Phosphorus Acquisition Efficiency and Transcriptomic Changes in Maize Plants Treated with Two Lignohumates
Abstract
:1. Introduction
2. Results
2.1. Characterization of Humates
2.1.1. Spectroscopic Characterization
2.1.2. Elemental Content and Acidity
2.1.3. Thermal Stability
2.1.4. Hormone Quantification
2.1.5. FRAP and Phenol Content
2.2. Effects of Humate Application on Plant Growth, SPAD, C and N Contents and FRAP
2.3. P accumulation, PAE and Activity of P-Mining Enzymes
2.4. Spectroscopic Characterization of Maize Leaves
2.5. Changes in Gene Expression as a Function of P Supply and H1 Treatment
3. Discussion
4. Materials and Methods
4.1. Humate Characterization
4.1.1. FT-IR and NMR Spectroscopy
4.1.2. Elemental Analysis and Total Acidity
4.1.3. Thermal Analysis
4.1.4. Hormone Quantification
4.1.5. Analysis of Antioxidant Capacity (FRAP = Ferric-Reducing Antioxidant Power Assay) and Total Phenols
4.2. Plant Growth and Experimental Design
4.3. Phosphorus Acquisition Efficiency (PAE)
4.4. Enzyme Activity
4.5. Transcriptomic Analysis and Gene Ontology (GO) Analyses
4.6. Statistics and Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Frossard, E.; Condron, L.M.; Oberson, A.; Sinaj, S.; Fardeau, J.C. Processes governing phosphorus availability in temperate soils. J. Environ. Qual. 2000, 29, 15–23. [Google Scholar] [CrossRef]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.E.; Simpson, R.J.; Richardson, A.E. The growth and phosphorus utilisation of plants in sterile media when supplied with inositol hexaphosphate, glucose 1-phosphate or inorganic phosphate. Plant Soil 2000, 220, 165–174. [Google Scholar] [CrossRef]
- Santoro, V.; Martin, M.; Persson, P.; Lerda, C.; Said-Pullicino, D.; Magnacca, G.; Celi, L. Inorganic and organic P retention by coprecipitation during ferrous iron oxidation. Geoderma 2019, 348, 168–180. [Google Scholar] [CrossRef]
- Celi, L.; Prati, M.; Magnacca, G.; Santoro, V.; Martin, M. Role of crystal-line iron oxides on stabilization of inositol phosphates in soil. Geoderma 2020, 374, 114442. [Google Scholar] [CrossRef]
- Vance, C.P. Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol. 2001, 127, 390–397. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Richardson, A.E.; Lynch, J.P.; Ryan, P.R.; Delhaize, E.; Smith, F.A.; Smith, S.E.; Harvey, P.R.; Ryan, M.H.; Veneklaas, E.J.; Lambers, H.; et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil 2011, 349, 121–156. [Google Scholar] [CrossRef]
- Chen, C.R.; Condron, L.M.; Davis, M.R.; Sherlock, R.R. Phosphorus dynamics in the rhizosphere of perennial ryegrass (Lolium perenne L.) and radiata pine (Pinus radiata D.Don.). Soil Biol. Biochem. 2002, 34, 487–499. [Google Scholar] [CrossRef]
- Turner, B.L.; Papházy, M.; Haygarth, P.; Mckelvie, J.D. Inositol phosphates in the environment. Biol. Sci. 2002, 357, 449–469. [Google Scholar] [CrossRef] [PubMed]
- Konietzny, U.; Greiner, R. Molecular and catalytic properties of phytate-degrading enzymes (phytases). Int. J. Food Sci. Technol. 2002, 37, 791–812. [Google Scholar] [CrossRef]
