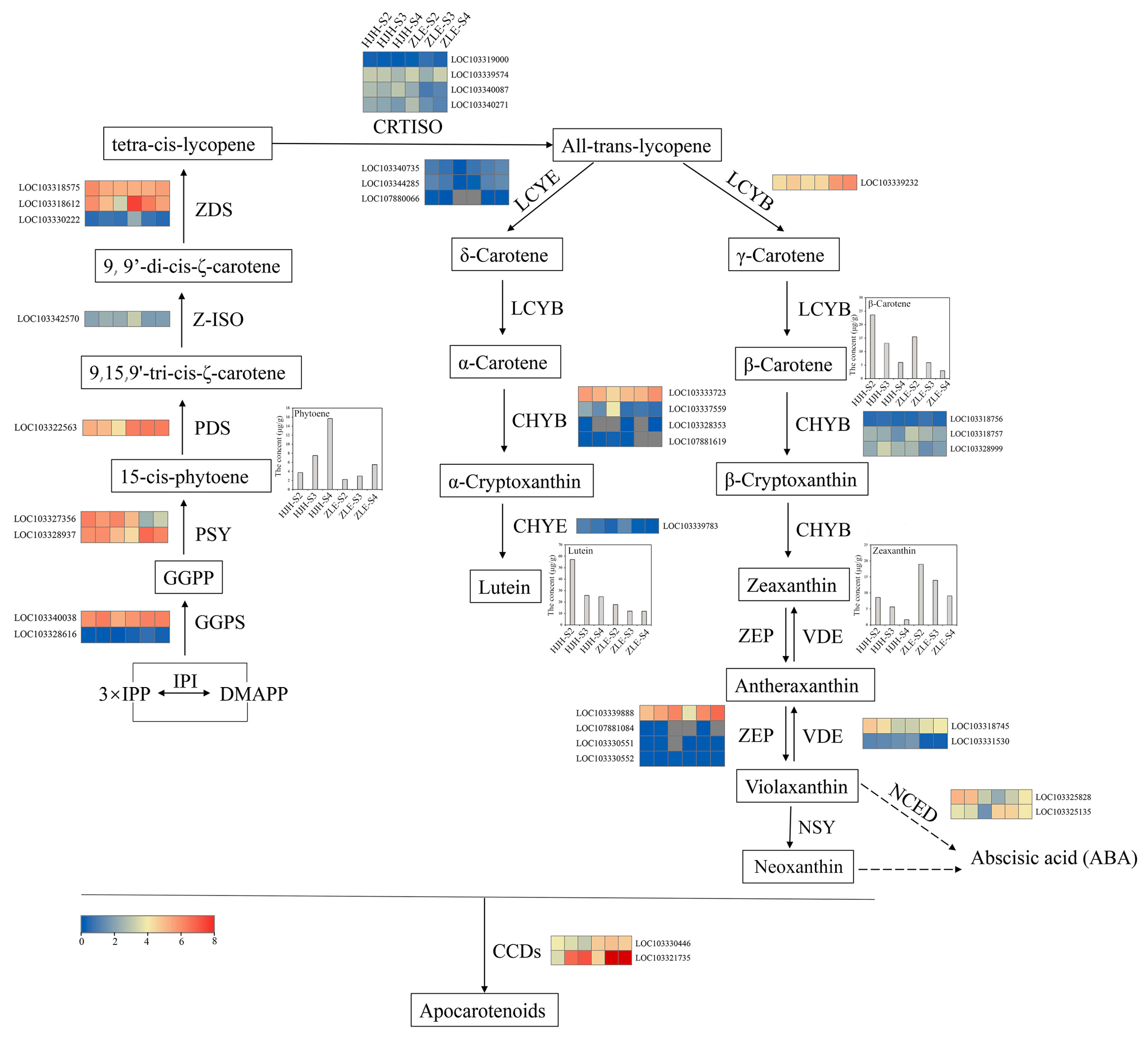
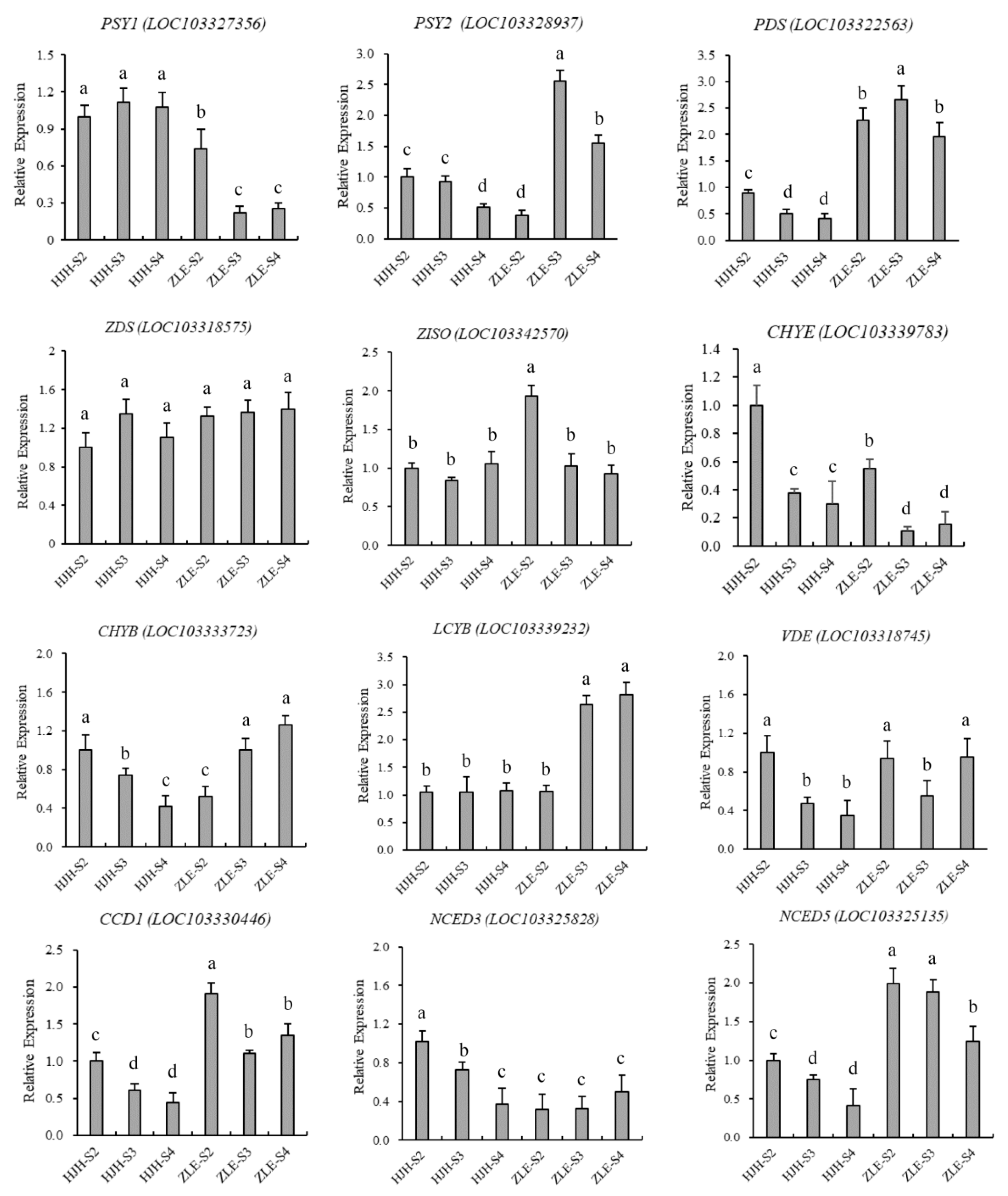
2.5. Structural Genes Associated with Carotenoid Biosynthesis
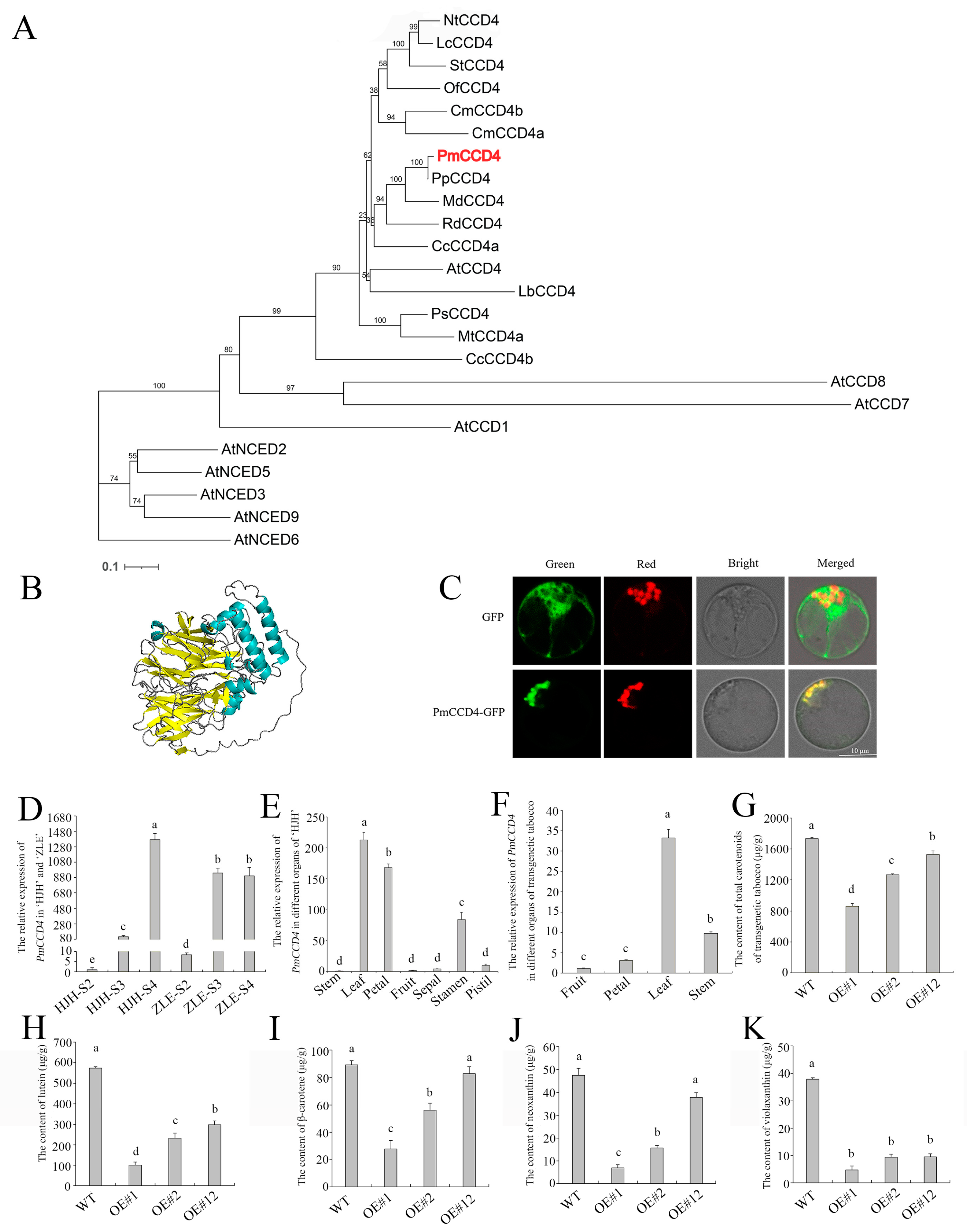
2.6. Potential Transcriptional Regulation Mechanisms
2.7. Overexpression of PmCCD4 Decreased Carotenoids Content in Tobacco
3. Discussion
3.1. Impact of Flavonoids and Carotenoids on Yellow Flowers Coloration in P. mume
3.2. Carotenoid Biosynthesis in P. mume
3.3. Potential Regulatory Genes Related to Carotenoids in P. mume
3.4. PmCCD4 Reduced Carotenoids in Transgenic Tobacco
4. Material and Methods
4.1. Plant Materials
4.2. Petal Color Measurement
4.3. Metabolome Analysis
4.4. RNA Isolation, Library Establishment, and Sequencing
4.5. Transcriptome Assembly and Gene Annotation
4.6. Differentially Expressed Genes Screening
4.7. Quantitative Real-Time PCR Analysis of Screened Gene
4.8. Gene Clone and Protein Analysis
4.9. Subcellular Localization Analysis
4.10. Determination of Carotenoids from PmCCD4-Overexpressing Transgenic Tobacco
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, R.; Owens, S.J.; Rørslett, B. Plants and colour: Flowers and pollination. Opt. Laser Technol. 2011, 43, 282–294. [Google Scholar] [CrossRef]
- Cui, C.; Zhang, S.; You, L.; Ren, J.; Luo, W.; Chen, W.; Zhao, M. Antioxidant capacity of anthocyanins from Rhodomyrtus tomentosa (Ait.) and identification of the major anthocyanins. Food Chem. 2013, 39, 1–8. [Google Scholar] [CrossRef]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Chemistry, Pharmacology and Health Benefits of Anthocyanins. Phytother. Res. 2016, 30, 1265–1286. [Google Scholar] [CrossRef]
- Tanaka, Y.; Tsuda, S.; Kusumi, T. Metabolic Engineering to Modify Flower Color. Plant Cell Physiol. 1998, 39, 1119–1126. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Strack, D.; Vogt, T.; Schliemann, W. Recent advances in betalain research. Phytochemistry 2003, 62, 247–269. [Google Scholar] [CrossRef]
- Xu, L.; Yang, P.; Feng, Y.; Xu, H.; Cao, Y.; Tang, Y.; Yuan, S.; Liu, X.; Ming, J. Spatiotemporal Transcriptome Analysis Provides Insights into Bicolor Tepal Development in Lilium “Tiny Padhye”. Front. Plant Sci. 2017, 8, 398. [Google Scholar] [CrossRef]
