14-3-3 Proteins and the Plasma Membrane H+-ATPase Are Involved in Maize (Zea mays) Magnetic Induction
Abstract
1. Introduction
2. Results
2.1. The GMF Is Required for Maize Root Development
2.2. The GMF Is Required for Proper Activity of the Plasma Membrane H+-ATPase
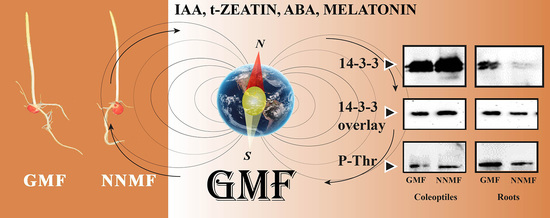
2.3. The GMF Is Required for the Phosphorylation of the 14-3-3 Protein Binding Site and the Binding of 14-3-3 Proteins
2.4. The GMF Induces an Oxidative State in Both Maize Coleoptiles and Roots
2.5. The GMF Is Required for Proper Production and Gene Expression of the Phytohormones Auxin and Cytokinin
2.6. Reduction of the GMF Triggers the Modulation of Stress-Related Phytohormones
2.7. Gene Expression of Calcium and Potassium Channels Requires the GMF
2.8. The GMF Is Necessary for Proper Expression of Some Genes Involved in Primary Metabolism
2.9. The Maize MagR Homolog ZmISCA2 Is Magnetically Induced along with CRY1
3. Discussion
3.1. GMF Modulates the 14-3-3-Mediated Activation of the Root PM H+-ATPase
3.2. Oxidative State, Second Messengers, and Phytohormone Homeostasis Play a Role in Plant Magnetic Induction
3.3. Maize Magnetic Induction Is Associated with ZmISCA and Photoreceptor Modulation
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Near Null Magnetic Field (NNMF) Generation System and Plant Exposure
4.3. Analysis of Shoot and Root Growth
4.4. Plasma Membrane Purification
4.5. H+-ATPase Activity
4.6. Immunoblotting
4.7. Overlay Assay
4.8. RNA Extraction and Quantitative Gene Expression
4.9. Protein and ROS Quantification
4.10. Phytohormone and Melatonin Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parmagnani, A.S.; Betterle, N.; Mannino, G.; D’Alessandro, S.; Nocito, F.F.; Ljumovic, K.; Vigani, G.; Ballottari, M.; Maffei, M.E. The Geomagnetic Field (GMF) Is Required for Lima Bean Photosynthesis and Reactive Oxygen Species Production. Int. J. Mol. Sci. 2023, 24, 2896. [Google Scholar] [CrossRef]
- Maffei, M.E. Magnetoreception in Plants. In Bioelectromagnetism: History, Foundations and Applications; Shoogo, U., Tsukasa, S., Eds.; CRC Press: Boca Raton, FL, USA, 2022; pp. 191–214. [Google Scholar]
- Erdmann, W.; Kmita, H.; Kosicki, J.Z.; Kaczmarek, Ł. How the Geomagnetic Field Influences Life on Earth—An Integrated Approach to Geomagnetobiology. Orig. Life Evol. Biosph. 2021, 51, 231–257. [Google Scholar] [CrossRef] [PubMed]
- Occhipinti, A.; De Santis, A.; Maffei, M.E. Magnetoreception: An unavoidable step for plant evolution? Trends Plant Sci. 2014, 19, 1–4. [Google Scholar] [CrossRef]
- Agliassa, C.; Narayana, R.; Bertea, C.M.; Rodgers, C.T.; Maffei, M.E. Reduction of the geomagnetic field delays Arabidopsis thaliana flowering time through downregulation of flowering-related genes. Bioelectromagnetics 2018, 39, 361–374. [Google Scholar] [CrossRef]
- Paponov, I.A.; Fliegmann, J.; Narayana, R.; Maffei, M.E. Differential root and shoot magnetoresponses in Arabidopsis thaliana. Sci. Rep. 2021, 11, 9195. [Google Scholar] [CrossRef]
