Signaling Cross-Talk between Salicylic and Gentisic Acid in the ‘Candidatus Phytoplasma Solani’ Interaction with Sangiovese Vines
Abstract
1. Introduction
2. Results
2.1. Plant Symptoms
2.2. Characterization of Phenylpropanoids Accumulated in Phytoplasma Infected Plants
2.2.1. Qualitative Analysis
| No. | Compound | RT a (min) | m/z exp. b | m/z calc. c | (M-H)− | Error (ppm) | Reference |
| 1A | Dihydroxybenzoic acid-glucoside is. 1 | 2.626 | 315.0736 | 315.0722 | C13H15O9 | −4.44 | [43,44,45] |
| 2A | Gentisic acid * | 3.703 | 153.0194 | 153.0193 | C7H5O4 | −0.65 | |
| 3A | Dihydroxybenzoic acid-glucoside is. 2 | 4.500 | 315.0754 | 315.0733 | C13H15O9 | −10.15 | [44,46] |
| 4A | Caffeic acid glucoside | 5.129 | 341.0896 | 341.0878 | C15H17O9 | −5.73 | [47] |
| 5A | 2,3 Dihydroxybenzoic acid * | 5.184 | 153.0208 | 153.0193 | C7H5O4 | −9.80 | |
| 6A | Hydroxybenzoic acid glucoside | 5.827 | 299.0789 | 299.0772 | C13H16O8 | −5.68 | [44] |
| 7A | Catechin * | 5.889 | 289.0721 | 289.0718 | C15H13O6 | −1.03 | |
| 8A | Coumaric-3-O- acid glucoside | 6.374 | 325.0940 | 325.0920 | C15H17O8 | −6.15 | [43,45] |
| 9A | Epicatechin * | 6.952 | 289.0737 | 289.0718 | C15H13O6 | −6.57 | |
| 10A | Ferulic acid-3-O-glucoside | 7.204 | 355.1049 | 355.1035 | C16H20O9 | −3.94 | [48] |
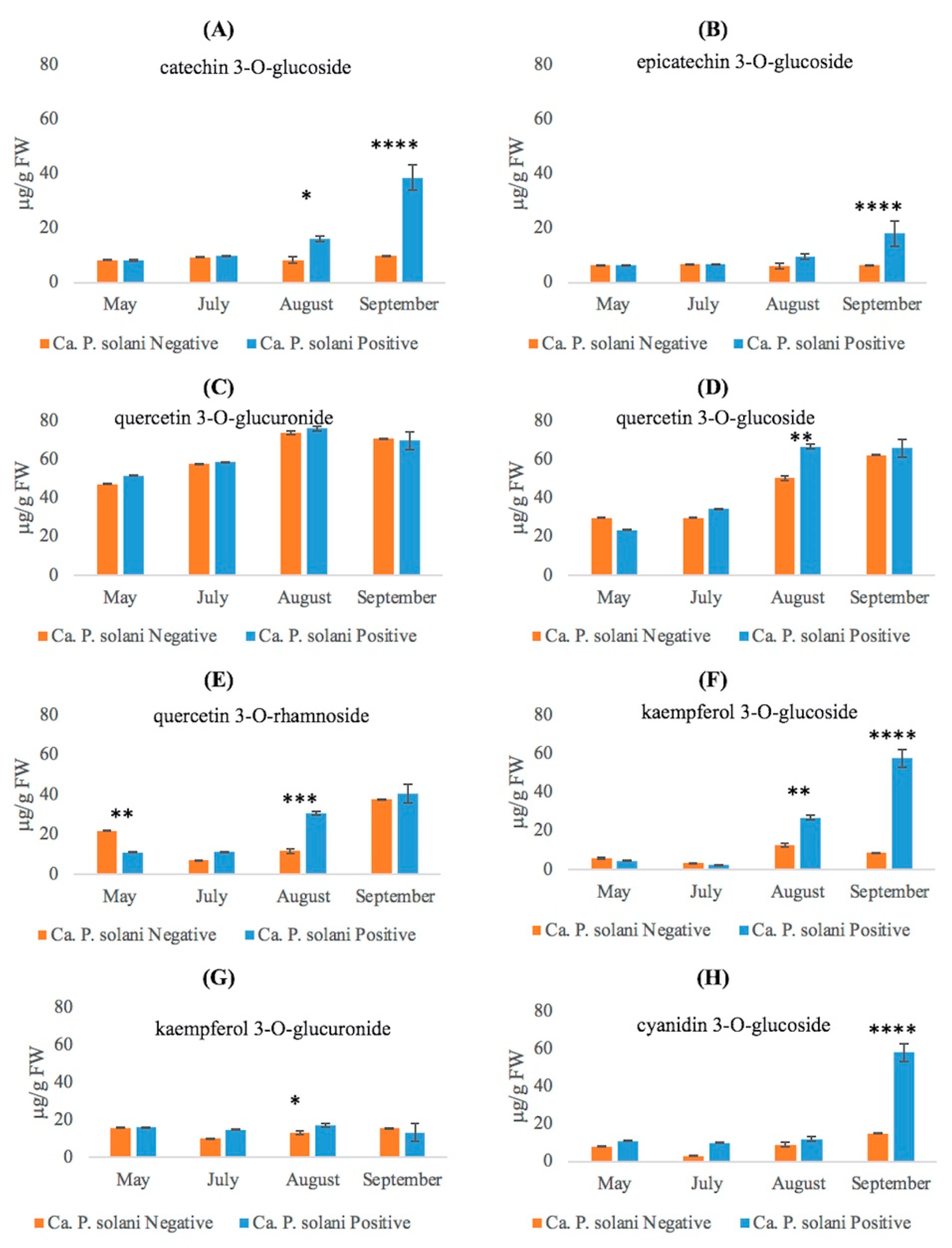
| 11A | Catechin-3-O-glucoside | 7.253 | 451.1324 | 451.1318 | C21H23O11 | −1.33 | [43] |
| 12A | Myricetin-3-O-glucuronide | 8.796 | 493.0640 | 493.0624 | C21H17O14 | −3.24 | [46] |
| 13A | Epicatechin- 3-O-glucoside | 8.819 | 451.1278 | 451.1318 | C21H23O11 | 8.86 | [43] |
| 14A | Myricetin-3-O-glucoside | 8.845 | 479.0842 | 479.0831 | C21H19O13 | −2.29 | [45] |
| 15A | Salicylic acid * | 9.426 | 137.0248 | 137.0244 | C7H5O3 | −3.04 | |
| 16A | Quercetin-3-O-glucoside * | 9.488 | 463.0908 | 463.0882 | C21H19O12 | −5.61 | [46] |
| 17A | Quercetin 3-O-glucuronide | 9.940 | 477.0726 | 477.0675 | C21H17O13 | −10.69 | [46] |
| 18A | Kaempferol 3-O-glucoside * | 10.438 | 447.0960 | 447.0933 | C21H19O11 | −6.03 | [46] |
| 19A | Kaempferol 3-O-rutinoside | 10.637 | 593.1513 | 593.1512 | C27H29O15 | −0.16 | [47] |
| 20A | Kaempferol 3-O-glucuronide | 10.808 | 461.0750 | 461.0725 | C21H17O12 | −5.42 | [46] |
| 21A | Quercetin 3-O-rhamnoside | 10.819 | 447.0968 | 447.0933 | C21H19O11 | −7.82 | [47] |
| 22A | Resveratrol 3-O-glucoside * | 11.167 | 389.1253 | 389.1242 | C20H21O8 | −2.82 | [47] |
| No. | Compound | RT a (min) | m/z exp. b | m/z calc. c | (M-H)+ | Error (ppm) | Reference |
| 23A | Delphinidin 3-O-glucoside * | 8.357 | 465.1033 | 465.1028 | C21H21O12 | −1.07 | [49] |
| 24A | Cyanidin 3-O-glucoside * | 9.637 | 449.1114 | 449.1078 | C21H21O11 | −8.01 | [49] |