- He, C.; Liu, X.; Da Silva, J.A.T.; Liu, N.; Zhang, M.; Duan, J. Transcriptome sequencing and metabolite profiling analyses provide comprehensive insight into molecular mechanisms of flower development in Dendrobium officinale (Orchidaceae). Plant Mol. Biol. 2020, 104, 529–548. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Tanaka, Y.; Ohmiya, A. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotech. 2008, 19, 190–197. [Google Scholar] [CrossRef]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2017, 11, 58–74. [Google Scholar] [CrossRef]
- Polívka, T.; Frank, H.A. Molecular Factors Controlling Photosynthetic Light Harvesting by Carotenoids. Acc. Chem. Res. 2010, 43, 1125–1134. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Xie, X.; Yoneyama, K.; Yoneyama, K. The Strigolactone Story. Annu. Rev. Phytopathol. 2010, 48, 93–117. [Google Scholar] [CrossRef]
- Marco Busch, A.S.A.R. Functional Analysis of the Early Steps of Carotenoid Biosynthesis in Tobacco. Plant Physiol. 2002, 128, 439–453. [Google Scholar] [CrossRef]
- Bai, C.; Capell, T.; Berman, J.; Medina, V.; Sandmann, G.; Christou, P.; Zhu, C. Bottlenecks in carotenoid biosynthesis and accumulation in rice endosperm are influenced by the precursor-product balance. Plant Biotechnol. J. 2016, 14, 195–205. [Google Scholar] [CrossRef]
- Gady, A.L.F.; Vriezen, W.H.; Van de Wal, M.H.B.J.; Huang, P.; Bovy, A.G.; Visser, R.G.F.; Bachem, C.W.B. Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol. Breed. 2012, 29, 801–812. [Google Scholar] [CrossRef]
- Giuliano, G.; Bartley, G.E.; Scolnik, P.A. Regulation of Carotenoid Biosynthesis during Tomato Development. Plant Cell 1993, 5, 379–387. [Google Scholar]
- Pogson, B.; McDonald, K.A.; Truong, M.; Britton, G.; DellaPenna, D. Arabidopsis Camtenoid Mutants Demonstrate That Lutein 1s Not Essential for Photosynthesis in Higher Plants. Plant Cell 1996, 8, 1627–1639. [Google Scholar]
- Ke, Q.; Kang, L.; Kim, H.S.; Xie, T.; Liu, C.; Ji, C.Y.; Kim, S.H.; Park, W.S.; Ahn, M.; Wang, S.; et al. Down-regulation of lycopene ε-cyclase expression in transgenic sweetpotato plants increases the carotenoid content and tolerance to abiotic stress. Plant Sci. 2019, 281, 52–60. [Google Scholar] [CrossRef]
- Ohmiya, A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. Nar. 2009, 26, 351–358. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Simkin, A.J.; Schwartz, S.H.; Auldridge, M.; Taylor, M.G.; Klee, H.J. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 2004, 40, 882–892. [Google Scholar] [CrossRef]
- Iuchi, S.S.; Kobayashi, M.M.; Yamaguchi-Shinozaki, K.K.; Shinozaki, K.K. A Stress-Inducible Gene for 9-cis-Epoxycarotenoid Dioxygenase Involved in Abscisic Acid Biosynthesis under Water Stress in Drought-Tolerant Cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef]
