Actin-Depolymerizing Factor Gene Family Analysis Revealed That CsADF4 Increased the Sensitivity of Sweet Orange to Bacterial Pathogens
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification and Chromosome Mapping of the ADF Gene in Sweet Orange
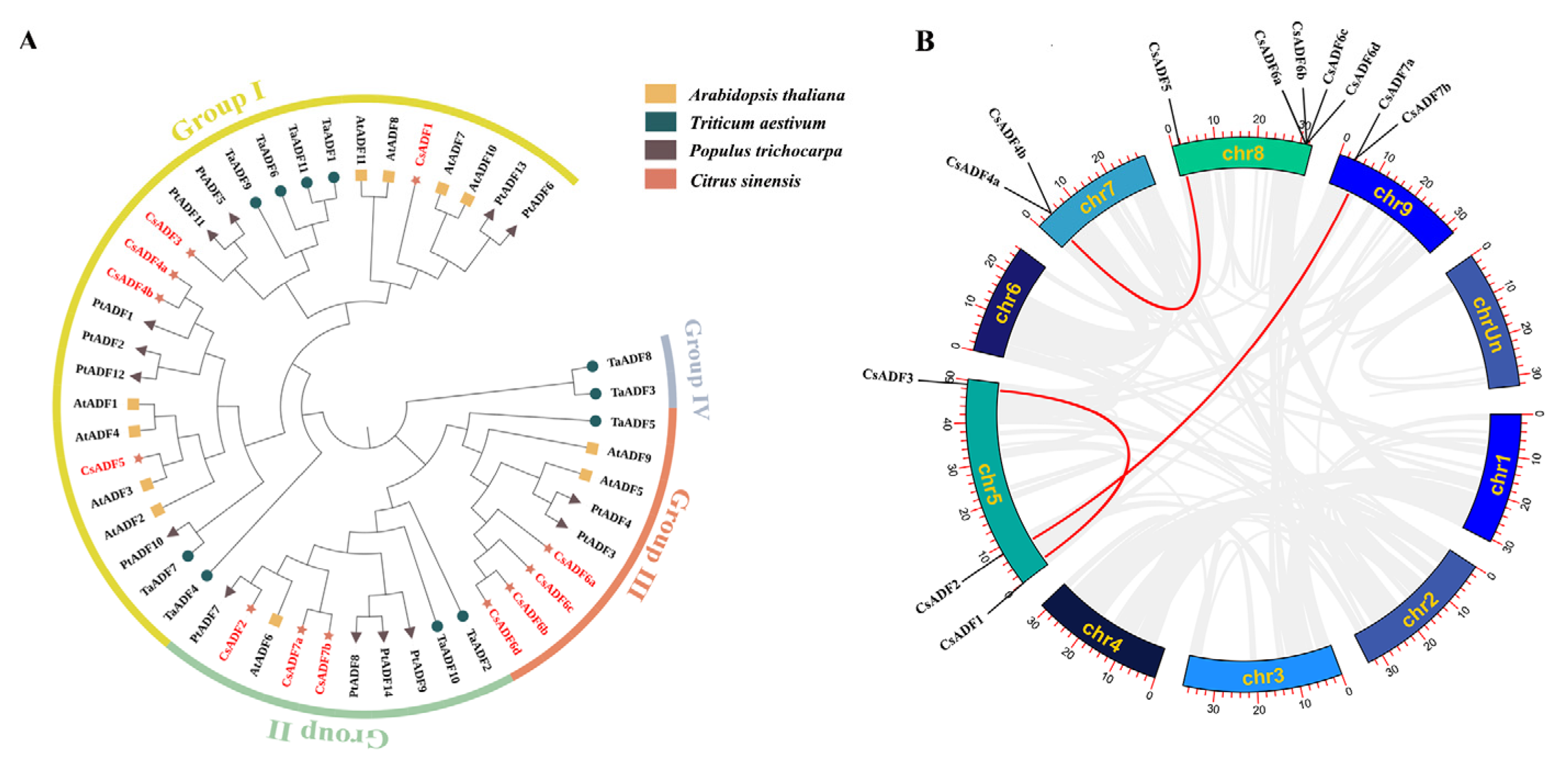
2.2. Phylogenetic Analysis and Collinear Analysis of CsADF Gene Family
2.3. Structural and Cis-Acting Regulatory Element Analysis of CsADF Genes
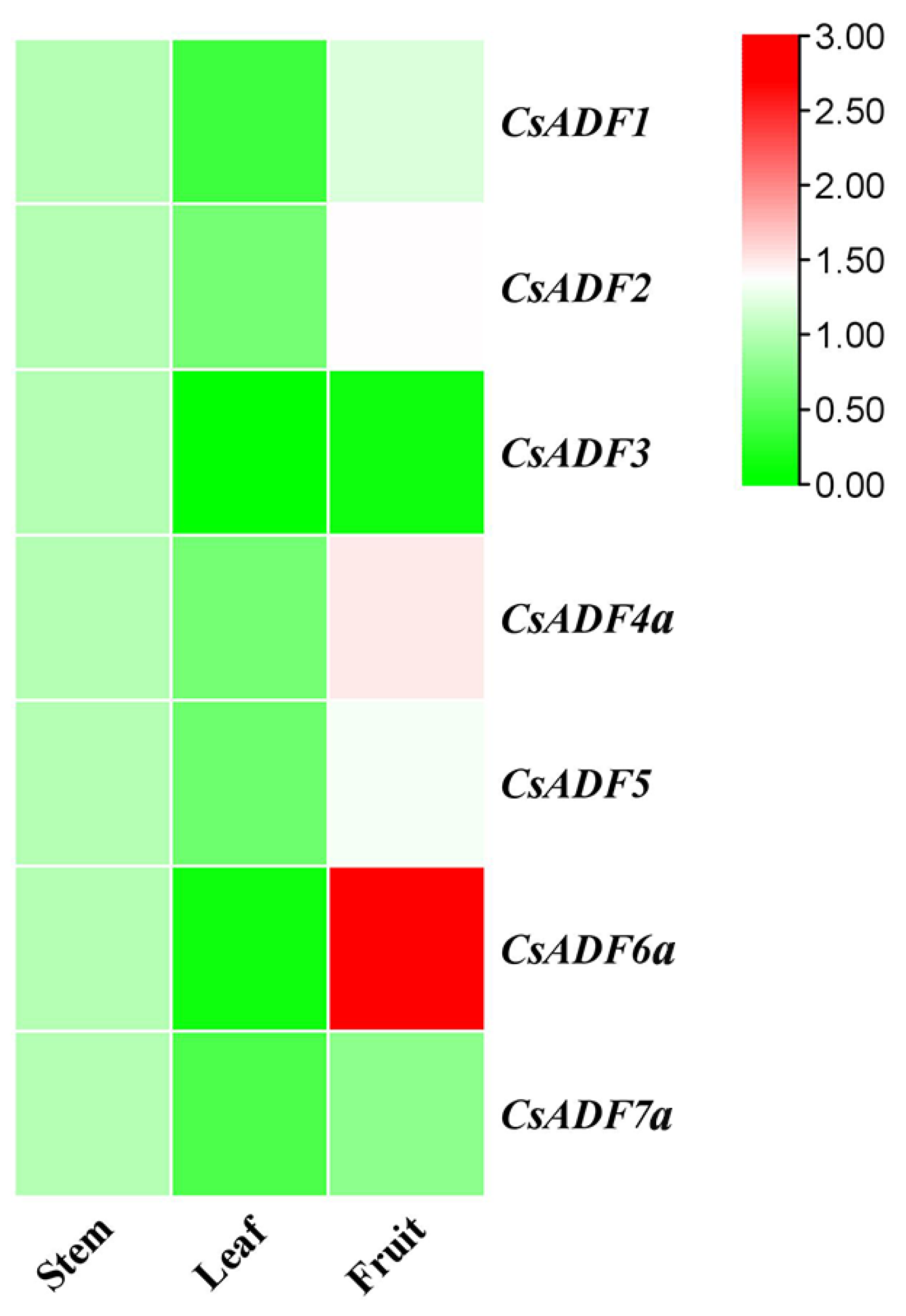
2.4. Expression Profile of CsADF Genes in Different Tissues of Sweet Orange
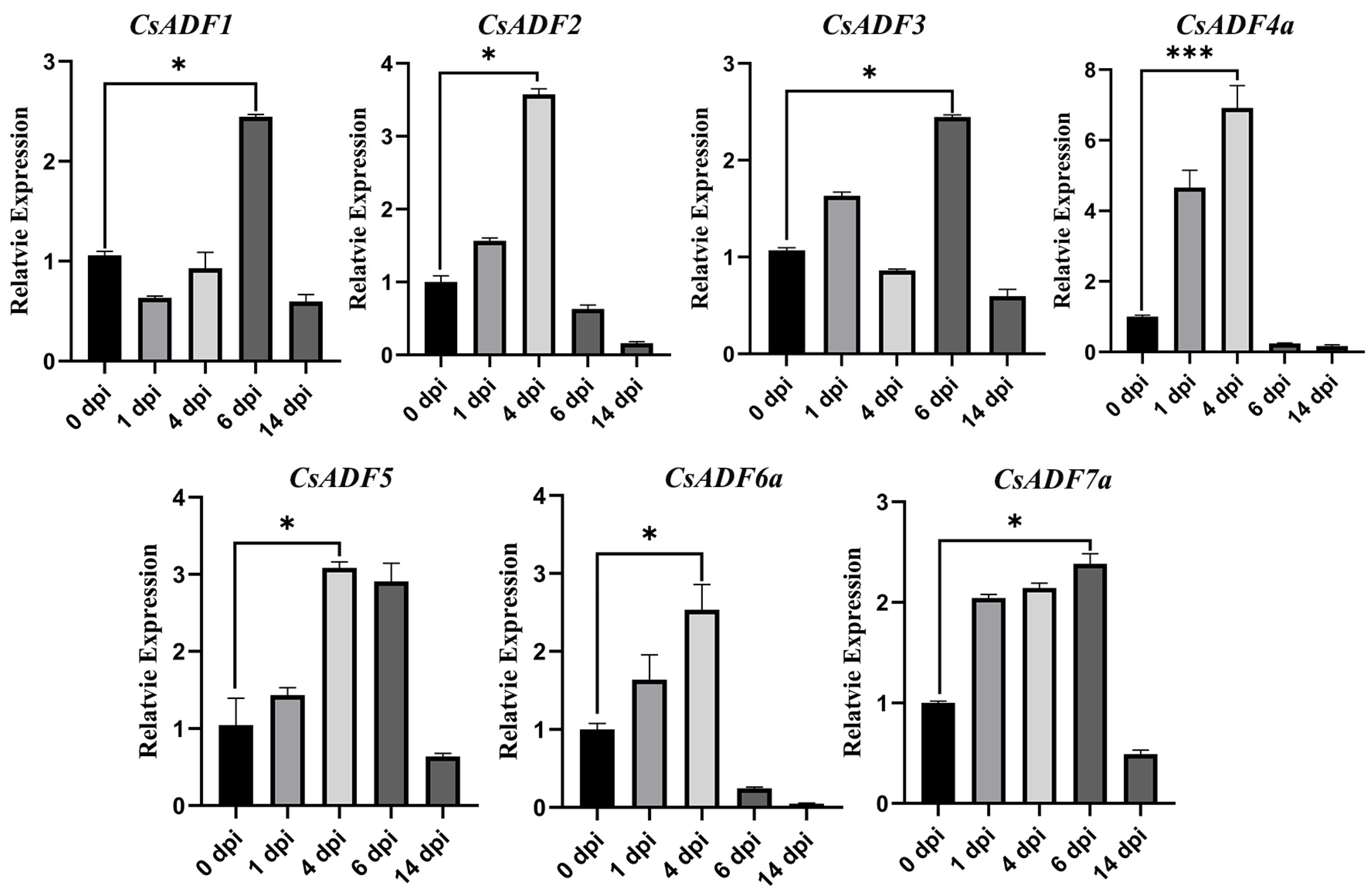
2.5. Expression of CsADF Gene Analysis in Sweet Orange after Xcc and CLas Infection
2.6. Subcellular Localization of CsADF Proteins
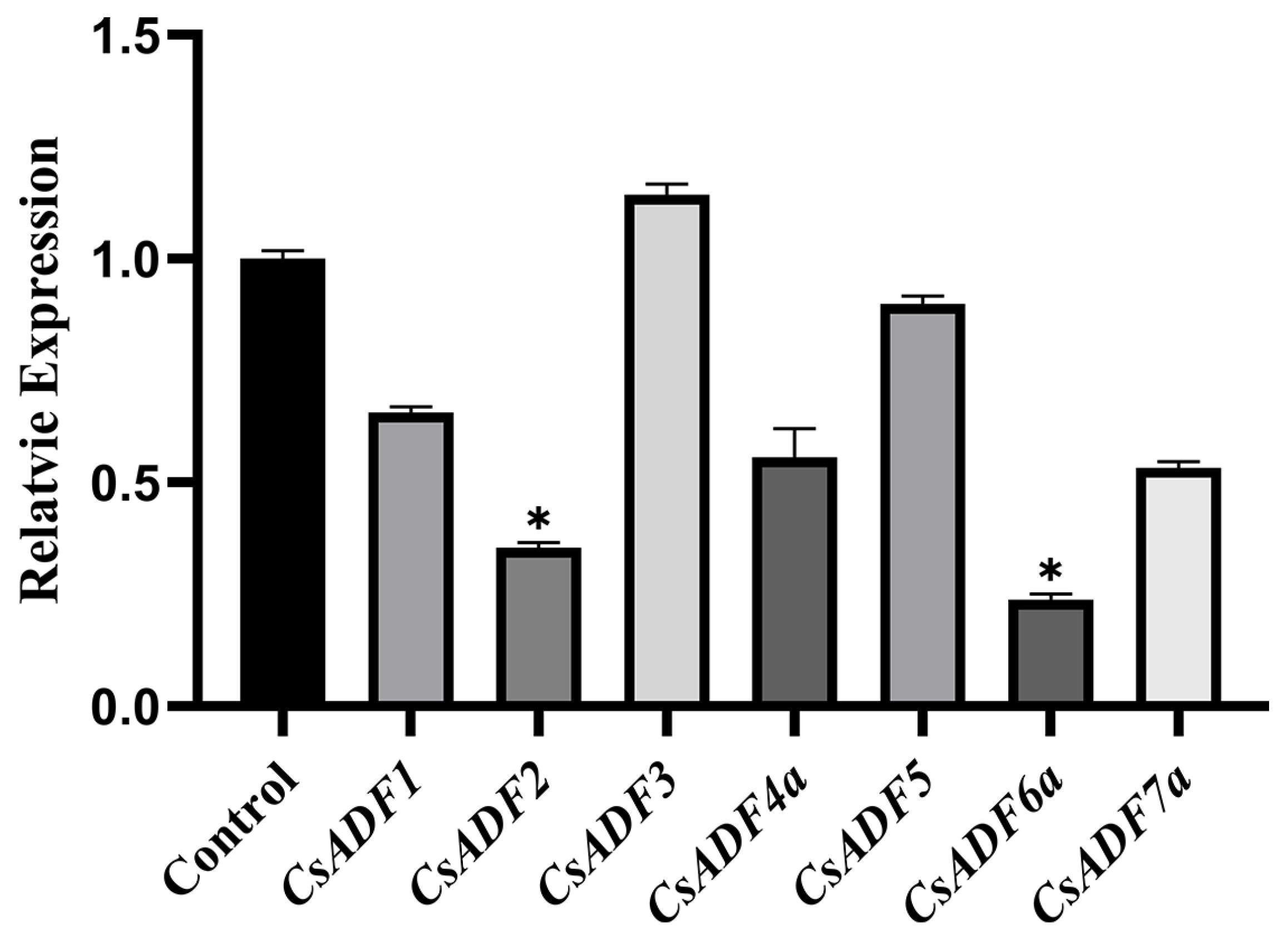
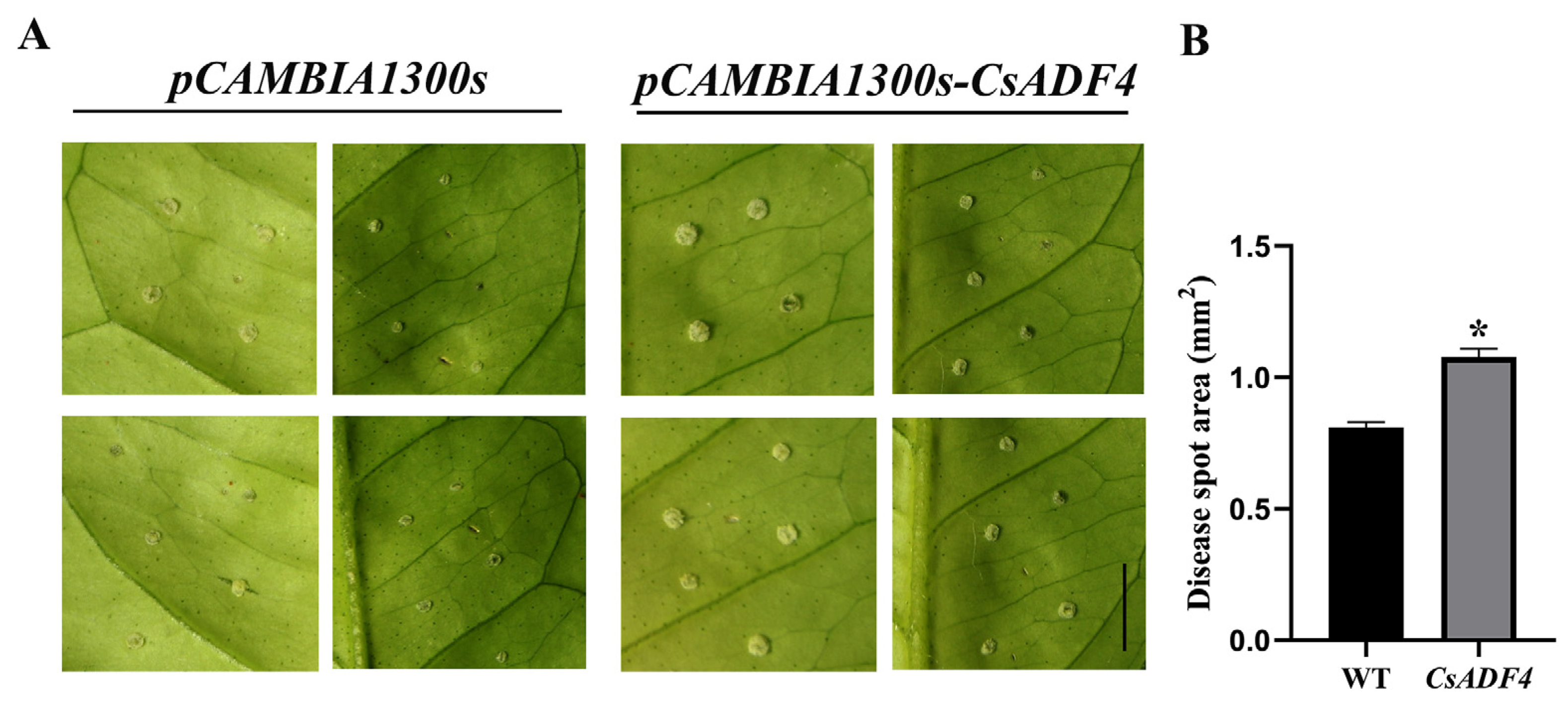
2.7. Transient Expression of CsADF4 Enhances Sweet Orange Susceptibility to Xcc
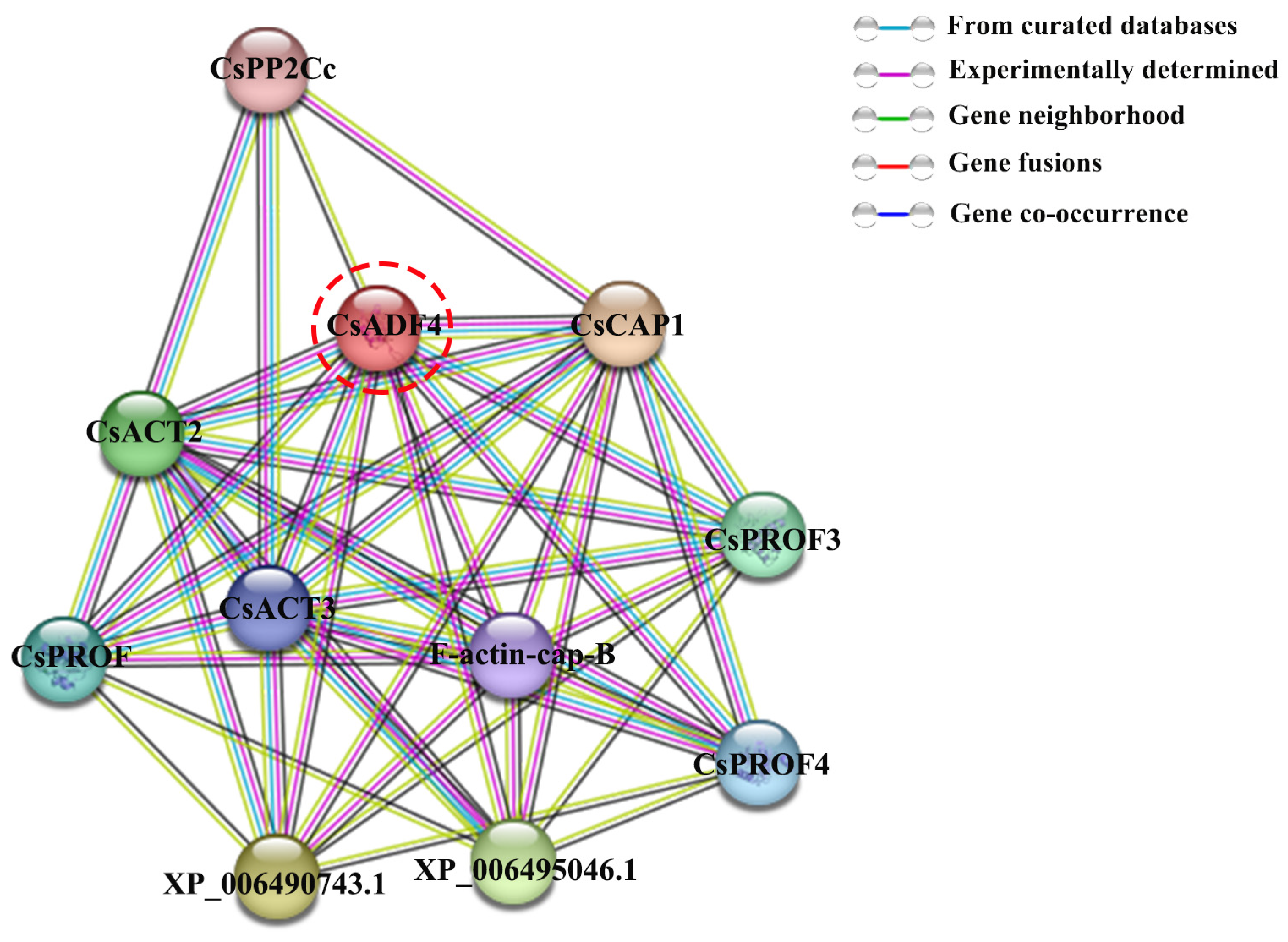
2.8. Analysis of CsADF4 Protein Interaction Network
3. Discussion
4. Materials and Methods
4.1. Gene Identification and Phylogenetic Analysis
4.2. Chromosome Distribution and Replication Events of CsADF Gene
4.3. Analysis of Gene Characteristics and Structure
4.4. Cis-Acting Regulatory Elements and Protein–Protein Interaction Network Prediction
4.5. Plant Materials and Treatments
4.6. RNA Extraction and Real-Time Quantitative PCR
4.7. Subcellular Localization
4.8. Expression Analysis of CsADF4
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tong, T. Global: The consumption of sweet oranges is increasing day by day. China Fruit News 2018, 35, 42. [Google Scholar]
- Hu, B.; Rao, M.J.; Deng, X.; Pandey, S.S.; Hendrich, C.; Ding, F.; Wang, N.; Xu, Q. Molecular signatures between citrus and Candidatus Liberibacter asiaticus. PLoS Pathog. 2021, 17, e1010071. [Google Scholar] [CrossRef]