- Raghothama, K.G. Phosphate transport and signaling. Curr. Opin. Plant Biol. 2000, 3, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; González, E.; Bustos, R.; Linhares, F.; Leyva, A.; Paz-Ares, J. The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 2004, 55, 285–293. [Google Scholar] [CrossRef]
- Ding, N.; Huertas, R.; Torres-Jerez, I.; Liu, W.; Watson, B.; Scheible, W.; Udvardi, M. Transcriptional, metabolic, physiological and developmental responses of switchgrass to phosphorus limitation. Plant Cell Environ. 2021, 44, 186–202. [Google Scholar] [CrossRef]
- Prodhan, M.A.; Pariasca-Tanaka, J.; Ueda, Y.; Hayes, P.E.; Wissuwa, M. Comparative transcriptome analysis reveals a rapid response to phosphorus deficiency in a phosphorus-efficient rice genotype. Sci. Rep. 2022, 12, 9460. [Google Scholar] [CrossRef]
- Wang, Y.; Duran, H.G.S.; van Haarst, J.C.; Schijlen, E.G.W.M.; Ruyter-Spira, C.; Medema, M.H.; Dong, L.; Bouwmeeste, H.J. The role of strigolactones in P deficiency induced transcriptional changes in tomato roots. BMC Plant Biol. 2021, 21, 349. [Google Scholar] [CrossRef]
- Nardi, S.; Pizzeghello, D.; Schiavon, M.; Ertani, A. Plant biostimulants: Physiological responses induced by protein hydrolyzed- based products and humic substances in plant metabolism. Sci. Agric. 2016, 73, 18–23. [Google Scholar] [CrossRef]
- Colla, G.; Hoagland, L.; Ruzzi, M.; Cardarelli, M.; Bonini, P.; Canaguier, R.; Rouphael, Y. Biostimulant action of protein hydrolysates: Unraveling their effects on plant physiology and microbiome. Front. Plant Sci. 2017, 22, 8. [Google Scholar] [CrossRef]
- Canellas, L.P.; Olivares, F.L.; Canellas, N.O.A.; Mazzei, P.; Piccolo, A. Humic acids increase the maize seedlings exudation yield. Chem. Biol. Technol. Agric. 2019, 6, 3. [Google Scholar] [CrossRef]
- Yakhin, O.I.; Lubyanov, A.A.; Yakhin, I.A.; Brown, P.H. Biostimulants in Plant Science: A Global Perspective. Front. Plant Sci. 2017, 7, 2049. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Freitas, H.; Dias, M.C. Strategies and prospects for biostimulants to alleviate abiotic stress in plants. Front. Plant Sci. 2022, 13, 1024243. [Google Scholar] [CrossRef]
- Olaetxea, M.; de Hita, D.; Calderin Garcia, A.; Fuentesa, M.; Baigorri, R.; Mora, V.; Garnica, M.; Urrutia, O.; Erro, J.; Zamarreño, A.M.; et al. Hypothetical framework integrating the main mechanisms involved in the promoting action of rhizospheric humic substances on plant root- and shoot growth. Appl. Soil Ecol. 2018, 123, 521–537. [Google Scholar] [CrossRef]
- Nardi, S.; Schiavon, M.; Francioso, O. Chemical structure and biological activity of humic substances define their role as plant growth promoters. Molecules 2021, 26, 2256. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Y.; Wang, L.; Song, G.; Ni, L.; Xu, M.; Nie, C.; Li, B.; Bai, Y. Analysis of the molecular composition of humic substances and their effects on physiological metabolism in maize based on untargeted metabolomics. Front. Plant Sci. 2023, 14, 1122621. [Google Scholar] [CrossRef]
- Jindo, K.; Sanches Soares, T.; Pereira Peres, L.E.; Azevedo, I.G.; Oliveira Aguiar, N.; Mazzei, P.; Spaccini, R.; Piccolo, A.; Olivares, F.L.; Canellas, L.P. Phosphorus speciation and high-affinity transporters are influenced by humic substances. J. Plant Nutr. Soil Sci. 2016, 179, 206–214. [Google Scholar] [CrossRef]