| 25A | Peonidin 3-O-glucoside * | 12.241 | 463.1253 | 463.1235 | C22H23O11 | −3.88 | [49] |
2.2.2. Identification of Phenylpropanoid Compounds
2.3. Phenolic Compounds Accumulate in ‘Ca. P. solani’-Positive Plants
2.4. Expression Analysis
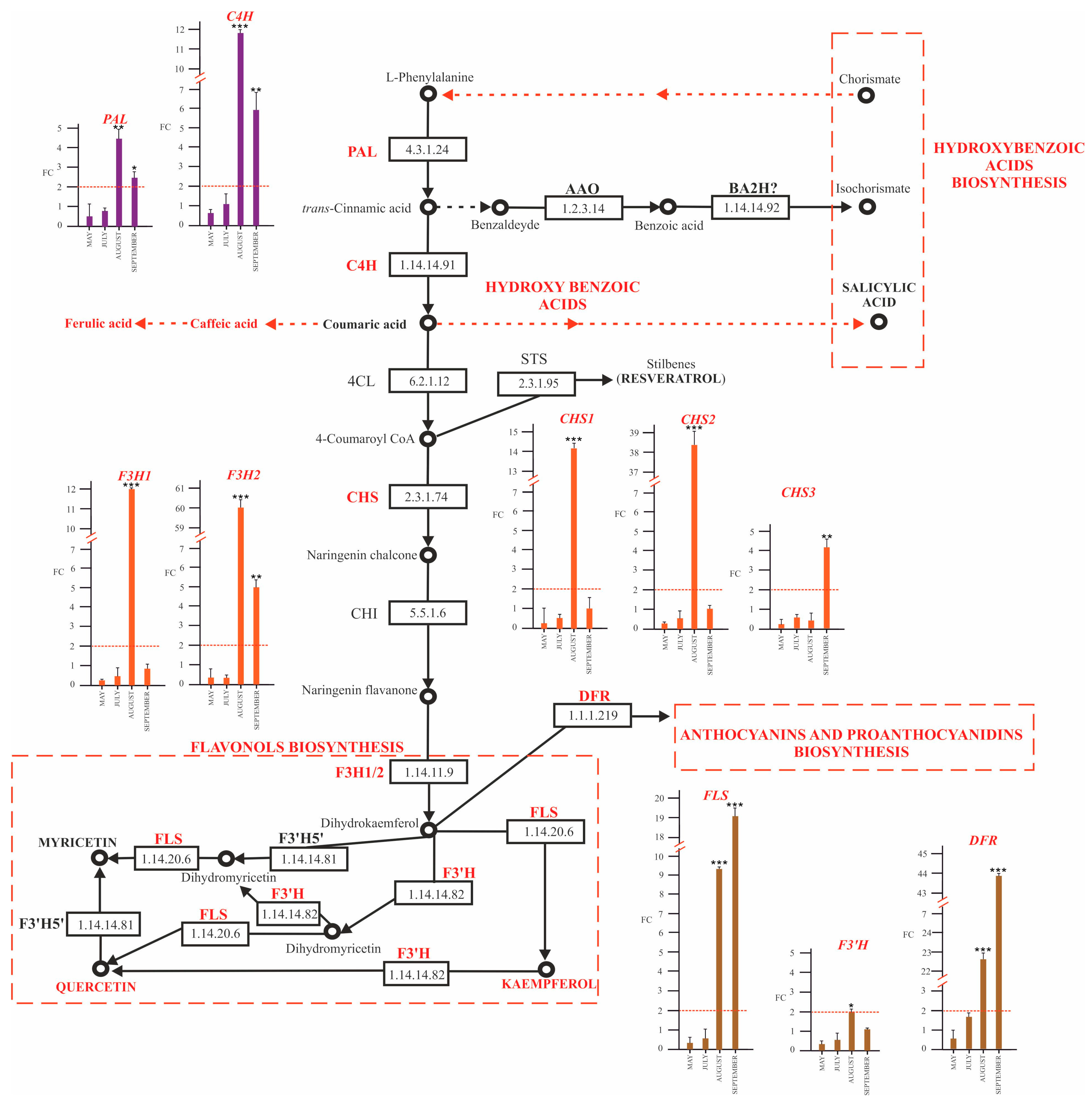
2.4.1. Modulation of Phenylpropanoid Biosynthetic Pathway Genes
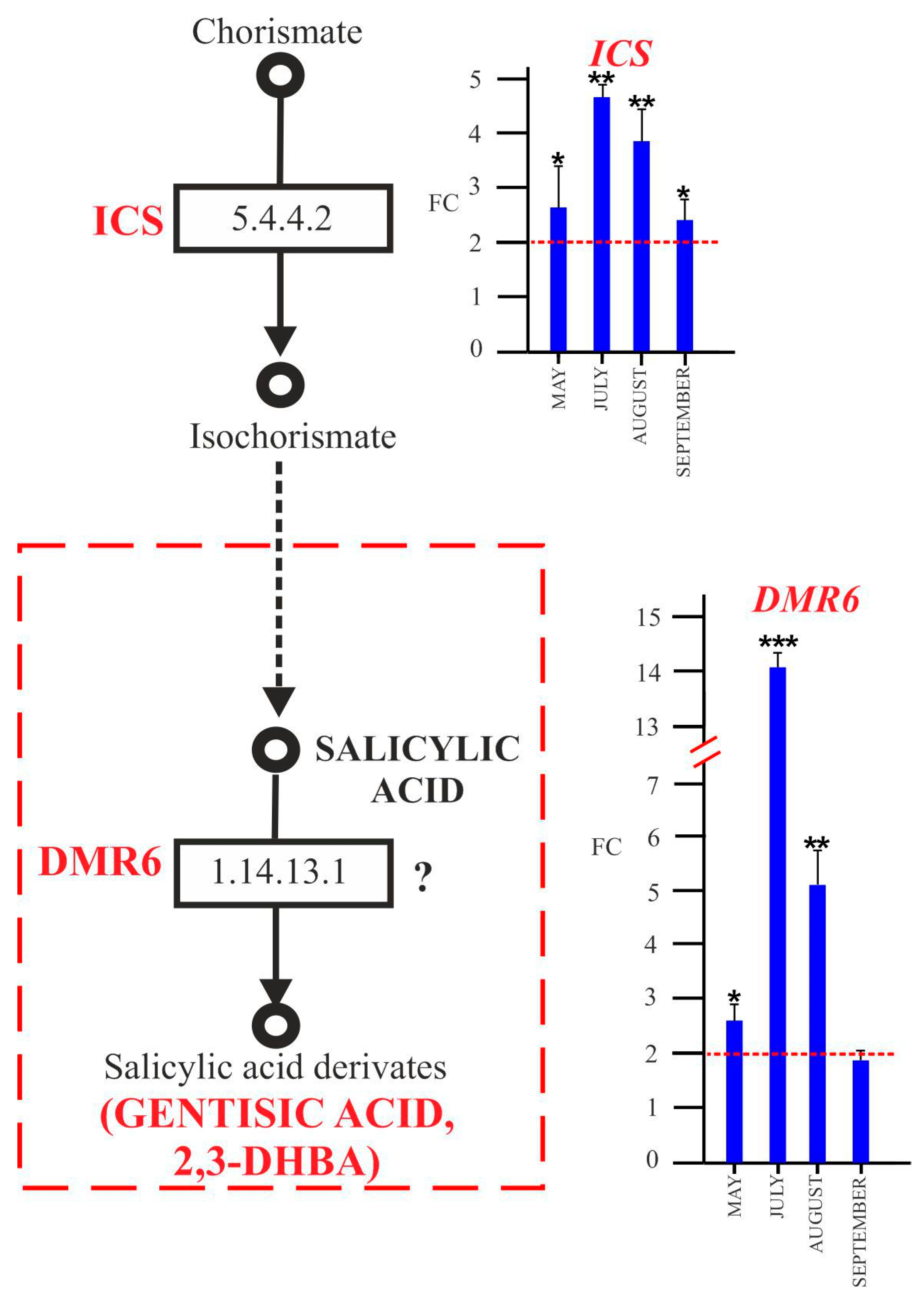
2.4.2. SA and Biosynthesis of Its Derivatives
2.4.3. Expression Patterns of SA-Dependent Defense-Related Genes
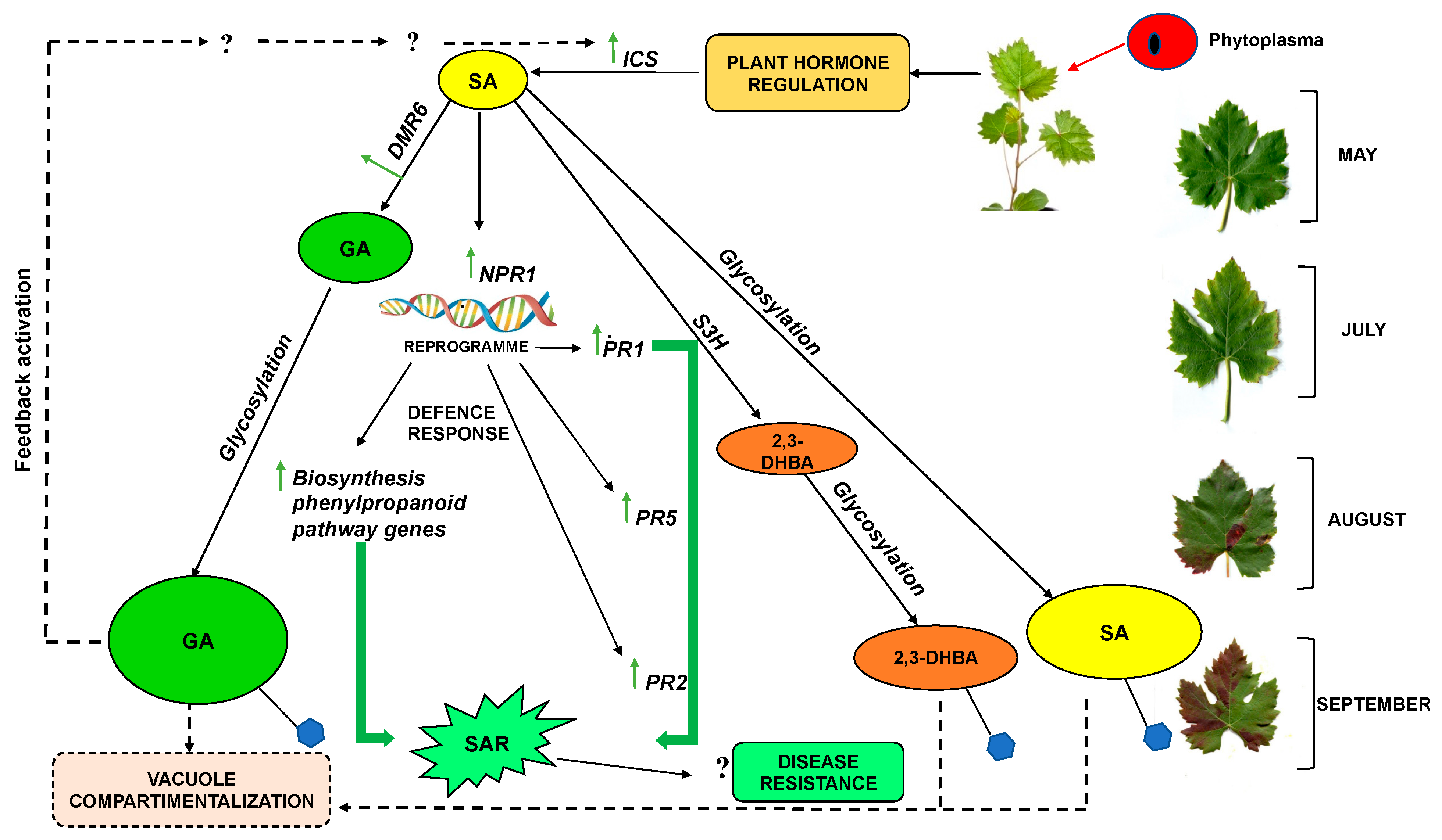
3. Discussion
4. Materials and Methods
4.1. Plant Samples and Phytoplasma Detection
4.2. Extraction of Phenylpropanoids
4.3. HPLC ESI/MS-TOF Analysis
4.4. RNA Extraction and qRT-PCR
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hogenhout, S.A.; Oshima, K.; Ammar, E.D.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Maejima, K.; Oshima, K.; Namba, S. Exploring the phytoplasmas, plant pathogenic bacteria. J. Gen. Plant Pathol. 2014, 80, 210–221. [Google Scholar] [CrossRef]
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, E.R. ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur and Bois noir-related diseases of plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2879–2894. [Google Scholar] [CrossRef]
- Starý, M.; Válová, P.; Šafářová, D.; Lauterer, P.; Ackermann, P.; Navrátil, M. Survey and molecular detection of Bois noir in vineyards of the Czech Republic. Hortic. Sci. 2013, 40, 83–87. [Google Scholar] [CrossRef]
- Belli, G.; Bianco, P.A.; Conti, M. Grapevine yellows in Italy: Past, present and future. J. Plant Pathology 2010, 92, 303–326. [Google Scholar]
- Cvrkovic, T.; Jovic, J.; Mitrovic, M.; Krstic, O.; Tosevski, I. Experimental and molecular evidence of Reptalus panzer as a natural vector of Bois noir. Plant Pathol. 2014, 63, 42–53. [Google Scholar] [CrossRef]
- Bertamini, M.; Nedunchezhian, N.; Tomasi, F.; Grando, M.S. Phytoplasma Stolbur-subgroup (Bois noir-BN) infection inhibits photosynthetic pigments, ribulose-1,5-bisphosphate carboxylase and photosynthetic activities in field grown grapevine (Vitis vinifera L. cv. Chardonnay) leaves. Physiol. Mol. Plant Pathol. 2002, 61, 357–366. [Google Scholar] [CrossRef]
- Rusjan, D.; Halbawirth, H.; Stich, K.; Mikulic-Petrovsek, M.; Veberic, R. Biochemical response of grapevine variety ‘Chardonnay (Vitis vinifera L.) to infection with grapevine yellows (Bois noir). Eur. J. Plant Pathol. 2012, 134, 231–237. [Google Scholar] [CrossRef]
- Maclean, A.M.; Sugio, A.; Makarova, O.V.; Findlay, K.C.; Grieve, V.M.; Toth, R.; Nicolaisen, M.; Hogenhout, S.A. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 2011, 157, 831–841. [Google Scholar] [CrossRef]