- Phadungsawat, B.; Watanabe, K.; Mizuno, S.; Kanekatsu, M.; Suzuki, S. Expression of CCD4 gene involved in carotenoid degradation in yellow-flowered Petunia × hybrida. Sci. Hortic. Amst. 2020, 261, 108916. [Google Scholar] [CrossRef]
- Drummond, R.S.M.; Martínez-Sánchez, N.M.; Janssen, B.J.; Templeton, K.R.; Simons, J.L.; Quinn, B.D.; Karunairetnam, S.; Snowden, K.C. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 Is Involved in the Production of Negative and Positive Branching Signals in Petunia. Plant Physiol. 2009, 151, 1867–1877. [Google Scholar] [CrossRef]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid Cleavage Dioxygenase (CmCCD4a) Contributes to White Color Formation in Chrysanthemum Petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef]
- Tian, X.; Ji, J.; Wang, G.; Jin, C.; Guan, C.; Wu, G. Molecular cloning and characterization of a novel carotenoid cleavage dioxygenase 1 from Lycium chinense. Biotechnol. Appl. Biochem. 2015, 62, 772–779. [Google Scholar] [CrossRef]
- Tian, X.; Ji, J.; Wang, G.; Jin, C.; Jia, C.; Li, Z. Cloning and Functional Characterisation of Carotenoid Cleavage Dioxygenase 4 from Wolfberry. Trans. Tianjin Univ. 2017, 23, 62–69. [Google Scholar] [CrossRef]
- Zheng, X.; Zhu, K.; Sun, Q.; Zhang, W.; Wang, X.; Cao, H.; Tan, M.; Xie, Z.; Zeng, Y.; Ye, J.; et al. Natural Variation in CCD4 Promoter Underpins Species-Specific Evolution of Red Coloration in Citrus Peel. Mol. Plant 2019, 12, 1294–1307. [Google Scholar] [CrossRef] [PubMed]
- Brandi, F.; Bar, E.; Mourgues, F.; Horvath, G.; Turcsi, E.; Giuliano, G.; Liverani, A.; Tartarini, S.; Lewinsohn, E.; Rosati, C. Study of ‘Redhaven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biol. 2011, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wu, M.; Cao, L.; Yuan, W.; Dong, M.; Wang, X.; Chen, W.; Shang, F. Characterization of OfWRKY3, a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene OfCCD4 in Osmanthus fragrans. Plant Mol. Biol. 2016, 91, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, H.; Wang, X.; Li, K.; Dong, M.; Li, Y.; Zhu, Q.; Shang, F. Mechanism of floral scent production in Osmanthus fragrans and the production and regulation of its key floral constituents, β-ionone and linalool. Hortic. Res. 2019, 6, 106. [Google Scholar] [CrossRef]