- Deng, X.L.; Zheng, Y.Q.; Zheng, Z. Research progress on Genomics of Citrus Huanglong pathogen. J. S. China Agric. Univ. 2019, 40, 137–148. [Google Scholar]
- Muhammad, F.R.; Md, S.A.; Kwang-Hyun, B. Endophyte Bacillus velezensis Isolated from Citrus spp. Controls Streptomy-cin-Resistant Xanthomonas citri subsp. citri That Causes Citrus Bacterial Canker. Agronomy 2019, 9, 470. [Google Scholar]
- Wang, N.; Lu, X.L.; Li, L.; Wu, J.F.; Shang, J.W.; Hao, Z.F.; Li, F.H. Research progress of plant ADF/Cofilin family ADF. Acta Bot. Boreali-Occident. Sin. 2015, 35, 2349–2354. [Google Scholar]
- Maciver, S.K.; Hussey, P.J. The ADF/cofilin family: Actin-remodeling proteins. Genome Biol. 2002, 3, reviews3007.1. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, Q.; Xue, Q. Comparative study of rice and Arabidopsis actin-depolymerizing factors gene families. J. Plant Physiol. 2006, 163, 69–79. [Google Scholar] [CrossRef]
- Ruzicka, D.R.; Kandasamy, M.K.; McKinney, E.C.; Burgos-Rivera, B.; Meagher, R.B. The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. Cell Mol. Biol. 2007, 52, 460–472. [Google Scholar] [CrossRef]
- Huang, J.; Sun, W.; Ren, J.; Yang, R.; Fan, J.; Li, Y.; Wang, X.; Joseph, S.; Deng, W.; Zhai, L. Genome-Wide Identification and Characterization of Actin-Depolymerizing Factor (ADF) Family Genes and Expression Analysis of Responses to Various Stresses in Zea mays L. Int. J. Mol. Sci. 2020, 21, 1751. [Google Scholar] [CrossRef]