- Bertea, C.M.; Narayana, R.; Agliassa, C.; Rodgers, C.T.; Maffei, M.E. Geomagnetic field (GMF) and plant evolution: Investigating the effects of GMF reversal on Arabidospis thaliana development and gene expression. J. Visual. Exp. 2015, 105, e53286. [Google Scholar]
- Parmagnani, A.S.; Mannino, G.; Maffei, M.E. Transcriptomics and Metabolomics of Reactive Oxygen Species Modulation in Near-Null Magnetic Field-Induced Arabidopsis thaliana. Biomolecules 2022, 12, 1824. [Google Scholar] [CrossRef]
- Vigani, G.; Islam, M.; Cavallaro, V.; Nocito, F.F.; Maffei, M.E. Geomagnetic Field (GMF)-Dependent Modulation of Iron-Sulfur Interplay in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Maffei, M.E.; Vigani, G. The Geomagnetic Field Is a Contributing Factor for an Efficient Iron Uptake in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Vigani, G.; Maffei, M.E. The Geomagnetic Field (GMF) Modulates Nutrient Status and Lipid Metabolism during Arabidopsis thaliana Plant Development. Plants 2020, 9, 1729. [Google Scholar] [CrossRef]
- Agliassa, C.; Maffei, M.E. Reduction of geomagnetic field (GMF) to near null magnetic field (NNMF) affects some Arabidopsis thaliana clock genes amplitude in a light independent manner. J. Plant Physiol. 2019, 232, 23–26. [Google Scholar] [CrossRef]
- Agliassa, C.; Narayana, R.; Christie, J.M.; Maffei, M.E. Geomagnetic field impacts on cryptochrome and phytochrome signaling. J. Photochem. Photobiol. B 2018, 185, 32–40. [Google Scholar] [CrossRef]
- Dhiman, S.K.; Wu, F.; Galland, P. Effects of weak static magnetic fields on the development of seedlings of Arabidopsis thaliana. Protoplasma 2022, 260, 767–786. [Google Scholar] [CrossRef]
- Parmagnani, A.S.; D’Alessandro, S.; Maffei, M.E. Iron-sulfur complex assembly: Potential players of magnetic induction in plants. Plant Sci. 2022, 325, 111483. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, H.B.; Lu, L.H.; Li, Y.Q. A mechanism of compass-free migratory navigation. J. Phys. D-Appl. Phys. 2022, 55, 9. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.S.; Yu, H.; Peng, J.H.; Hu, Z.R.; Chen, L. The role of 14-3-3 proteins in plant growth and response to abiotic stress. Plant Cell Rep. 2022, 41, 833–852. [Google Scholar] [CrossRef]
- Pallucca, R.; Visconti, S.; Camoni, L.; Cesareni, G.; Melino, S.; Panni, S.; Torreri, P.; Aducci, P. Specificity of ε and Non-ε Isoforms of Arabidopsis 14-3-3 Proteins Towards the H+-ATPase and Other Targets. PLoS ONE 2014, 9, e90764. [Google Scholar] [CrossRef]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 Proteins in Plant Hormone Signaling: Doing Several Things at Once. Front. Plant Sci. 2018, 9, 297. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Iori, V.; Marra, M.; Aducci, P. Phosphorylation-dependent interaction between plant plasma membrane H+-ATPase and 14-3-3 proteins. J. Biol. Chem. 2000, 275, 9919–9923. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, D.; Morris, E.R.; Walker, J.C. 14-3-3 and FHA Domains Mediate Phosphoprotein Interactions. Annu. Rev. Plant Biol. 2009, 60, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Oecking, C.; Jaspert, N. Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 2009, 12, 760–765. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef]