- Shafi, M.I.; Adnan, M.; Fahad, S.; Wahid, F.; Khan, A.; Yue, Z. Application of single superphosphate with humic acid improves the growth, yield and phosphorus uptake of wheat (Triticum aestivum L.) in calcareous soil. Agronomy 2020, 10, 1224. [Google Scholar] [CrossRef]
- Yuan, Y.; Gai, S.; Tang, C.; Jin, Y.; Cheng, K.; Antonietti, M.; Yang, F. Artificial humic acid improves maize growth and soil phosphorus utilization efficiency. Appl. Soil Ecol. 2022, 179, 104587. [Google Scholar] [CrossRef]
- Canellas, L.P.; Oliveras, F.L. Physiological responses to humic substances as plant growth promoter. Chem. Biol. Technol. Agric. 2014, 1, 3. [Google Scholar] [CrossRef]
- Lyons, G.; Genc, Y. Commercial humates in agriculture: Real substance or smoke and mirrors? Agronomy 2016, 6, 50. [Google Scholar] [CrossRef]
- Sadiq, S.A.; Baloch, D.M.; Hidayatullah, A.N. Role of coal-derived humic acid in the availability of nutrients and growth of sunflower under calcareous soil. J. Anim. Plant Sci. 2014, 24, 1737–1742. [Google Scholar]
- Schiavon, M.; Ertani, A.; Francioso, O.; Nardi, S. Manure fertilization gives high-quality earthworm coprolites with positive effects on plant growth and N metabolism. Agronomy 2019, 9, 659. [Google Scholar] [CrossRef]
- Muscolo, A.; Sidari, M.; Cozzolino, V.; Nuzzo, A.; Nardi, S.; Piccolo, A. Molecular characteristics of humic substances from different origins and their effects on growth and metabolism of Pinus laricio callus. Chem. Biol. Technol. Agric. 2022, 9, 72. [Google Scholar] [CrossRef]
- Iakimenko, O.S. Commercial humates from coal and their influence on soil properties and initial plant development. In Use of Humic Substances to Remediate Polluted Environments: From Theory to Practice; Springer: Dordrecht, The Netherlands, 2005; pp. 365–378. [Google Scholar]
- Zaccone, C.; Cocozza, C.; D’Orazio, V.; Plaza, C.; Cheburkin, A.K.; Miano, T.M. Influence of extractant on quality and trace elements content of peat humic acids. Talanta 2007, 73, 820–830. [Google Scholar] [CrossRef]
- Ertani, A.; Francioso, O.; Tugnoli, V.; Righi, V.; Nardi, S. Effect of commercial lignosulfonate-humate on Zea mays L. metabolism. J. Agric. Food Chem. 2011, 59, 11940–11948. [Google Scholar] [CrossRef]
- Tinti, A.; Tugnoli, V.; Bonora, S.; Francioso, O. Recent applications of vibrational mid-Infrared (IR) spectroscopy for studying soil components: A review. J. Centr. Eur. Agric. 2015, 16, 1–22. [Google Scholar] [CrossRef]
- Ferrari, E.; Francioso, O.; Nardi, S.; Saladini, M.; Dal Ferro, N.; Morari, F. DRIFT and HR MAS NMR characterization of humic substances from a soil treated with different organic and mineral fertilizers. J. Mol. Struc. 2011, 998, 216–224. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Chang, L.; Zi, C.; Liang, G.; Zhang, D.; Su, Y. A comparative study on the structural features of humic acids extracted from lignites using comprehensive spectral analyses. RSC Adv. 2020, 10, 22002–22009. [Google Scholar] [CrossRef]