- Roy-Zokan, E.M.; Dyer, K.A.; Meagher, R.B. Phylogenetic Patterns of Codon Evolution in the ACTIN-DEPOLYMERIZING FACTOR/COFILIN (ADF/CFL) Gene Family. PLoS ONE 2015, 10, e145917. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, W.; Hong, C. Comprehensive analysis of differentiallyexpressed rice actin depolymerizing factor gene-family and heterologous overexpression ofOsADF3 confers Arabidopsis Thaliana droughttolerance. Rice 2012, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Mun, J.; Kim, H.; Lee, S.; Kim, S. An upstream region in the first intron of petunia actin-depolymerizing factor 1 affects tis-sue-specific expression in transgenic Arabidopsis (Arabidopsis thaliana). Plant J. Cell Mol. Biol. 2007, 50, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Yu, H.J.; Lee, H.S.; Kwon, Y.M.; Lee, J.S.; Lee, I.; Kim, S.G. Two closely related cDNAs encoding actin-depolymerizing factors of petunia are mainly expressed in vegetative tissues. Gene 2000, 257, 167–176. [Google Scholar] [CrossRef]
- Smertenko, A.P.; Jiang, C.J.; Simmons, N.J.; Weeds, A.G.; Davies, D.R.; Hussey, P.J. Ser6 in the maize actin-depolymerizing factor, ZmADF3, is phosphorylated by a calcium-stimulated protein kinase and is essential for the control of functional activity. Plant J. Cell Mol. Biol. 1998, 14, 187–193. [Google Scholar] [CrossRef]
- Daniel, B.S.; Daniel, J.C. Dynamic Coordination of Cytoskeletal and Cell Wall Systems during Plant Cell Morphogenesis. Curr Biol. 2009, 19, R800–R811. [Google Scholar]
- Zhu, J.; Nan, Q.; Qin, T.; Qian, D.; Mao, T.; Yuan, S.; Wu, X.; Niu, Y.; Bai, Q.; An, L.; et al. Higher-Ordered Actin Structures Remodeled by Arabidopsis ACTIN-DEPOLYMERIZING FACTOR5 Are Important for Pollen Germination and Pollen Tube Growth. Mol. Plant 2017, 10, 1065–1081. [Google Scholar] [CrossRef] [PubMed]
- Lopez, I.; Anthony, R.G.; Maciver, S.K.; Jiang, C.J.; Khan, S.; Weeds, A.G.; Hussey, P.J. Pollen specific expression of maize genes encoding actin depolymerizing fac-tor-like proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 7415–7420. [Google Scholar] [CrossRef]