- Minato, N.; Himeno, M.; Hoshi, A.; Maejima, K.; Komatsu, K.; Takebayashi, Y.; Kasahara, H.; Yusa, A.; Oshima, K.; Kamiya, Y.; et al. The phytoplasmal virulence factor TENGU causes plant sterility by downregulating of the jasmonic acid and auxin pathways. Sci. Rep. 2014, 4, 7399. [Google Scholar] [CrossRef]
- Sugio, A.; Maclean, A.M.; Hogenhout, S.A. The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN-TCP binding and destabilization. New Phytol. 2014, 202, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Boccacci, P.; Margaria, P.; Palmano, S.; Gribaudo, I. Hydrogen peroxide accumulation and transcriptional changes in Grapevines recovered from Flavescence Dorée disease. Phytopathology 2013, 103, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Landi, L.; Romanazzi, G. Seasonal variation of defense-related gene expression in leaves from Bois noir affected and recovered grapevines. J. Agr. Food Chem. 2011, 59, 6628–6637. [Google Scholar] [CrossRef] [PubMed]
- Mou, H.Q.; Lu, J.; Zhu, S.F.; Lin, C.L.; Tian, G.Z.; Xu, X.; Zhao, W.J. Transcriptomic analysis of Paulownia infected by Paulownia witches’-broom Phytoplasma. PLoS ONE 2013, 8, e77217. [Google Scholar] [CrossRef]
- Margaria, P.; Ferrandino, A.; Caciagli, P.; Kedrina, O.; Schubert, A.; Palmano, S. Metabolic and transcript analysis of the flavonoid pathway in diseased and recovered Nebbiolo and Barbera grapevines (Vitis vinifera L.) following infection by Flavescence dorée phytoplasma. Plant Cell Environ. 2014, 37, 2183–2200. [Google Scholar] [CrossRef]
- Dixon, R.; Achnine, L.; Kota, P.; Liu, C.J.; Srinivasa Reddy, M.S. The phenylpropanoid pathway and plant defence. A genomics prospective. Mol. Mol. Plant Pathol. Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Sung, Y.C.; Lin, C.P.; Hsu, H.J.; Chen, Y.L.; Chen, J.C. Silencing of CrNPR1 and CrNPR3 a Alters plant susceptibility to periwinkle leaf yellowing phytoplasma. Front. Plant Sci. 2019, 10, 1183. [Google Scholar] [CrossRef]
- Patui, S.; Bertolini, A.; Clincon, L.; Ermacora, P.; Braidot, E.; Vianello, A.; Zancani, M. Involvement of plasma membrane peroxidases and oxylipin pathway in the recovery from phytoplasma disease in apple (Malus domestica). Physiol. Plant 2013, 148, 200–213. [Google Scholar] [CrossRef]
- Ahmad, J.N.; Eveillard, S. Study of the expression of defense related protein genes in stolbur C and stolbur PO phytoplasma-infected tomato. Bull. Insectol. 2011, 64, S159–S160. [Google Scholar]
- Punelli, F.; Al Hassan, M.; Fileccia, V.; Uva, P.; Pasquini, G.; Martinelli, F. A microarray analysis highlights the role of tetrapyrrole pathways in grapevine responses to “stolbur” phytoplasma, phloem virus infections and recovered status. Physiol. Mol. Plant Pathol. 2016, 93, 129–137. [Google Scholar] [CrossRef]