- Schnitzer, M.; Hoffman, J. A thermogravimetric approach to the classification of organic soils. Soil Sci. Soc. Am. J. 1966, 30, 63–66. [Google Scholar] [CrossRef]
- Plante, A.F.; Fernández, J.M.; Leifeld, J. Application of thermal analysis techniques in soil science. Geoderma 2009, 153, 1–10. [Google Scholar] [CrossRef]
- Provenzano, M.R.; Cavallo, O.; Malerba, A.D.; Fabbri, C.; Zaccone, C. Unravelling (maize silage) digestate features throughout a full-scale plant: A spectroscopic and thermal approach. J. Clean. Prod. 2018, 193, 372–378. [Google Scholar] [CrossRef]
- Zaccone, C.; Plaza, C.; Ciavatta, C.; Miano, T.M.; Shotyk, W. Advances in the determination of humification degree in peat since Achard (1786): Applications in geochemical and paleoenvironmental studies. Earth-Sci. Rev. 2018, 185, 163–178. [Google Scholar] [CrossRef]
- Ertani, A.; Francioso, O.; Ferrari, E.; Schiavon, M.; Nardi, S. Spectroscopic-chemical fingerprint and biostimulant activity of a protein-based product in solid form. Molecules 2018, 23, 1031. [Google Scholar] [CrossRef] [PubMed]
- Vujinović, T.; Zanin, L.; Venuti, S.; Contin, M.; Ceccon, P.; Tomasi, N.; Pinton, R.; Cesco, S.; De Nobili, M. Biostimulant Action of dissolved humic substances from a conventionally and an organically managed soil on nitrate acquisition in maize plants. Front. Plant Sci. 2020, 10, 1652. [Google Scholar] [CrossRef]
- Ji, J.; Li, N.; Cui, H.; Li, Y.; Zhao, X.; Zhang, H.; Ma, H. Study on monitoring SPAD values for multispatial spatial vertical scales of summer maize based on UAV multispectral remote sensing. Agriculture 2023, 13, 1004. [Google Scholar] [CrossRef]
- Carstensen, A.; Herdean, A.; Schmidt, S.B.; Sharma, A.; Spetea, C.; Pribil, M.; Husted, S. The impacts of phosphorus deficiency on the photosynthetic electron transport chain. Plant Physiol. 2018, 177, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Kayoumu, M.; Iqbal, A.; Muhammad, N.; Li, X.; Li, L.; Wang, X.; Gui, H.; Qi, Q.; Ruan, S.; Guo, R.; et al. Phosphorus availability affects the photosynthesis and antioxidant system of contrasting low-P-tolerant cotton genotypes. Antioxidants 2023, 12, 466. [Google Scholar] [CrossRef]
- Hamed, S.B.; Lefi, E.; Chaieb, M. Effect of phosphorus concentration on the photochemical stability of PSII and CO2 assimilation in Pistacia vera L. and Pistacia atlantica Desf. Plant Physiol. Biochem. 2019, 142, 283–291. [Google Scholar] [CrossRef]
- Garrone, A.; Archipowa, N.; Zipfel, P.F.; Hermann, G.; Dietzek, B. Plant protochlorophyllide oxidoreductases A and B: Catalytic efficiency and initial reaction steps. J. Biol. Chem. 2015, 290, 28530–28539. [Google Scholar] [CrossRef]
- Spohn, M.; Kuzyakov, Y. Phosphorus mineralization can be driven by microbial need for carbon. Soil Biol. Biochem. 2013, 61, 69–75. [Google Scholar] [CrossRef]
- Santoro, V.; Schiavon, M.; Visentin, I.; Constán-Aguilar, C.; Cardinale, F.; Celi, L. Strigolactones affect phosphorus acquisition strategies in tomato plants. Plant Cell Environ. 2021, 44, 3628–3642. [Google Scholar] [CrossRef] [PubMed]