- Dong, B.; Yang, Q.; Song, Z.; Niu, L.; Cao, H.; Liu, T.; Du, T.; Yang, W.; Qi, M.; Chen, T.; et al. Hyperoside promotes pollen tube growth by regulating the depolymerization effect of ac-tin-depolymerizing factor 1 on microfilaments in okra. Hortic Res-Engl. 2021, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Burgos-Rivera, B.; Ruzicka, D.R.; Deal, R.B.; McKinney, E.C.; King-Reid, L.; Meagher, R.B. ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 2008, 68, 619–632. [Google Scholar] [CrossRef]
- Peng, S.Q.; Huang, D.F. Expression of Arabidopsis ADF/Cofilin family 4 gene (AtADF4) in tobacco causes morphological changes of plants. J. Plant Physiol. Mol. Biol. 2006, 32, 52–56. [Google Scholar]
- Xu, K.; Zhao, Y.; Zhao, S.; Liu, H.; Wang, W.; Zhang, S.; Yang, X. Genome-Wide Identification and Low Temperature Responsive Pattern of Actin Depolymer-izing Factor (ADF) Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 618984. [Google Scholar] [CrossRef] [PubMed]
- Jean, D.; Eric, C.; Fathey, S. Identification and characterization of a low temperature regulated gene encoding an actin-binding protein from wheat. Febs Lett. 1996, 389, 324–327. [Google Scholar]
- Li, P.G.; Li, H.M.; Zhang, S. Cloning and Stress Resistance Analysis of the ADF Gene from Medicinal Wild Rice. Acta Ag-Riculturae Boreali-Sin. 2017, 32, 37–44. [Google Scholar]
- Zhang, P.; Qian, D.; Luo, C.; Niu, Y.; Li, T.; Li, C.; Xiang, Y.; Wang, X.; Niu, Y. Arabidopsis ADF5 Acts as a Downstream Target Gene of CBFs in Response to Low-Temperature Stress. Front Cell Dev. Biol. 2021, 9, 635533. [Google Scholar] [CrossRef]