- Manosalva, P.M.; Bruce, M.; Leach, J.E. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 2011, 68, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.Y.; Zhang, W.R.; Xu, T.Y.; Tang, M. Effect of arbuscular mycorrhizal fungi and phosphorus on drought-induced oxidative stress and 14-3-3 proteins gene expression of Populus cathayana. Front. Microbiol. 2022, 13, 20. [Google Scholar] [CrossRef]
- Visconti, S.; D’Ambrosio, C.; Fiorillo, A.; Arena, S.; Muzi, C.; Zottini, M.; Aducci, P.; Marra, M.; Scaloni, A.; Camoni, L. Overexpression of 14-3-3 proteins enhances cold tolerance and increases levels of stress-responsive proteins of Arabidopsis plants. Plant Sci. Int. J. Exp. Plant Biol. 2019, 289, 110215. [Google Scholar] [CrossRef]
- He, Y.; Zhang, Y.; Chen, L.H.; Wu, C.L.; Luo, Q.C.; Zhang, F.; Wei, Q.H.; Li, K.X.; Chang, J.L.; Yang, G.X.; et al. A Member of the 14-3-3 Gene Family in Brachypodium distachyon, BdGF14d, Confers Salt Tolerance in Transgenic Tobacco Plants. Front. Plant Sci. 2017, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zeng, H.; Xu, F.; Yan, F.; Xu, W. H+-ATPases in Plant Growth and Stress Responses. Annu. Rev. Plant Biol. 2022, 73, 495–521. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. From plant physiology to pharmacology: Fusicoccin leaves the leaves. Planta 2019, 249, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma Membrane H(+)-ATPase Regulation in the Center of Plant Physiology. Mol. Plant 2016, 9, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Jahn, T.; Fuglsang, A.T.; Olsson, A.; Bruntrup, I.M.; Collinge, D.B.; Volkmann, D.; Sommarin, M.; Palmgren, M.G.; Larsson, C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 1997, 9, 1805–1814. [Google Scholar] [PubMed]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. Cell Mol. Biol. 2002, 32, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Svennelid, F.; Olsson, A.; Piotrowski, M.; Rosenquist, M.; Ottman, C.; Larsson, C.; Oecking, C.; Sommarin, M. Phosphorylation of Thr-948 at the C Terminus of the Plasma Membrane H+-ATPase Creates a Binding Site for the Regulatory 14-3-3 Protein. Plant Cell 1999, 11, 2379–2391. [Google Scholar]
- Shanko, A.V.; Mesenko, M.M.; Klychnikov, O.I.; Nosov, A.V.; Ivanov, V.B. Proton pumping in growing part of maize root: Its correlation with 14-3-3 protein content and changes in response to osmotic stress. Biochemistry 2003, 68, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Di Lucente, C.; Pallucca, R.; Visconti, S.; Aducci, P. Binding of phosphatidic acid to 14-3-3 proteins hampers their ability to activate the plant plasma membrane H+-ATPase. IUBMB Life 2012, 64, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Frías, I.; Caldeira, M.T.; Pérez-Castiñeira, J.R.; Navarro-Aviñó, J.P.; Culiañez-Maciá, F.A.; Kuppinger, O.; Stransky, H.; Pagés, M.; Hager, A.; Serrano, R. A major isoform of the maize plasma membrane H(+)-ATPase: Characterization and induction by auxin in coleoptiles. Plant Cell 1996, 8, 1533–1544. [Google Scholar] [PubMed]