- Santoro, V.; Schiavon, M.; Visentin, I.; Martin, M.; Said-Pullicino, D.; Cardinale, F.; Celi, L. Tomato plant responses induced by sparingly available inorganic and organic phosphorus forms are modulated by strigolactones. Plant Soil 2022, 474, 355–372. [Google Scholar] [CrossRef]
- García, C.; Ceccanti, B.; Masciandaro, G.; Hernández, T. Phosphatase and β-glucosidase activities in humic substances from animal wastes. Biores. Technol. 1995, 53, 79–87. [Google Scholar] [CrossRef]
- Bessman, M.J. A cryptic activity in the Nudix hydrolase superfamily. Protein Sci. 2019, 28, 1494–1500. [Google Scholar] [CrossRef]
- Jindo, K.; Canellas, L.P.; Albacete, A.; dos Santos, L.F.; Frinani Roca, R.L.; Baia, D.C.; Aguiar Canellas, N.O.; Goron, T.L.; Olivares, F.L. Interaction between humic substances and plant hormones for phosphorous acquisition. Agronomy 2020, 10, 640. [Google Scholar] [CrossRef]
- Souza, A.C.; Olivares, F.L.; Peres, L.E.P.; Piccolo, A.; Canellas, L.P. Plant hormone crosstalk mediated by humic acids. Chem. Biol. Technol. Agric. 2022, 9, 29. [Google Scholar] [CrossRef]
- Canellas, L.P.; Dobbss, L.B.; Oliveira, A.L.; Chagas, J.G.; Aguiar, N.O.; Rumjanek, V.M.; Novotny, E.H.; Olivares, F.L.; Spaccini, R.; Piccolo, A. Chemical properties of humic matter as related to induction of plant lateral roots. Eur. J. Soil Sci. 2012, 63, 315–324. [Google Scholar] [CrossRef]
- Olaetxea, M.; Mora, V.; Bacaicoa, E.; Baigorri, R.; Garnica, M.; Fuentes, M.; Zamarreño, A.M.; Spíchal, L.; García-Mina, J.M. Root ABA and H+-ATPase are key players in the root and shoot growth-promoting action of humic acids. Plant Direct 2019, 3, 10. [Google Scholar] [CrossRef]
- Mora, V.; Baigorri, R.; Bacaicoa, E.; Zamarreño, A.M.; García-Mina, J.M. The humic acid-induced changes in the root concentration of nitric oxide, IAA and ethylene do not explain the changes in root architecture caused by humic acid in cucumber. Environ. Exp. Bot. 2012, 76, 24–32. [Google Scholar] [CrossRef]
- Jia, Z.; Giehl, R.F.H.; von Wirén, N. Nutrient–hormone relations: Driving root plasticity in plants. Mol. Plant 2022, 15, 86–103. [Google Scholar] [CrossRef]
- Pizzeghello, D.; Schiavon, M.; Francioso, O.; Dalla Vecchia, F.; Ertani, A.; Nardi, S. Bioactivity of size-fractionated and unfractionated humic substances from two forest soils and comparative effects on N and S metabolism, nutrition and root anatomy of Allium sativum L. Front. Plant Sci. 2020, 11, 1203. [Google Scholar] [CrossRef]
- Schiavon, M.; Pizzeghello, D.; Muscolo, A.; Nardi, S. High molecular size humic substances enhance phenylpropanoid metabolism in maize (Zea mays L.). J. Chem. Ecol. 2010, 36, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Muscolo, A.; Sidari, M.; Nardi, S. Humic substance: Relationship between structure and activity. Deeper information suggests univocal findings. J. Geochem. Explor. 2013, 129, 103–111. [Google Scholar] [CrossRef]
- Nardi, S.; Pizzeghello, D.; Ertani, A. Hormone-like activity of the soil organic matter. Appl. Soil Ecol. 2018, 123, 517–520. [Google Scholar] [CrossRef]
- Canellas, L.P.; Canellas, N.O.A.D.S.; Irineu, L.E.S.; Olivares, F.L.; Piccolo, A. Plant chemical priming by humic acids. Chem. Biol. Technol. Agric. 2020, 7, 12. [Google Scholar] [CrossRef]