- Han, Y.; Lu, M.; Yue, S.; Li, K.; Dong, M.; Liu, L.; Wang, H.; Shang, F. Comparative methylomics and chromatin accessibility analysis in Osmanthus fragrans uncovers regulation of genic transcription and mechanisms of key floral scent production. Hortic. Res. 2022, 9, uhac096. [Google Scholar] [CrossRef]
- Lu, C.; Qu, J.; Deng, C.; Liu, F.; Zhang, F.; Huang, H.; Dai, S. The transcription factor complex CmAP3-CmPI-CmUIF1 modulates carotenoid metabolism by directly regulating the carotenogenic gene CmCCD4a-2 in chrysanthemum. Hortic. Res. 2022, 9, uhac020. [Google Scholar] [CrossRef]
- Zhao, C.; Guo, W.; Chen, J. Molecular Structures of the Anthocyanins from the Flower Color Pigment of Prunus mume ‘Nanjing Hong’ (Nanjing red). Acta Bot. Yunnanica 2004, 26, 549–557. [Google Scholar]
- Zhao, C.; Guo, W.; Chen, J. Isolation and Structural Identification of the Anthocyanins from the Flower Color Pigment of Prunus mume ‘Nanjing Hongxu’. Sci. Silvae Sin. 2006, 1, 29–36. [Google Scholar]
- Zhang, Q.; Hao, R.; Xu, Z.; Yang, W.; Wang, J.; Cheng, T.; Pan, H.; Zhang, Q. Isolation and functional characterization of a R2R3-MYB regulator of Prunus mume anthocyanin biosynthetic pathway. Plant Cell Tissue Organ Cult. 2017, 131, 417–429. [Google Scholar] [CrossRef]
- Yue, F.; Chen, Y.; Liu, B.; Gan, D.; Wang, H.; Wang, D. Identification of WD40 gene family in Prunus mume and its expression level under light illumination. PeerJ Prepr. 2018, 6, e27101v–e27106v. [Google Scholar] [CrossRef]
- Ma, K.; Zhang, Q.; Cheng, T.; Yan, X.; Pan, H.; Wang, J. Substantial Epigenetic Variation Causing Flower Color Chimerism in the Ornamental Tree Prunus mume Revealed by Single Base Resolution Methylome Detection and Transcriptome Sequencing. Int. J. Mol. Sci. 2018, 19, 2315. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, P.K.; Beyer, P.; Wunn, J.; Kloti, A.; Armstrong, G.A.; Schledz, M.; von Lintig, J.; Potrykus, I. Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J. 1997, 11, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; McCarty, D.R.; Klee, H.J. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 2006, 9, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Itoh, Y.; Ozeki, Y.; Iwashina, T.; Yamaguchi, M. Variation in chalcononaringenin 2′-O-glucoside content in the petals of carnations (Dianthus caryophyllus) bearing yellow flowers. Sci. Hortic. Amst. 2004, 99, 175–186. [Google Scholar] [CrossRef]