- Fu, Y.; Duan, X.; Tang, C.; Li, X.; Voegele, R.T.; Wang, X.; Wei, G.; Kang, Z. TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. Cell Mol. Biol. 2014, 78, 16–30. [Google Scholar] [CrossRef]
- Tang, C.; Deng, L.; Chang, D.; Chen, S.; Wang, X.; Kang, Z. TaADF3, an Actin-Depolymerizing Factor, Negatively Modulates Wheat Resistance Against Puccinia striiformis. Front. Plant Sci. 2016, 6, 1214. [Google Scholar] [CrossRef]
- Zhang, B.; Hua, Y.; Wang, J.; Huo, Y.; Shimono, M.; Day, B.; Ma, Q. TaADF4, an actin-depolymerizing factor from wheat, is required for resistance to the stripe rust pathogen Puccinia striiformis f. sp. tritici. Plant J. Cell Mol. Biol. 2017, 89, 1210–1224. [Google Scholar] [CrossRef]
- Tian, M.; Chaudhry, F.; Ruzicka, D.R.; Meagher, R.B.; Staiger, C.J.; Day, B. Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 2009, 150, 815–824. [Google Scholar] [CrossRef]
- Porter, K.; Shimono, M.; Tian, M.; Day, B. Arabidopsis Actin-Depolymerizing Factor-4 links pathogen perception, defense acti-vation and transcription to cytoskeletal dynamics. PLoS Pathog. 2012, 8, e1003006. [Google Scholar] [CrossRef]
- Ontivero, M.; Zamora, G.M.; Salazar, S.; Ricci, J.C.D.; Castagnaro, A.P. Isolation of a strawberry gene fragment encoding an actin depolymerizing factor-like protein from genotypes resistant to Colletotrichum acutatum. Genome 2011, 54, 1041–1044. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews. Genetics 2011, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Q.; Du, S.Y.; Jiang, A.N. Visualization analysis of collinearity between poplar and willow genomes. Jiangsu Agric. Sci. 2018, 46, 22–27. [Google Scholar]
- Dong, C.H.; Kost, B.; Xia, G.; Chua, N.H. Molecular identification and characterization of the Arabidopsis AtADF1, AtADFS and AtADF6 genes. Plant Mol. Biol. 2001, 45, 517–527. [Google Scholar] [CrossRef]