- Elmayan, T.; Fromentin, J.; Riondet, C.; Alcaraz, G.; Blein, J.-P.; Simon-Plas, F. Regulation of reactive oxygen species production by a 14-3-3 protein in elicited tobacco cells. Plant Cell Environ. 2007, 30, 722–732. [Google Scholar] [CrossRef]
- Manchester, L.C.; Tan, D.X.; Reiter, R.J.; Park, W.; Monis, K.; Qi, W. High levels of melatonin in the seeds of edible plants: Possible function in germ tissue protection. Life Sci. 2000, 67, 3023–3029. [Google Scholar] [CrossRef]
- Wiltschko, R.; Dehe, L.; Gehring, D.; Thalau, P.; Wiltschko, W. Interactions between the visual and the magnetoreception system: Different effects of bichromatic light regimes on the directional behavior of migratory birds. J. Physiol.-Paris 2013, 107, 137–146. [Google Scholar] [CrossRef]
- Mannino, G.; Casacci, L.P.; Bianco Dolino, G.; Badolato, G.; Maffei, M.E.; Barbero, F. The Geomagnetic Field (GMF) Is Necessary for Black Garden Ant (Lasius niger L.) Foraging and Modulates Orientation Potentially through Aminergic Regulation and MagR Expression. Int. J. Mol. Sci. 2023, 24, 4387. [Google Scholar] [CrossRef]
- Pooam, M.; Jourdan, N.; El Esawi, M.; Sherrard, R.M.; Ahmad, M. HEK293 cell response to static magnetic fields via the radical pair mechanism may explain therapeutic effects of pulsed electromagnetic fields. PLoS ONE 2020, 15, e0243038. [Google Scholar] [CrossRef]
- Pooam, M.; Arthaut, L.D.; Burdick, D.; Link, J.; Martino, C.F.; Ahmad, M. Magnetic sensitivity mediated by the Arabidopsis blue-light receptor cryptochrome occurs during flavin reoxidation in the dark. Planta 2019, 249, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, K.J. A candidate magnetoreceptor. Nat. Mater. 2016, 15, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Hayashi, K.-I.; Kinoshita, T. Auxin Activates the Plasma Membrane H+-ATPase by Phosphorylation during Hypocotyl Elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef]
- Poeggeler, B.; Saarela, S.; Reiter, R.J.; Tan, D.X.; Chen, L.D.; Manchester, L.C.; Barlow-Walden, L.R. Melatonin--a highly potent endogenous radical scavenger and electron donor: New aspects of the oxidation chemistry of this indole accessed in vitro. Ann. N. Y. Acad. Sci. 1994, 738, 419–420. [Google Scholar] [CrossRef]
- Wang, H.Z.; Zhang, X. Magnetic Fields and Reactive Oxygen Species. Int. J. Mol. Sci. 2017, 18, 20. [Google Scholar] [CrossRef]
- Kaya, H.; Nakajima, R.; Iwano, M.; Kanaoka, M.M.; Kimura, S.; Takeda, S.; Kawarazaki, T.; Senzaki, E.; Hamamura, Y.; Higashiyama, T.; et al. Ca2+-Activated Reactive Oxygen Species Production by Arabidopsis RbohH and RbohJ Is Essential for Proper Pollen Tube Tip Growth. Plant Cell 2014, 26, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.L.; Cao, X.Y.; Wang, X.Y.; Zhang, W.; Li, W.X.; Wang, X.Q.; Liu, S.Q.; Lyu, D.G. RBOH-dependent hydrogen peroxide signaling mediates melatonin-induced anthocyanin biosynthesis in red pear fruit. Plant Sci. 2021, 313, 11. [Google Scholar] [CrossRef]
- Camoni, L.; Harper, J.F.; Palmgren, M.G. 14-3-3 proteins activate a plant calcium-dependent protein kinase (CDPK). FEBS Lett. 1998, 430, 381–384. [Google Scholar] [CrossRef]