- Togami, J.; Okuhara, H.; Nakamura, N.; Ishiguro, K.; Hirose, C.; Ochiai, M.; Fukui, Y.; Yamaguchi, M.; Tanaka, Y. Isolation of cDNAs encoding tetrahydroxychalcone 2′-glucosyltransferase activity from carnation, cyclamen, and catharanthus. Plant Biotechnol. Nar. 2011, 28, 231–238. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Li, B.; Feng, C.; Wu, Q.; Wang, Q.; Li, S.; Yu, X.; Wang, L. Chemical Mechanism of Flower Color Microvariation in Paeonia with Yellow Flowers. Hortic. Plant J. 2020, 6, 179–190. [Google Scholar] [CrossRef]
- Xue, Q.; Fan, H.; Yao, F.; Cao, X.; Liu, M.; Sun, J.; Liu, Y. Transcriptomics and targeted metabolomics profilings for elucidation of pigmentation in Lonicera japonica flowers at different developmental stages. Ind. Crop. Prod. 2020, 145, 111981. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, S.; Zhang, H.; Chen, X.; Liang, F.; Qin, H.; Zhang, Y.; Cong, R.; Xin, H.; Zhang, Z. Carotenoid metabolite and transcriptome dynamics underlying flower color in marigold (Tagetes erecta L.). Sci. Rep. 2020, 10, 16835. [Google Scholar] [CrossRef]
- Wan, H.; Yu, C.; Han, Y.; Guo, X.; Ahmad, S.; Tang, A.; Wang, J.; Cheng, T.; Pan, H.; Zhang, Q. Flavonols and Carotenoids in Yellow Petals of Rose Cultivar (Rosa ‘Sun City’): A Possible Rich Source of Bioactive Compounds. J. Agr. Food Chem. 2018, 66, 4171–4181. [Google Scholar] [CrossRef]
- Wan, H.; Yu, C.; Han, Y.; Guo, X.; Luo, L.; Pan, H.; Zheng, T.; Wang, J.; Cheng, T.; Zhang, Q. Determination of Flavonoids and Carotenoids and Their Contributions to Various Colors of Rose Cultivars (Rosa spp.). Front. Plant Sci. 2019, 10, 123. [Google Scholar] [CrossRef]
- Hao, Z.; Liu, S.; Hu, L.; Shi, J.; Chen, J. Transcriptome analysis and metabolic profiling reveal the key role of carotenoids in the petal coloration of Liriodendron tulipifera. Hortic. Res. 2020, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Hirschberg, J.; Cohen, M.; Harker, M.; Lotan, T.; Mann, V.; Pecker, I. Pecker Molecular genetics of the carotenoid biosynthesis pathway in plants and algae. Pure Appl. Chem. 1997, 69, 2151–2158. [Google Scholar] [CrossRef]
- Moehs, C.P.; Tian, L.; Osteryoung, K.W.; Dellapenna, D. Analysis of carotenoid biosynthetic gene expression during marigold petal development. Plant Mol. Biol. 2001, 45, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Pecker, I.; Gabbay, R.; Cunningham, F.J.; Hirschberg, J. Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol. Biol. 1996, 30, 807–819. [Google Scholar] [CrossRef]
- Ronen, G.; Cohen, M.; Zamir, D.; Hirschberg, J. Regulation of carotenoid biosynthesis during tomato fruit development: Expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 1999, 17, 341–351. [Google Scholar] [CrossRef]