- Jiang, C.J.; Weeds, A.G.; Hussey, P.J. The maize actin-depolymerizing factor, ZmADF3, redistributes to the growing tip of elongating root hairs and can be induced to translocate into the nucleus with actin. Plant J. Cell Mol. Biol. 1997, 12, 1035–1043. [Google Scholar] [CrossRef]
- Wang, Y.K.; Gao, J.G.; Yuan, G.L. Cloning and Expression Analysis of TaADF8 Gene in Common Wheat. J. Wheat Crops 2015, 35, 143–150. [Google Scholar]
- Wang, L.; Qiu, T.; Yue, J.; Guo, N.; He, Y.; Han, X.; Wang, Q.; Jia, P.; Wang, H.; Li, M.; et al. Arabidopsis ADF1 is Regulated by MYB73 and is Involved in Response to Salt Stress Affecting Actin Filament Organization. Plant Cell Physiol. 2021, 62, 1387–1395. [Google Scholar] [CrossRef]
- Byun, M.Y.; Cui, L.H.; Lee, A.; Oh, H.G.; Yoo, Y.H.; Lee, J.; Kim, W.T.; Lee, H. Abiotic Stress-Induced Actin-Depolymerizing Factor 3 From Deschampsia antarctica Enhanced Cold Tolerance When Constitutively Expressed in Rice. Front. Plant Sci. 2021, 12, 734500. [Google Scholar] [CrossRef]
- Chen, X.B.; Wang, D.M.; Liu, J. Response of Plant Cell Microfilament Skeleton to Fungal Infection. J. Agric. Univ. Hebei 2001, 3, 100–104. [Google Scholar]
- Ma, Q.H.; Chen, X.F.; Mu, J.H.; Hou, X.L. Expression Analysis of ADF7 and ADF10 Genes in Napa cabbage Infected by Botrytis cinerea. J. Nanjing Agric. Univ. 2015, 38, 742–747. [Google Scholar]
- Kandasamy, M.K.; Gilliland, L.U.; McKinney, E.C.; Meagher, R.B. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 2001, 13, 1541–1554. [Google Scholar] [CrossRef]