- Gondor, O.K.; Janda, T.; Soós, V.; Pál, M.; Majláth, I.; Adak, M.K.; Balázs, E.; Szalai, G. Salicylic Acid Induction of Flavonoid Biosynthesis Pathways in Wheat Varies by Treatment. Front. Plant Sci. 2016, 7, 1447. [Google Scholar] [CrossRef] [PubMed]
- Hren, M.; Nikolic, P.; Rotter, P.; Blejec, A.; Terrier, N.; Ravnikar, M.; Dermastia, M.; Gruden, K. ‘Bois noir’ phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genom. 2009, 10, 460. [Google Scholar] [CrossRef] [PubMed]
- Albertazzi, G.; Milc, J.; Caffagni, A.; Francia, E.; Roncaglia, E.; Ferrari, F.; Tagliafico, E.; Stefani, E.; Pecchioni, N. Gene expression in grapevine cultivars in response to Bois Noir phytoplasma infection. Plant Sci. 2009, 176, 792–804. [Google Scholar] [CrossRef]
- Dermastia, M.; Nikolic, P.; Chersicola, M.; Gruden, K. Transcriptional profiling in infected and recovered grapevine plant response to “Candidatus phytoplasma solani”. Phytopath. Moll. 2015, 5, S123. [Google Scholar] [CrossRef]
- Paolacci, A.R.; Catarcione, G.; Ederli, L.; Zadra, C.; Pasqualini, S.; Badiani, M.; Musetti, R.; Santi, S.; Ciaffi, M. Jasmonate-mediated defence responses, unlike salicylate-mediated responses, are involved in the recovery of grapevine from Bois noir disease. BMC Plant Biol. 2017, 17, 118. [Google Scholar] [CrossRef]
- Frías, M.; Brito, N.; González, C. The Botrytis cinerea cerato-platanin BcSpl1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol. Plant Pathol. 2013, 14, 191–196. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance turning local infection into global defense. Ann. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Conrath, U. Systemic acquired resistance. Plant Sign. Behav. 2006, 1, 179–184. [Google Scholar] [CrossRef]
- Dean, J.V.; Delaney, S.P. Metabolism of salicylic acid in wild- type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 2008, 132, 417–425. [Google Scholar] [CrossRef]
- Bartsch, M.; Bednarek, P.; Vivancos, P.D.; Schneider, B.; von Roepenack-Lahaye Foyer, C.H.; Kombrink, E.; Scheel, D.; Parker, J.E. Accumulation of isochorismate-derived 2,3-dihydroxybenzoic 3-O-beta-D-xyloside in Arabidopsis resistance to pathogens and ageing of leaves. J. Biol. Chem. 2010, 285, 25654–25665. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.A.; Klessig, D.F. How does the multifaced plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol. 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Abedi, F.; Razavi, B.M.; Hosseinzadeh, H. A review on gentisic acid as a plant derived phenolic and metabolite of aspirin: Comprehensive pharmacology, toxicology, and some pharmaceutical aspects. Phytother. Res. 2019, 34, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Prezelj, N.; Fragner, L.; Weckwerth, W.; Dermastia, M. Metabolic consequencea of infection of graopevine (Vitis vinifera L.) cv. “Modra frankinja” with Flavescence Dorée phytoplasma. Front. Plant Sci. 2016, 7, 711. [Google Scholar] [CrossRef] [PubMed]