- Zhang, C.H.; Nie, X.; Kong, W.L.; Deng, X.X.; Sun, T.; Liu, X.H.; Li, Y.S. Genome-Wide Identification and Evolution Analysis of the Gibberellin Oxidase Gene Family in Six Gramineae Crops. Genes 2022, 13, 16. [Google Scholar] [CrossRef]
- Avalbaev, A.M.; Somov, K.A.; Yuldashev, R.A.; Shakirova, F.M. Cytokinin oxidase is key enzyme of cytokinin degradation. Biochemistry 2012, 77, 1354–1361. [Google Scholar] [CrossRef]
- Ostrowski, M.; Jakubowska, A. Auxin receptors. Postep. Biol. Komorki 2008, 35, 79–95. [Google Scholar]
- Ramireddy, E.; Nelissen, H.; Leuendorf, J.E.; Van Lijsebettens, M.; Inze, D.; Schmulling, T. Root engineering in maize by increasing cytokinin degradation causes enhanced root growth and leaf mineral enrichment. Plant Mol. Biol. 2021, 106, 555–567. [Google Scholar] [CrossRef]
- Friero, I.; Alarcon, M.V.; Gordillo, L.; Salguero, J. Abscisic acid is involved in several processes associated with root system architecture in maize. Acta Physiol. Plant. 2022, 44, 12. [Google Scholar] [CrossRef]
- Jiang, L.; Chen, X.; Gu, X.; Deng, M.; Li, X.; Zhou, A.; Suo, M.; Gao, W.; Lin, Y.; Wang, Y.; et al. Light Quality and Sucrose-Regulated Detached Ripening of Strawberry with Possible Involvement of Abscisic Acid and Auxin Signaling. Int. J. Mol. Sci. 2023, 24, 5681. [Google Scholar] [CrossRef]
- Holtman, W.L.; Roberts, M.R.; Oppedijk, B.J.; Testerink, C.; van Zeijl, M.J.; Wang, M. 14-3-3 proteins interact with a 13-lipoxygenase, but not with a 9-lipoxygenase. FEBS Lett. 2000, 474, 48–52. [Google Scholar] [CrossRef]
- Yang, X.; Wang, W.; Coleman, M.; Orgil, U.; Feng, J.; Ma, X.; Ferl, R.; Turner, J.G.; Xiao, S. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 2009, 60, 539–550. [Google Scholar] [CrossRef]
- Gutjahr, C.; Riemann, M.; Muller, A.; Duchting, P.; Weiler, E.W.; Nick, P. Cholodny-Went revisited: A role for jasmonate in gravitropism of rice coleoptiles. Planta 2005, 222, 575–585. [Google Scholar] [CrossRef]
- Ronzan, M.; Piacentini, D.; Fattorini, L.; Federica, D.; Caboni, E.; Eiche, E.; Ziegler, J.; Hause, B.; Riemann, M.; Betti, C.; et al. Auxin jasmonate crosstalk in Oryza sativa L. root system formation after cadmium and/or arsenic exposure. Environ. Exp. Bot. 2019, 165, 59–69. [Google Scholar] [CrossRef]
- Garrido, I.; Espinosa, F.; Alvarez-Tinaut, M. Oxidative defence reactions in sunflower roots induced by methyl-jasmonate and methyl-salicylate and their relation with calcium signalling. Protoplasma 2009, 237, 27–39. [Google Scholar] [CrossRef]
- Narayana, R.; Fliegmann, J.; Paponov, I.; Maffei, M.E. Reduction of geomagnetic field (GMF) to near null magnetic field (NNMF) affects Arabidopsis thaliana root mineral nutrition. Life Sci. Space Res. 2018, 19, 43–50. [Google Scholar] [CrossRef]
- Ahmad, M.; Galland, P.; Ritz, T.; Wiltschko, R.; Wiltschko, W. Magnetic intensity affects cryptochrome-dependent responses in Arabidopsis thaliana. Planta 2007, 225, 615–624. [Google Scholar] [CrossRef]