- Rodrigues de Queiroz, A.; Hines, C.; Brown, J.; Sahay, S.; Vijayan, J.; Stone, J.M.; Bickford, N.; Wuellner, M.; Glowacka, K.; Buan, N.R.; et al. The effects of exogenously applied antioxidants on plant growth and resilience. Phytochem. Rev. 2023, 22, 407–447. [Google Scholar] [CrossRef]
- Hernández, I.; Munné-Bosch, S. Linking phosphorus availability with photo-oxidative stress in plants. J. Exp. Bot. 2015, 66, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Schnitzer, M.; Gupta, U.C. Determination of acidity in soil organic Matter. Soil Sci. Am. J. 1965, 29, 274–277. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “Antioxidant Power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Nicoletto, C.; Santagata, S.; Bona, S.; Sambo, P. Influence of cut number on qualitative traits in different cultivars of sweet basil. Ind. Crop Prod. 2013, 44, 465–472. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. The Water Culture Method for Growing Plants without Soil; California Agricultural Experiment Station Circular 347: Davis, CA, USA, 1938; pp. 1–39. [Google Scholar]
- Ohno, T.; Zibilske, L.M. Determination of low concentration of phosphorus in soil extracts using malachite green. Soil Sci. Soc. Am. J. 1991, 55, 892–895. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
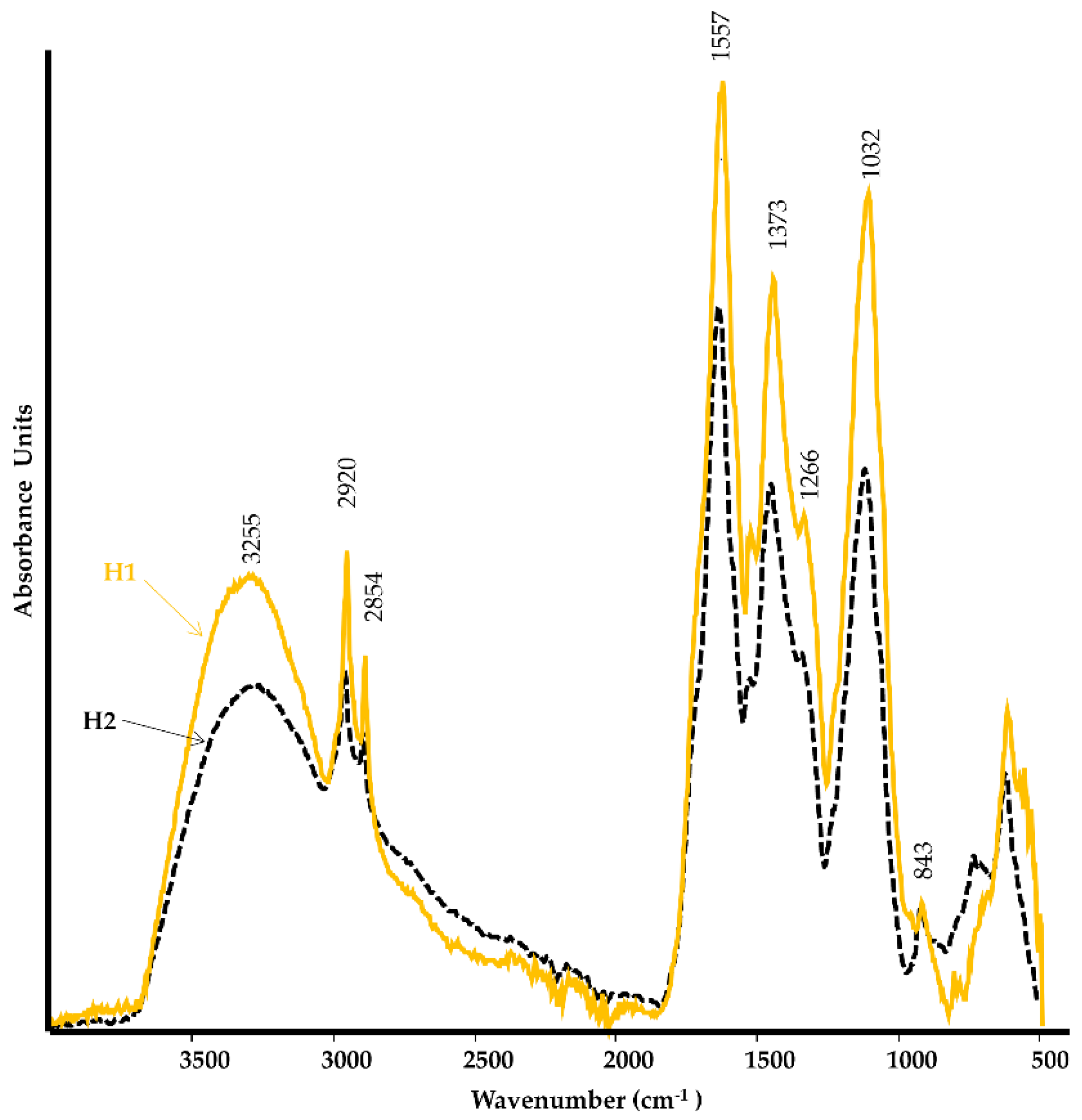

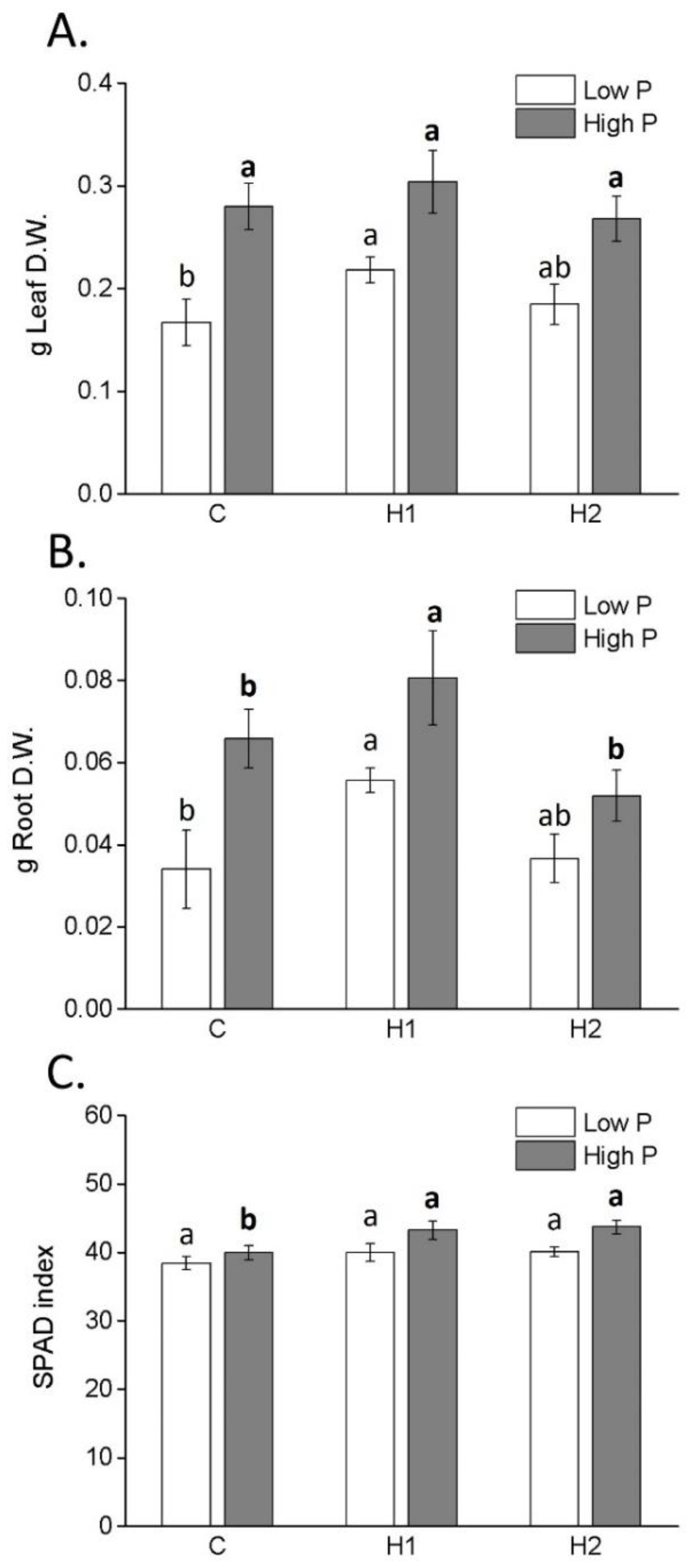
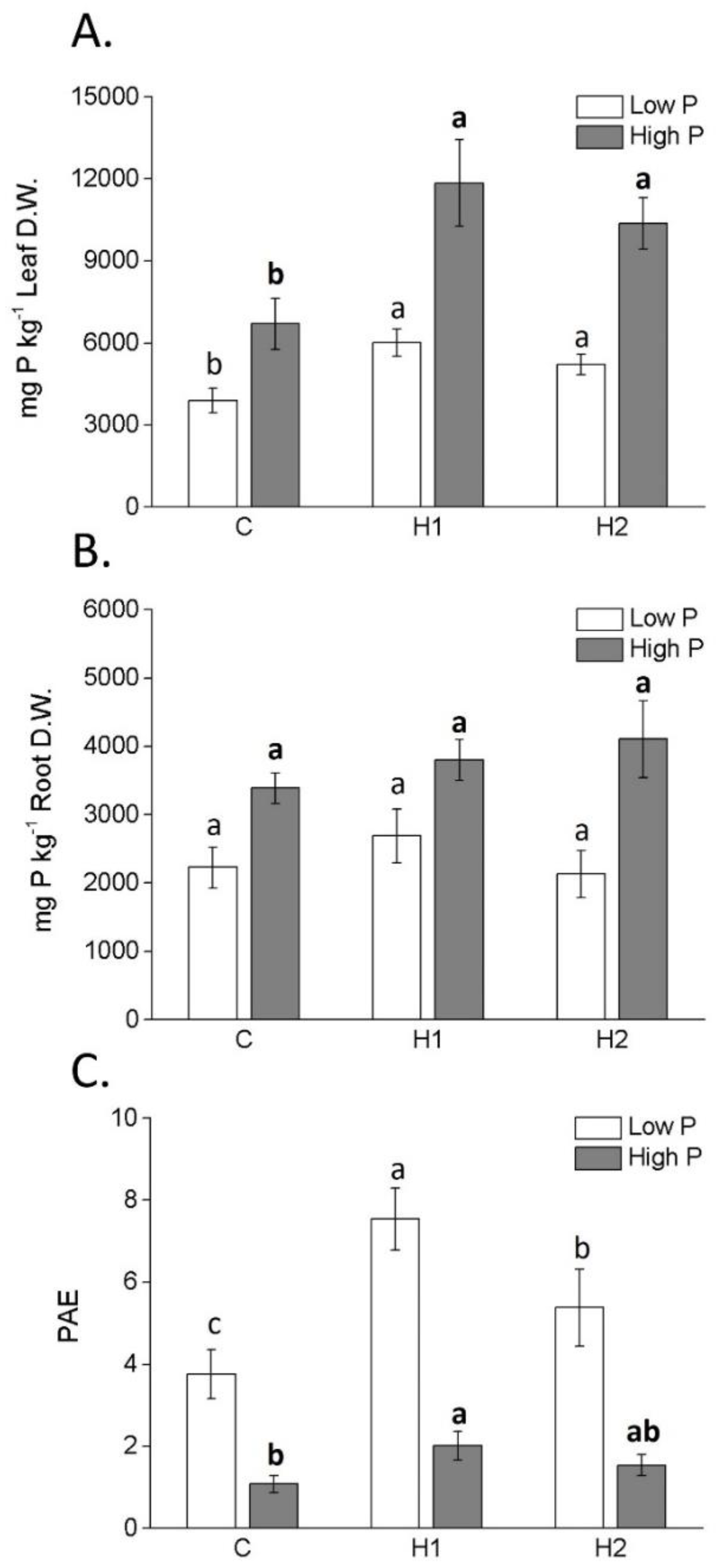
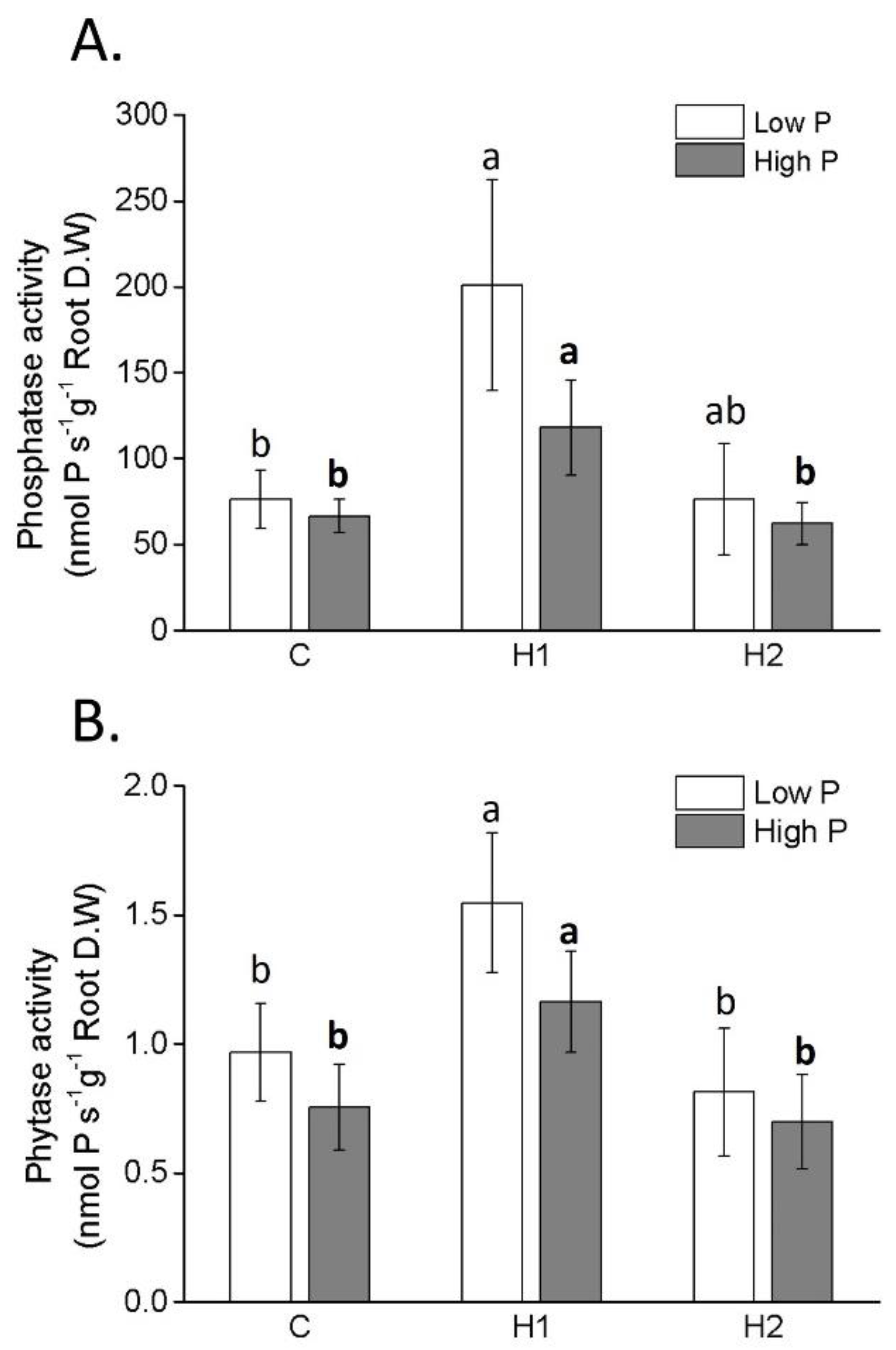

| Parameter | H1 | H2 |
|---|---|---|
| N (%) | 1.64 ± 0.03 a | 1.49 ± 0.04 b |
| C (%) | 42.02 ± 0.07 a | 39.52 ± 0.10 b |
| H (%) | 5.36 ± 0.07 a | 4.96 ± 0.11 b |
| S (%) | 0.07 + 0.00 b | 0.14 + 0.01 a |
| O (%) | 50.97 + 0.07 b | 54.01 + 0.09 a |
| C/N | 25.66 + 0.37 a | 26.50 + 0.66 a |
| C/H | 7.84 + 0.12 a | 7.97 + 0.19 a |
| C/O | 0.82 + 0.00 a | 0.73 + 0.00 b |
| Ash (%) | 28.8 | 30.8 |
| R1 | 1.8 | 1.1 |
| R2 | 0.25 | 0.36 |
| R3 | 0.25 | 0.20 |
| WL400–550/200–300 | 0.459 | 0.567 |
| WL400–550/300–400 | 0.916 | 0.947 |
| TG-T50 (°C) | 364 | 407 |
| IAA (ng mg C−1) | 0.13 ± 0.06 a | 0.13 ± 0.04 a |
| Zeatin riboside (ng mg C−1) | 1.86 ± 0.16 a | 1.56 ± 0.19 b |
| ABA (ng mg C−1) | 9.59 ± 2.36 a | 6.99 ± 1.93 a |
| GA (ng mg C−1) | 0.013 ± 0.004 a | 0.104 ± 0.015 b |
| Acidity (meq H+ mg−1) | 2.50 ± 0.05 b | 3.33 ± 0.09 a |
| TP (mg GAE kg−1) | 89.31 ± 0.16 a | 59.43 ± 0.07 b |
| FRAP (mg Fe2+ kg−1) | 536.5 + 0.10 a | 72.11 + 0.01 b |
| Leaves | Roots | |||
|---|---|---|---|---|
| N (%) | C (%) | N (%) | C (%) | |
| Low P | ||||
| Control | 4.75 ± 0.47 b | 34.00 ± 1.51 a | 3.98 ± 0.31 b | 31.51 ± 1.58 a |
| H1 | 5.27 ± 0.34 a | 33.35 ± 1.48 a | 4.61 ± 0.10 a | 31.87 ± 1.19 a |
| H2 | 5.11 ± 0.26 a | 32.94 ± 1.26 a | 4.71 ± 0.20 a | 32.11 ± 0.57 a |
| High P | ||||
| Control | 5.00 ± 0.01 b | 33.82 ± 0.76 a | 4.93 ± 0.32 b | 28.91 ± 1.80 c |
| H1 | 5.72 ± 0.36 a | 34.50 ± 0.78 a | 5.35 ± 0.39 ab | 32.21 ± 0.38 b |
| H2 | 5.52 ± 0.17 a | 33.10 ± 3.11 a | 5.69 ± 0.40 a | 34.41 ± 0.08 a |
| Leaves | Roots | Leaves | Roots | |
|---|---|---|---|---|
| mg Fe2+ kg−1 F.W. | mg Fe2+ kg−1 F.W. | |||
| Low P | High P | |||
| Control | 0.41 ± 0.05 b | 0.51 ± 0.13 b | 0.51 ± 0.13 b | 0.26 ± 0.05 b |