- Rotter, A.; Nikolić, P.; Turnšec, N.; Kogovšek, P.; Blejec, A.; Gruden, K.; Dermastia, M. Statistical modeling of long-term grapevine response to ‘Candidatus Phytoplasma solani’ infection in the field. Eur. J. Plant Physiol. 2018, 150, 653–668. [Google Scholar] [CrossRef]
- Dermastia, M. Plant hormones in Phytoplasma Infected Plants. Front. Plant Sci. 2019, 10, 477. [Google Scholar] [CrossRef]
- Zhang, K.; Halitschke, R.; Yin, C.; Liu, C.; Gan, S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812. [Google Scholar] [CrossRef]
- Negro, C.; Sabella, E.; Nicolì, F.; Pierro, R.; Materazzi, A.; Panattoni, A.; Aprile, A.; Nutricati, E.; Vergine, M.; Miceli, A. Biochemical changes in leaves of Vitis vinifera cv. Sangiovese infected by Bois noir phytoplasma. Pathogens 2020, 9, 269. [Google Scholar] [CrossRef]
- Pierro, R.; Passera, A.; Panattoni, A.; Casati, P.; Luvisi, A.; Rizzo, D.; Bianco, P.A.; Quaglino, F.; Materazzi, A. Molecular typing of ‘Bois noir’ phytoplasma strains in the Chianti Classico area (Tuscany, Central Italy) and their association with symptom severity in Vitis vinifera L. cv. Sangiovese. Phytopathology 2018, 108, 362–373. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.S.; Liu, C.J.; Zhang, K. S5/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, G.; Liu, Q.; Chen, L.; Li, Y.; Hou, B. Modulation of plant salicylic acid-associated immune responses via glycosylation of Dihydroxybenzoic acids. Plant Physiol. 2018, 176, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defence Hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Perestrelo, R.; Lu, Y.; Santos, S.A.O.; Silvestre, A.J.D.; Neto, C.P.; Camara, J.S.; Rocha, S.M. Phenolic profile of Sercial and Tinta Negra Vitis vinifera L. grape skins by HPLC–DAD–ESI-MS novel phenolic compounds in Vitis vinifera L. grape. Food Chem. 2012, 135, 94–104. [Google Scholar] [CrossRef]
- Fang, N.; Yu, S.; Prior, R.L. LC/MS/MS characterization of phenolic constituents in dried plums. J. Agric. Food Chem. 2002, 50, 3579–3585. [Google Scholar] [CrossRef]
- Sun, J.; Liang, F.; Bin, Y.; Li, P.; Duan, C. Screening non-colored phenolics in red wines using liquid chromatography/ultraviolet and mass spectrometry/mass spectrometry libraries. Molecules 2007, 12, 679–693. [Google Scholar] [CrossRef]
- Goufo, P.; Kumar Singh, R.; Cortez, I. A reference list of phenolic compounds (including stilbenes) in grapevine (Vitis vinifera L.) roots, woods, canes, stems, and leaves. Antioxidants 2020, 9, 398. [Google Scholar] [CrossRef]