- Qin, S.; Yin, H.; Yang, C.; Dou, Y.; Liu, Z.; Zhang, P.; Yu, H.; Huang, Y.; Feng, J.; Hao, J.; et al. A magnetic protein biocompass. Nat. Mater. 2016, 15, 217–226. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Serrano, R. H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: Purification and reconstitution. Methods Enzymol. 1988, 157, 533–544. [Google Scholar]
- Fiorillo, A.; Manai, M.; Visconti, S.; Camoni, L. The Salt Tolerance-Related Protein (STRP) Is a Positive Regulator of the Response to Salt Stress in Arabidopsis thaliana. Plants 2023, 12, 1704. [Google Scholar] [CrossRef]
- Muzi, C.; Camoni, L.; Visconti, S.; Aducci, P. Cold stress affects H(+)-ATPase and phospholipase D activity in Arabidopsis. Plant Physiol. Biochem. PPB 2016, 108, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Barbero, F.; Aducci, P.; Maffei, M.E. Spodoptera littoralis oral secretions inhibit the activity of Phaseolus lunatus plasma membrane H+-ATPase. PLoS ONE 2018, 13, e0202142. [Google Scholar] [CrossRef] [PubMed]
- Paiardini, A.; Aducci, P.; Cervoni, L.; Cutruzzolà, F.; Di Lucente, C.; Janson, G.; Pascarella, S.; Rinaldo, S.; Visconti, S.; Camoni, L. The phytotoxin fusicoccin differently regulates 14-3-3 proteins association to mode III targets. IUBMB Life 2014, 66, 52–62. [Google Scholar] [CrossRef]
- Visconti, S.; Camoni, L.; Fullone, M.R.; Lalle, M.; Marra, M.; Aducci, P. Mutational analysis of the interaction between 14-3-3 proteins and plant plasma membrane H+-ATPase. J. Biol. Chem. 2003, 278, 8172–8178. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; Yoshida, T.; Perez de Souza, L.; Alseekh, S.; Bajdzienko, K.; Fernie, A.R.; Giavalisco, P. An improved extraction method enables the comprehensive analysis of lipids, proteins, metabolites and phytohormones from a single sample of leaf tissue under water-deficit stress. Plant J. 2020, 103, 1614–1632. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Binhi, V.N.; Prato, F.S. Rotations of macromolecules affect nonspecific biological responses to magnetic fields. Sci. Rep. 2018, 8, 13495. [Google Scholar] [CrossRef] [PubMed]
- Bunney, T.D.; van Walraven, H.S.; de Boer, A.H. 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc. Natl. Acad. Sci. USA 2001, 98, 4249–4254. [Google Scholar] [CrossRef] [PubMed]
- Klychnikov, O.I.; Li, K.W.; Lill, H.; de Boer, A.H. The V-ATPase from etiolated barley (Hordeum vulgare L.) shoots is activated by blue light and interacts with 14-3-3 proteins. J. Exp. Bot. 2007, 58, 1013–1023. [Google Scholar] [CrossRef]
- Grinberg, M.; Mudrilov, M.; Kozlova, E.; Sukhov, V.; Sarafanov, F.; Evtushenko, A.; Ilin, N.; Vodeneev, V.; Price, C.; Mareev, E. Effect of extremely low-frequency magnetic fields on light-induced electric reactions in wheat. Plant Signal. Behav. 2022, 17, 2021664. [Google Scholar] [CrossRef] [PubMed]








| Gene | Coleoptiles | Roots |
|---|---|---|
| RBOH1 | 0.83 ± 0.09 | 0.22 ± 0.02 |
| SOD1 | 1.36 ± 0.12 | 0.79 ± 0.08 |
| CAT1 | 0.46 ± 0.09 | 0.55 ± 0.08 |
| APX1 | 0.29 ± 0.08 | 0.42 ± 0.06 |
| GSR1 | 0.67 ± 0.08 | 0.53 ± 0.05 |
| GMF | NNMF | |||
|---|---|---|---|---|