| H1 | 0.64 ± 0.05 a | 0.75 ± 0.03 a | 0.75 ± 0.03 a | 0.41 ± 0.04 a |
| H2 | 0.52 ± 0.04 a | 0.63 ± 0.05 b | 0.63 ± 0.05 b | 0.33 ± 0.06 ab |
| Number of DEGs | ||
|---|---|---|
| Comparisons | Overexpressed | Underexpressed |
| HP vs. LP | 528 | 239 |
| HP vs. HP_H1 | 245 | 278 |
| LP vs. LP_H1 | 179 | 164 |
| Gene ID | Log2FC | p-Value | Description |
|---|---|---|---|
| DEG in HP–LP | |||
| Zm00001d049554 | −2.4766 | 0.0001 | Putative glycerol-3-phosphate transporter 1 |
| Zm00001d026156 | −1.6923 | 0.0007 | glycerol 3-phosphate permease |
| Zm00001d008310 | −1.8869 | 0.0176 | inositol-1-monophosphatase |
| Zm00001d043267 | −1.3904 | 0.0325 | putative 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 |
| Zm00001d011734 | −1.9039 | 0.0406 | phosphatase phospho 1 |
| Zm00001d031653 | −1.6914 | 1.9 × 10−5 | uncharacterized LOC100216744 |
| Zm00001d040519 | −2.1226 | 0.015557 | rhicadhesin receptor |
| Zm00001d049958 | −1.6753 | 0.009485 | Putative peptidyl-prolyl cis-trans isomerase WD40 repeat domain family protein |
| Zm00001d053952 | −1.4504 | 0.038968 | Bax inhibitor 1 |
| Zm00001d039439 | −1.517 | 0.0272 | Protein BRASSINAZOLE-RESISTANT 1 |
| Zm00001d046538 | −1.1071 | 0.024709 | SEC14-like protein 1 |
| Zm00001d013869 | −1.2837 | 0.03877 | SAUR56-auxin-responsive SAUR family member |
| Zm00001d028370 | −1.1231 | 0.017965 | protein OBERON 3 |
| DEG in HP-HP_H1 | |||
| Zm00001d014564 | −1.3433 | 0.042067 | sulfate transporter 6 |
| Zm00001d012913 | −1.0042 | 0.025655 | methionine adenosyltransferase |
| Zm00001d039138 | −1.1375 | 0.016453 | methionine aminopeptidase |
| Zm00001d012537 | −1.223 | 0.029047 | somatic embryogenesis receptor-like kinase 3 |
| Zm00001d003306 | −1.7652 | 0.003905 | putative LRR receptor-like serine/threonine-protein kinase |
| Zm00001d053967 | −1.0378 | 0.045439 | auxin response factor 21 |
| Zm00001d038508 | −1.3427 | 0.014187 | auxin response factor |
| Zm00001d008893 | −1.7031 | 0.005967 | auxin response factor 10 |
| Zm00001d021526 | −1.2159 | 0.017747 | SNARE-interacting protein KEULE |
| Zm00001d032380 | −1.0691 | 0.041987 | SCARECROW-like protein |
| Zm00001d050810 | −1.5393 | 0.007842 | WAT1-related protein |
| Zm00001d024854 | −1.2124 | 0.015484 | trehalose-6-phosphate synthase |
| Zm00001d014811 | −1.3976 | 0.000873 | alkaline alpha galactosidase 3 |
| Zm00001d042025 | −1.525 | 0.00095 | probable galactinol--sucrose galactosyltransferase 1 |
| Zm00001d039029 | −1.0826 | 0.033409 | 60 kDa jasmonate-induced protein |
| Zm00001d018797 | −1.4042 | 0.034359 | putative glycogen synthase kinase family protein |
| Zm00001d044644 | −1.6404 | 0.020783 | Anamorsin homolog |
| Zm00001d031941 | −1.6483 | 6 × 10−5 | chaperone DNA J2 |
| Zm00001d011454 | −2.734 | 0.000183 | Zinc finger CCCH domain-containing protein 19 |
| Zm00001d013202 | −9.4918 | 0.03132 | DNA-directed RNA polymerase subunit beta’-like |
| Zm00001d014937 | −2.1111 | 0.005352 | CBF1-interacting co-repressor CIR N-terminal |
| Zm00001d003306 | −1.7652 | 0.003905 | LRR receptor-like serine/threonine-protein kinase |
| DEG in LP-LP_H1 | |||