- Sandhu, A.K.; Gu, L. Antioxidant capacity, phenolic content, and profiling of phenolic compounds in the seeds, skin, and pulp of Vitis rotundifolia (Muscadine Grapes) as determined by HPLC-DAD-ESI-MS. J. Agric. Food Chem. 2010, 58, 4681–4692. [Google Scholar] [CrossRef]
- Tang, J.; Dunshea, F.R.; Suleria, H.A.R. LC_ESI_QTOF/MS characterization of phenolic compounds from medicinal plants (Hops and Juniper Berries) and their antioxidant activity. Foods 2019, 9, 7. [Google Scholar] [CrossRef]
- Lopes-Lutz, D.; Dettmann, J.; Nimalaratne, C.; Schieber, A. Characterization and quantification of polyfenols in Amazon grape (Pourouma cecropiifolia Martius). Molecules 2010, 15, 8543–8552. [Google Scholar] [CrossRef]
- Belles, J.M.; Garro, R.; Fayos, J.; Navarro, P.; Primo, J.; Conejero, V. Gentisic acid as pathogen-inducible signal, additional to salicylic acid for activation of plant defenses in tomato. MPMI 1999, 12, 227–235. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Vlot, A.C.; Wildrmuth, M.C.; Klessig, D.F. Salicylic acid biosynthesis and metabolism. Arab. Book 2011, 9, e0156. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Palmano, S. Response of Vitis vinifera L. cv. ’Nebbiolo’ proteome to Flavescence dorée phytoplasma infection. Proteomics 2011, 11, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Abbà, S.; Palmano, S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phosphor-proteomic approach with data integration into functional network. BMC Genom. 2013, 14, 38. [Google Scholar] [CrossRef]
- López-Gresa, M.P.; Lisón, P.; Yenush, L.; Conejero, V.; Rodrigo, I.; Bellés, J.M. Salicylic acid is involved in the basal resistance of Tomato plants to Citrus Exocortis virois and Tomato Spotted Wilt virus. PLoS ONE 2016, 11, e0166938. [Google Scholar] [CrossRef] [PubMed]
- Fayos, J.; Belles, J.M.; López-Gresa, M.P.; Jaime Primo, J.; Vicente Conejero, V. Induction of gentisic acid 5-O-B-xylopyranoside in tomato and cucumber plant infected by different pathogens. Phytochemistry 2006, 67, 142–148. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef]
- Jones, P.; Vogt, T. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 2001, 213, 164–174. [Google Scholar] [CrossRef]
- Bellés, J.M.; Garro, R.; Pallaás, V.; Fayos, J.; Rodrigo, I.; Conejero, V. Accumulation of gentisic acid as associated with systemic infections but not with the hypersensitive response in plant-pathogen interactions. Planta 2006, 223, 500–511. [Google Scholar] [CrossRef]
- Campos, L.; Lisón, P.; López-Gresa, M.P.; Rodrigo, I.; Zacarés, L.; Conejero, V. Transgenic tomato plants overexpressing tyramine N-hydroxycinnamoyl transferase exhibit elevated hydroxycinnamic acid amide levels and enhanced resistance to Pseudomonas syringae. MPMI 2014, 27, 1159–1169. [Google Scholar] [CrossRef]