- Ureshino, K.; Nakayama, M.; Miyajima, I. Contribution made by the carotenoid cleavage dioxygenase 4 gene to yellow colour fade in azalea petals. Euphytica 2016, 207, 401–417. [Google Scholar] [CrossRef]
- Hai, N.T.L.; Masuda, J.; Miyajima, I.; Thien, N.Q.; Mojtahedi, N.; Hiramatsu, M.; Kim, J.; Okubo, H. Involvement of Carotenoid Cleavage Dioxygenase 4 Gene in Tepal Color Change in Lilium brownii var. colchesteri. J. Jpn. Soc. Hortic. Sci. 2012, 81, 366–373. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Zhang, R.; Shi, D.; Lin, N.; Guo, P.; Wang, Y.; Shang, F.; Liu, Y. Transcriptome and carotenoids profiling of flowers in different Osmanthus fragrans cultivars provide insight into transcriptional control network of carotenoid-related genes expression. Sci. Hortic. 2022, 303, 111201. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney-Smith, C.; Mila, I.; Bouzayen, M.; Magallanes-Lundback, M.; DellaPenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef]
- Lin, W.; Li, Y.; Lu, Q.; Lu, H.; Li, J. Combined Analysis of the Metabolome and Transcriptome Identified Candidate Genes Involved in Phenolic Acid Biosynthesis in the Leaves of Cyclocarya paliurus. Int. J. Mol. Sci. 2020, 21, 1337. [Google Scholar] [CrossRef]
- Lu, S.; Ye, J.; Zhu, K.; Zhang, Y.; Zhang, M.; Xu, Q.; Deng, X. A fruit ripening-associated transcription factor CsMADS5 positively regulates carotenoid biosynthesis in citrus. J. Exp. Bot. 2021, 72, 3028–3043. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The Citrus Transcription Factor CsMADS6 Modulates Carotenoid Metabolism by Directly Regulating Carotenogenic Genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Sun, Q.; Chen, H.; Mei, X.; Lu, S.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J. Exp. Bot. 2021, 72, 3137–3154. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, M.; Wang, Y.; Shen, S. The R2R3-MYB transcription factor MYB44 modulates carotenoid biosynthesis in Ulva prolifera. Algal Res. 2022, 62, 102578. [Google Scholar] [CrossRef]
- Dang, Q.; Sha, H.; Nie, J.; Wang, Y.; Yuan, Y.; Jia, D. An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation. Hortic. Res. 2021, 8, 223. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A New Tomato NAC (NAM/ATAF1/2/CUC2) Transcription Factor, SlNAC4, Functions as a Positive Regulator of Fruit Ripening and Carotenoid Accumulation. Plant Cell Physiol. 2014, 55, 119–135. [Google Scholar] [CrossRef]