- Zhu, W.; Xu, X.; Peng, F.; Yan, D.Z.; Zhang, S.; Xu, R.; Wu, J.; Li, X.; Wei, W.; Chen, W. The cyclase-associated protein ChCAP is important for regulation of hyphal growth, appressorial development, penetration, pathogenicity, conidiation, intracellular cAMP level, and stress tolerance in Colletotrichum higginsianum. Plant Sci. Int. J. Exp. Plant Biol. 2019, 283, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, P.; Kessels, M.M.; Cope, M.J.; Drubin, D.G. The ADF homology (ADF-H) domain: A highly exploited actin-binding module. Mol. Biol. Cell. 1998, 9, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- de Paula Santos Martins, C.; Pedrosa, A.M.; Du, D.; Goncalves, L.P.; Yu, Q.; Gmitter, F.G., Jr.; Costa, M.G. Genome-Wide Characterization and Expression Analysis of Major Intrinsic Proteins during Abiotic and Biotic Stresses in Sweet Orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e138786. [Google Scholar] [CrossRef]
- Du, J.; Wang, Q.; Shi, H.; Zhou, C.; He, J.; Wang, X. A prophage-encoded effector from “Candidatus Liberibacter asiaticus” targets ASCORBATE PEROXIDASE6 in citrus to facilitate bacterial infection. Mol. Plant Pathol. 2023, 24, 302–316. [Google Scholar] [CrossRef]

| Gene Name | Sequence ID | Chromosomal | Protein | ||||
|---|---|---|---|---|---|---|---|
| MW (kDa) | Theoretical pI | Instability Index | Aliphatic Index | GRAVY | |||
| CsADF1 | Cs_ont_5g002950.1 | chr5:1964451-1965092 | 16.94 | 8.89 | 43.08 | 74.8 | −0.429 |
| CsADF2 | Cs_ont_5g011920.1 | chr5:7702323-7704616 | 16.87 | 7.75 | 45.64 | 62.19 | −0.734 |
| CsADF3 | Cs_ont_5g047810.1 | chr5:48291662-48292851 | 16.06 | 5.29 | 51.16 | 70.86 | −0.522 |
| CsADF4a | Cs_ont_7g004070.1 | chr7:3085584-3089118 | 15.96 | 5.31 | 42.83 | 70.86 | −0.519 |
| CsADF4b | Cs_ont_7g004070.2 | chr7:3085584-3089118 | 15.96 | 5.31 | 42.83 | 70.86 | −0.519 |
| CsADF5 | Cs_ont_8g003160.1 | chr8:1684426-1686707 | 19.73 | 6.31 | 50.57 | 67.95 | −0.429 |