- Chen, H.Y.; Li, X. Identification of a residue responsible for UDP-sugar donor selectivity of a dihydroxybenzoic acid glycosyltransferase from Arabidopsis natural accessions. Plant J. 2017, 89, 195–203. [Google Scholar] [CrossRef]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic acid biosynthesis in plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef] [PubMed]
- de Toledo Thomazella, D.P.; Brail, Q.; Douglas, D.; Staskawicz, B. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv 2016, 064824. [Google Scholar] [CrossRef]
- Shigenaga, A.M.; Argueso, C.T. No hormone to rule them all: Interactions of plant hormones during the responses of plant to pathogens. J. Semin. Cell Dev. Biol. 2016, 56, 174–189. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Li, Q.; Zhao, Y.; Wie, H.; Wang, J.; Baker, C.J. Integration of metabolomics and existing omics data reveals new insights into phytoplasma-induced metabolic reprogramming in host plants. PLoS ONE 2021, 16, e0246203. [Google Scholar] [CrossRef]
- Rusjan, D.; Veberic, R.; Mikukic-Petkovsek, M.M. The response of phenolic compounds in grapes of the variety ‘Chardonnay’ (Vitis vinifera L.) to the infection by phytoplasma Bois noir. Eur. J. Plant Pathol. 2012, 133, 965–974. [Google Scholar] [CrossRef]
- Slatnar, A.; Mikulic Petkovsek, M.; Halbwirth, H.; Stampar, F.; Stich, K.; Veberic, R. Response of the phenylpropanoid pathway to Venturia inaequalis infection in maturing fruit of ‘Braeburn’ apple. J. Hortic. Sci. Biotechnol. 2010, 85, 465–472. [Google Scholar] [CrossRef]
- Geny, L.; Darrieumerlou, A.; Doneche, B. Conjugate polyamines and hydroxycinnamic acids in grape berries during Botrytis cinerea disease development: Differences between ‘noble rot’ and ‘grey mould’. Aust. J. Grape Wine Res. 2003, 9, 102–106. [Google Scholar] [CrossRef]
- Rusjan, D.; Petkovsek, M.M. Phenolic Responses in 1-Year-Old Canes of Vitis Vinifera cv. Chardonnay Induced by Grapevine Yellows (Bois noir); Australian Society of Viticulture and Oenology Inc.: Glen Osmond, Australia, 2014; pp. 123–134. [Google Scholar] [CrossRef]
- Dixon, R.A.; Xie, D.Y.; Sharma, S.B. Proanthocyanidins—A final frontier in flavonoid research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef]
- Mellway, R.D.; Tran, L.T.; Prouse, M.B.; Campbell, M.M.; Constabel, C.P. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 2009, 150, 924–941. [Google Scholar] [CrossRef]
- Ullah, C.; Unsicker, S.B.; Fellenberg, C.; Constabel, C.P.; Schmidt, A.; Gershenzon, J.; Hammerbacher, A. Flavan-3-ols are an effective chemical defense against rust infection. Plant Physiol. 2017, 175, 1560–1578. [Google Scholar] [CrossRef]