| Coleoptiles | Roots | Coleoptiles | Roots | |
| IAA | 1.793 ± 0.105 a | 1.306 ± 0.042 b | 1.269 ± 0.041 b | 1.135 ± 0.075 b |
| ZEA | 24.926 ± 1.729 c | 21.916 ± 1.338 bc | 27.355 ± 1.888 b | 33.675 ± 2.67 a |
| ABA | 8.101 ± 0.421 c | 11.809 ± 1.049 b | 5.068 ± 0.734 d | 19.513 ± 1.124 a |
| OPDA | 364.44 ± 21.803 a | 85.567 ± 6.509 c | 315.183 ± 13.064 b | 70.691 ± 2.151 c |
| JA | 181.856 ± 13.554 a | 41.338 ± 3.051 c | 154.197 ± 9.129 b | 34.767 ± 2.313 c |
| MeJA | 13.691 ± 0.685 a | 13.126 ± 0.385 a | 10.514 ± 0.598 b | 13.145 ± 0.648 a |
| SA | 143.533 ± 3.902 a | 86.798 ± 2.88 c | 124.669 ± 8.216 b | 85.21 ± 1.697 c |
| MeSA | 36.097 ± 1.36 a | 6.841 ± 0.149 d | 32.413 ± 1.416 b | 11.145 ± 0.767 c |
| Gene | Coleoptiles | Roots |
|---|---|---|
| CKO3 | 0.89 ± 0.19 | 0.41 ± 0.07 |
| CRR1 | 1.77 ± 0.32 | 0.23 ± 0.03 |
| PIN1 | 1.17 ± 0.19 | 0.57 ± 0.03 |
| PIN3 | 0.75 ± 0.07 | 0.65 ± 0.06 |
| ABP1 | 0.71 ± 0.02 | 0.46 ± 0.04 |
| Gene | Coleoptiles | Roots |
|---|---|---|
| GA20OX3 | 3.82 ± 0.62 | 0.43 ± 0.05 |
| GA2OX1 | 1.37 ± 0.38 | 0.64 ± 0.09 |
| EREB10 | 1.17 ± 0.18 | 0.57 ± 0.06 |
| BRL1 | 0.41 ± 0.11 | 2.22 ± 0.12 |
| BSK1 | 0.83 ± 0.08 | 0.61 ± 0.06 |
| Gene | Coleoptiles | Roots |
|---|---|---|
| CDPK1 | 1.08 ± 0.25 | 0.49 ± 0.02 |
| CAL1 | 2.01 ± 0.14 | 0.24 ± 0.07 |
| CAP1 | 0.62 ± 0.04 | 0.52 ± 0.06 |
| CAX1 | 0.65 ± 0.03 | 0.51 ± 0.06 |
| KCH1 | 2.21 ± 0.32 | 0.28 ± 0.01 |
| PIP1b | 1.46 ± 0.09 | 1.03 ± 0.09 |
| SLAC1 | 0.94 ± 0.22 | 0.61 ± 0.05 |
| Gene | Coleoptiles | Roots |
|---|---|---|
| SUS1 | 0.53 ± 0.08 | 1.97 ± 0.21 |
| IDH1 | 0.88 ± 0.11 | 0.38 ± 0.07 |
| MDH1 | 0.81 ± 0.13 | 0.53 ± 0.03 |
| PEP1 | 3.14 ± 0.38 | 2.13 ± 0.33 |
| GOGAT1 | 0.67 ± 0.04 | 0.41 ± 0.02 |
| Gene | Coleoptiles | Roots |
|---|---|---|
| ZmISCA1 | 0.83 ± 0.07 | 0.84 ± 0.11 |
| ZmISCA2 | 0.79 ± 0.11 | 1.21 ± 0.11 |
| CRY1 | 0.76 ± 0.17 | 1.28 ± 0.16 |
| CRY2 | 1.21 ± 0.16 | 0.98 ± 0.07 |
| PHYA1 | 0.63 ± 0.11 | 0.99 ± 0.06 |
| PHYB1 | 0.63 ± 0.04 | 0.64 ± 0.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiorillo, A.; Parmagnani, A.S.; Visconti, S.; Mannino, G.; Camoni, L.; Maffei, M.E. 14-3-3 Proteins and the Plasma Membrane H+-ATPase Are Involved in Maize (Zea mays) Magnetic Induction. Plants 2023, 12, 2887. https://doi.org/10.3390/plants12152887
Fiorillo A, Parmagnani AS, Visconti S, Mannino G, Camoni L, Maffei ME. 14-3-3 Proteins and the Plasma Membrane H+-ATPase Are Involved in Maize (Zea mays) Magnetic Induction. Plants. 2023; 12(15):2887. https://doi.org/10.3390/plants12152887
Chicago/Turabian StyleFiorillo, Anna, Ambra S. Parmagnani, Sabina Visconti, Giuseppe Mannino, Lorenzo Camoni, and Massimo E. Maffei. 2023. "14-3-3 Proteins and the Plasma Membrane H+-ATPase Are Involved in Maize (Zea mays) Magnetic Induction" Plants 12, no. 15: 2887. https://doi.org/10.3390/plants12152887
APA StyleFiorillo, A., Parmagnani, A. S., Visconti, S., Mannino, G., Camoni, L., & Maffei, M. E. (2023). 14-3-3 Proteins and the Plasma Membrane H+-ATPase Are Involved in Maize (Zea mays) Magnetic Induction. Plants, 12(15), 2887. https://doi.org/10.3390/plants12152887