| Zm00001d039644 | −5.3403 | 0.005928 | cytokinin-O-glucosyltransferase 3 |
| Zm00001d040011 | −3.5851 | 0.045957 | Protein ABA DEFICIENT 4 chloroplastic |
| Zm00001d016132 | −4.3379 | 0.016201 | AT-hook motif nuclear-localized protein 26 |
| Zm00001d013655 | −4.1118 | 0.021104 | nicotianamine synthase 3 |
| Zm00001d017501 | −3.8926 | 0.030827 | peroxisomal biogenesis factor 19 |
| Zm00001d025296 | −3.43 | 0.014938 | nudix hydrolase 13 |
| Zm00001d052702 | −3.4285 | 0.012467 | biotin synthase |
| Zm00001d003522 | −3.1687 | 0.000711 | Pyridoxal phosphate (PLP)-dependent transferase superfamily protein |
| Zm00001d012508 | −3.1634 | 0.017243 | WRKY22-superfamily of TFs having WRKY and zinc finger domains |
| Zm00001d032692 | −1.7173 | 0.047759 | MATH domain containing protein |
| Zm00001d032576 | −2.7589 | 0.009631 | protochlorophyllide reductase B |
| Zm00001d049886 | −2.6671 | 0.039014 | grx_S12-glutaredoxin subgroup I |
| Zm00001d044606 | −1.3068 | 0.033098 | grx_S16-glutaredoxin subgroup II |
| Zm00001d037993 | −2.6568 | 0.006811 | ran-binding protein 1 |
| Zm00001d029457 | −2.4776 | 0.006301 | nucleoredoxin 1 |
| Zm00001d029361 | −2.3294 | 0.020461 | zinc finger CCCH domain-containing protein 15 homolog |
| Zm00001d047802 | −1.553 | 0.045089 | Dof zinc finger protein DOF2.2 |
| Zm00001d037221 | −1.2931 | 0.014043 | Transcription factor TCP14 |
| Zm00001d050107 | −2.1979 | 0.017464 | 12-oxophytodienoate reductase 8 |
| Zm00001d047105 | −3.037 | 0.035362 | Oxysterol-binding protein-related protein 2A |
| Zm00001d045298 | −1.4759 | 0.024132 | Lipase-like |
| Zm00001d033905 | −2.0678 | 0.041084 | growth-regulating-factor-interacting factor 1 |
| Zm00001d034035 | −1.4166 | 0.031865 | oligopeptide transporter 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santoro, V.; Della Lucia, M.C.; Francioso, O.; Stevanato, P.; Bertoldo, G.; Borella, M.; Ferrari, E.; Zaccone, C.; Schiavon, M.; Pizzeghello, D.; et al. Phosphorus Acquisition Efficiency and Transcriptomic Changes in Maize Plants Treated with Two Lignohumates. Plants 2023, 12, 3291. https://doi.org/10.3390/plants12183291
Santoro V, Della Lucia MC, Francioso O, Stevanato P, Bertoldo G, Borella M, Ferrari E, Zaccone C, Schiavon M, Pizzeghello D, et al. Phosphorus Acquisition Efficiency and Transcriptomic Changes in Maize Plants Treated with Two Lignohumates. Plants. 2023; 12(18):3291. https://doi.org/10.3390/plants12183291
Chicago/Turabian StyleSantoro, Veronica, Maria Cristina Della Lucia, Ornella Francioso, Piergiorgio Stevanato, Giovanni Bertoldo, Matteo Borella, Erika Ferrari, Claudio Zaccone, Michela Schiavon, Diego Pizzeghello, and et al. 2023. "Phosphorus Acquisition Efficiency and Transcriptomic Changes in Maize Plants Treated with Two Lignohumates" Plants 12, no. 18: 3291. https://doi.org/10.3390/plants12183291
APA StyleSantoro, V., Della Lucia, M. C., Francioso, O., Stevanato, P., Bertoldo, G., Borella, M., Ferrari, E., Zaccone, C., Schiavon, M., Pizzeghello, D., & Nardi, S. (2023). Phosphorus Acquisition Efficiency and Transcriptomic Changes in Maize Plants Treated with Two Lignohumates. Plants, 12(18), 3291. https://doi.org/10.3390/plants12183291