- Chung, M.; Vrebalov, J.; Alba, R.; Lee, J.; McQuinn, R.; Chung, J.; Klein, P.; Giovannoni, J. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010, 64, 936–947. [Google Scholar] [CrossRef]
- Xiong, C.; Luo, D.; Lin, A.; Zhang, C.; Shan, L.; He, P.; Li, B.; Zhang, Q.; Hua, B.; Yuan, Z.; et al. A tomato B-box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome-mediated degradation. New Phytol. 2019, 221, 279–294. [Google Scholar] [CrossRef]
- Ledent, V.; Vervoort, M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef]
- Zhou, D.; Shen, Y.; Zhou, P.; Fatima, M.; Lin, J.; Yue, J.; Zhang, X.; Chen, L.; Ming, R. Papaya CpbHLH1/2 regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Glazińska, P.; Zienkiewicz, A.; Wojciechowski, W.; Kopcewicz, J. Author links open overlay the putative miR172 target gene In APETALA2-like is involved in the photoperiodic flower induction of Ipomoea nil. J. Plant Physiol. 2009, 166, 1801–1813. [Google Scholar] [CrossRef] [PubMed]
- García-Limones, C.; Schnäbele, K.; Blanco-Portales, R.; Luz Bellido, M.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J. Functional Characterization of FaCCD1: A Carotenoid Cleavage Dioxygenase from Strawberry Involved in Lutein Degradation during Fruit Ripening. J. Agr. Food Chem. 2008, 56, 9277–9285. [Google Scholar] [CrossRef]
- Gao, J.; Yang, S.; Tang, K.; Li, G.; Gao, X.; Liu, B.; Wang, S.; Feng, X. GmCCD4 controls carotenoid content in soybeans. Plant Biotechnol. J. 2021, 19, 801–813. [Google Scholar] [CrossRef]
- Kloer, D.P.; Ruch, S.; Al-Babili, S.; Beyer, P.; Schulz, G.E. The Structure of a Retinal-Forming Carotenoid Oxygenase. Science 2005, 308, 267–269. [Google Scholar] [CrossRef]
- Huang, F.; Molnár, P.; Schwab, W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. [Google Scholar] [CrossRef]
- Ma, J.; Li, J.; Zhao, J.; Zhou, H.; Ren, F.; Wang, L.; Gu, C.; Liao, L.; Han, Y. Inactivation of a Gene Encoding Carotenoid Cleavage Dioxygenase (CCD4) Leads to Carotenoid-Based Yellow Coloration of Fruit Flesh and Leaf Midvein in Peach. Plant Mol. Biol. Rep. 2014, 32, 246–257. [Google Scholar] [CrossRef]