| CsADF6a | Cs_ont_8g027730.1 | chr8:31643742-31645476 | 16.32 | 6.73 | 23.24 | 70.28 | −0.214 |
| CsADF6b | Cs_ont_8g027730.2 | chr8:31643742-31645476 | 13.25 | 6.08 | 13.1 | 80.52 | 0.148 |
| CsADF6c | Cs_ont_8g027730.3 | chr8:31643742-31645476 | 13.25 | 6.08 | 13.1 | 80.52 | 0.148 |
| CsADF6d | Cs_ont_8g027730.4 | chr8:31643742-31645476 | 13.25 | 6.08 | 13.1 | 80.52 | 0.148 |
| CsADF7a | Cs_ont_9g008180.1 | chr9:5690959-5694031 | 16.58 | 6.84 | 34.24 | 65.45 | −0.669 |
| CsADF7b | Cs_ont_9g008180.2 | chr9:5690959-5694031 | 16.58 | 6.84 | 34.24 | 65.45 | −0.669 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Dai, S.; Wang, X.; Gentile, A.; Zhang, Z.; Xie, Q.; Su, Y.; Li, D.; Wang, B. Actin-Depolymerizing Factor Gene Family Analysis Revealed That CsADF4 Increased the Sensitivity of Sweet Orange to Bacterial Pathogens. Plants 2023, 12, 3054. https://doi.org/10.3390/plants12173054
Xu J, Dai S, Wang X, Gentile A, Zhang Z, Xie Q, Su Y, Li D, Wang B. Actin-Depolymerizing Factor Gene Family Analysis Revealed That CsADF4 Increased the Sensitivity of Sweet Orange to Bacterial Pathogens. Plants. 2023; 12(17):3054. https://doi.org/10.3390/plants12173054
Chicago/Turabian StyleXu, Jing, Suming Dai, Xue Wang, Alessandra Gentile, Zhuo Zhang, Qingxiang Xie, Yajun Su, Dazhi Li, and Bing Wang. 2023. "Actin-Depolymerizing Factor Gene Family Analysis Revealed That CsADF4 Increased the Sensitivity of Sweet Orange to Bacterial Pathogens" Plants 12, no. 17: 3054. https://doi.org/10.3390/plants12173054
APA StyleXu, J., Dai, S., Wang, X., Gentile, A., Zhang, Z., Xie, Q., Su, Y., Li, D., & Wang, B. (2023). Actin-Depolymerizing Factor Gene Family Analysis Revealed That CsADF4 Increased the Sensitivity of Sweet Orange to Bacterial Pathogens. Plants, 12(17), 3054. https://doi.org/10.3390/plants12173054