- Gonnet, J. Colour effects of co-pigmentation of anthocyanins revisited-1. A calorimetric definition using the CIELAB scale. Food Chem. 1998, 63, 409–415. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Deng, Y.J.; Liu, J.X.; Duan, A.Q.; Liu, H.; Xiong, A.S. DcCCD4 catalyzes the degradation of α-carotene and β-carotene to affect carotenoid accumulation and taproot color in carrot. Plant J. 2021, 108, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Ding, A.; Zhang, T.; Luo, L.; Wang, J.; Cheng, T.; Zhang, Q. Expansion of PmBEAT genes in the Prunus mume genome induces characteristic floral scent production. Hortic. Res. 2019, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Cong, T.; Yao, Y.; Cheng, T.; Yuan, C.; Zhang, Q. KNOX Genes Were Involved in Regulating Axillary Bud Formation of Chrysanthemum × morifolium. Int. J. Mol. Sci. 2023, 24, 7081. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools—An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]


| Samples and Stages | RHSCC | L* | a* | b* | C* | h |
|---|---|---|---|---|---|---|
| HJH-S1 | 5D | 82.45 | −6.97 | 32.69 | 33.43 | −77.97 |
| HJH-S2 | 5D | 83.84 | −6.72 | 33.49 | 34.16 | −78.65 |
| HJH-S3 | 2D | 89.97 | −6.53 | 26.42 | 27.22 | −76.12 |
| HJH-S4 | 155D | 94.75 | −5.78 | 15.50 | 16.55 | −69.55 |
| HJH-S5 | 155B | 92.76 | −7.17 | 20.28 | 21.51 | −70.52 |
| ZLE-S1 | 4D | 91.95 | −8.33 | 23.57 | 25.00 | −70.53 |
| ZLE-S2 | 155A | 93.88 | −7.35 | 17.10 | 18.61 | −66.73 |
| ZLE-S3 | 155D | 95.82 | −6.97 | 12.44 | 14.26 | −60.75 |
| ZLE-S4 | 155D | 95.95 | −6.85 | 12.47 | 14.23 | −61.20 |
| ZLE-S5 | 155D | 95.30 | −6.47 | 12.95 | 14.47 | −63.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, A.; Bao, F.; Yuan, X.; Wang, J.; Cheng, T.; Zhang, Q. Integrative Analysis of Metabolome and Transcriptome Revealed Lutein Metabolism Contributed to Yellow Flower Formation in Prunus mume. Plants 2023, 12, 3333. https://doi.org/10.3390/plants12183333
Ding A, Bao F, Yuan X, Wang J, Cheng T, Zhang Q. Integrative Analysis of Metabolome and Transcriptome Revealed Lutein Metabolism Contributed to Yellow Flower Formation in Prunus mume. Plants. 2023; 12(18):3333. https://doi.org/10.3390/plants12183333
Chicago/Turabian StyleDing, Aiqin, Fei Bao, Xi Yuan, Jia Wang, Tangren Cheng, and Qixiang Zhang. 2023. "Integrative Analysis of Metabolome and Transcriptome Revealed Lutein Metabolism Contributed to Yellow Flower Formation in Prunus mume" Plants 12, no. 18: 3333. https://doi.org/10.3390/plants12183333
APA StyleDing, A., Bao, F., Yuan, X., Wang, J., Cheng, T., & Zhang, Q. (2023). Integrative Analysis of Metabolome and Transcriptome Revealed Lutein Metabolism Contributed to Yellow Flower Formation in Prunus mume. Plants, 12(18), 3333. https://doi.org/10.3390/plants12183333