- Ullah, C.; Unsicker, S.B.; Reichet, M.; Gershenzon, J.; Hammerbacher, A. Accumulation of Catechin and Proanthocyanidins in Black Poplar Stems After Infection by Plectosphaerella populi: Hormonal Regulation, Biosynthesis and Antifungal Activity. Front. Plant Sci. 2019, 10, 1441. [Google Scholar] [CrossRef] [PubMed]
- Junqueira, A.; Bedendo, I.; Pascholati, S. Biochemical changes in corn plants infected by the maize bushy stunt phytoplasma. Physiol. Mol. Plant Pathol. 2004, 65, 181–185. [Google Scholar] [CrossRef]
- Gutha, L.R.; Casassa, L.F.; Harbertson, F.; Naidu, R.A. Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biol. 2010, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Nicoliì, F.; Negro, C.; Nutricati, E.; Vergine, M.; Aprile, A.; Sabella, E.; Damiano, G.; De Bellis, L.; Luvisi, A. Accumulation of azelaic acid in Xylella fastidiosa-infected olive trees: A mobile metabolite for health screening. Phytopathology 2019, 109, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Angelini, E.; Bianchi, G.I.; Filippini, L.; Morassutti, C.; Borgo, M. A new TaqMan method the identification of phytoplasma associated with grapevine yellows by real time PCR assay. J. Microbiol. Methods 2007, 68, 613–622. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Realtime RT-PCR (TaqMan®) assays for the detection of Grapevine leafroll associated viruses 1–5 and 9. J. Virol. Methods 2007, 141, 22–29. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Comparison of low-density arrays, RT-PCR and real-time TaqMan RT-PCR in detection of grapevine viruses. J. Virol. Methods 2008, 149, 292–299. [Google Scholar] [CrossRef]
- Wei, T.; Lebas, B.S.M.; Shiller, J.B.; Quinn, B.D.; Clover, G.R.G. Detection of five viruses infecting dormant bulbs by TaqMan-based real-time RT-PCR. Australas. Plant Path. 2012, 41, 93–98. [Google Scholar] [CrossRef]
- Vergine, M.; Nicolì, F.; Negro, C.; Luvisi, A.; Nutricati, E.; Accogli, R.A.; Sabella, E.; Miceli, A. Phytochemical profiles and antioxidant activity of Salvia species from Southern Italy. Rec. Nat. Prod. 2019, 13, 205–215. [Google Scholar] [CrossRef]
- Aprile, A.; Negro, C.; Sabella, E.; Luvisi, A.; Nicolì, F.; Nutricati, E.; Vergine, M.; Blando, F.; De Bellis, L. Antioxidant activity and anthocyanin contents in olives (cv Cellina di Nardò) during ripening and after fermentation. Antioxidants 2019, 8, 138. [Google Scholar] [CrossRef]
- Bertazzon, N.; Bagnaresi, P.; Forte, V.; Mazzucotelli, E.; Filippin, L.; Guerra, D.; Zechini, A.; Cattivelli, L.; Angelini, E. Grapevine comparative early transcriptomic profiling suggests that Flavescence dorée phytoplasma represses plant responses induced by vector feeding in susceptible varieties. BMC Genom. 2019, 20, 526. [Google Scholar] [CrossRef] [PubMed]
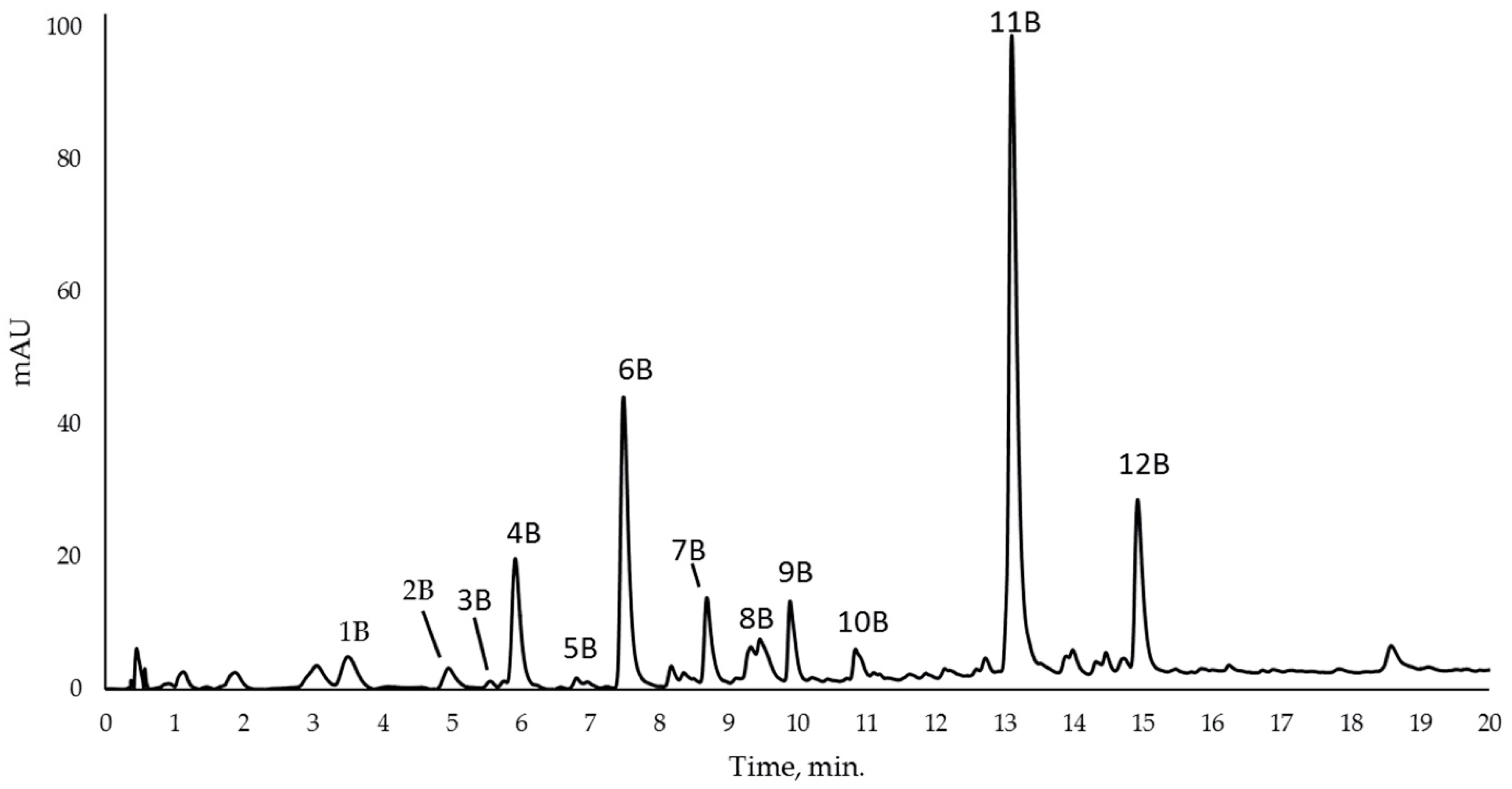
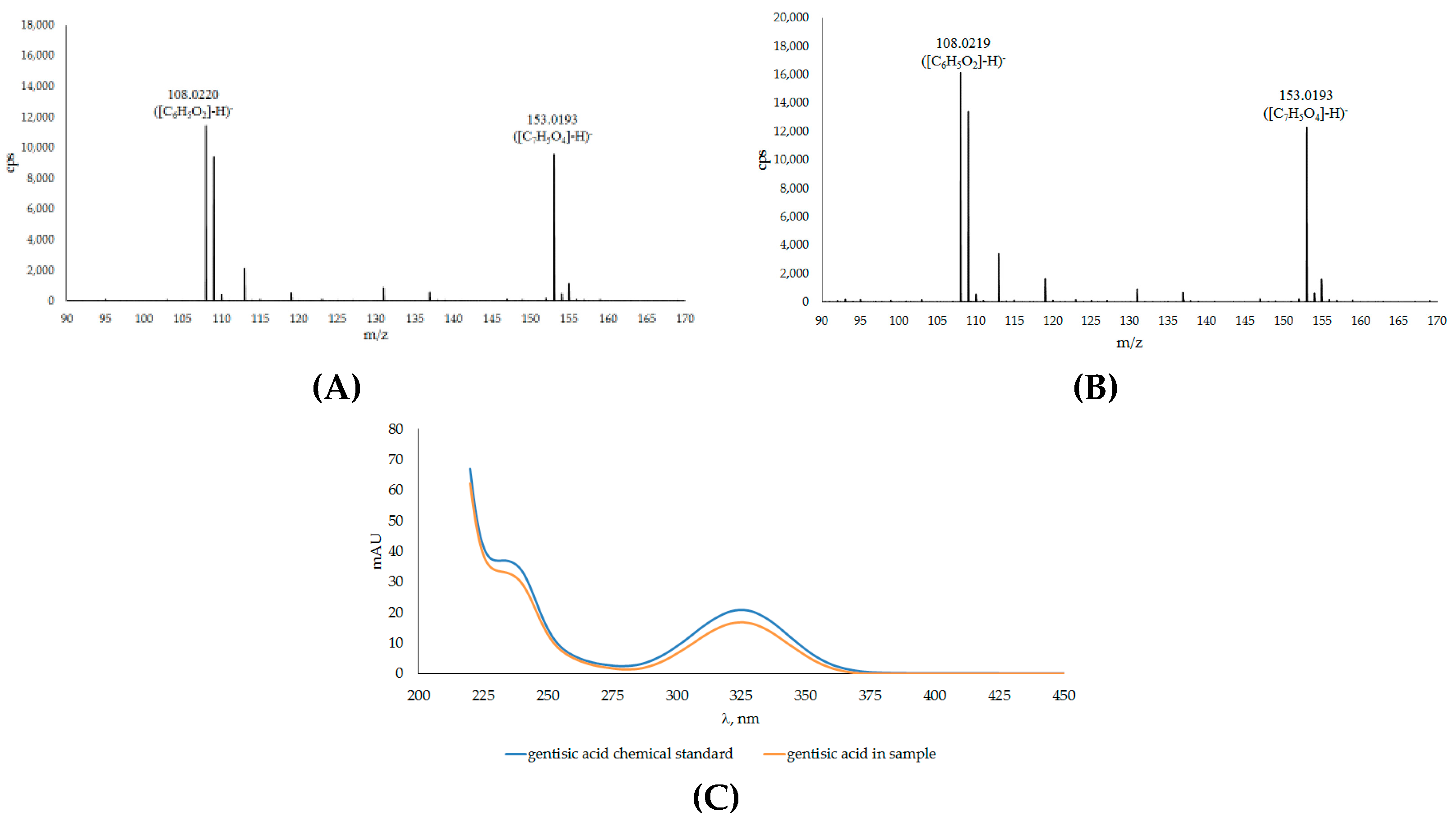
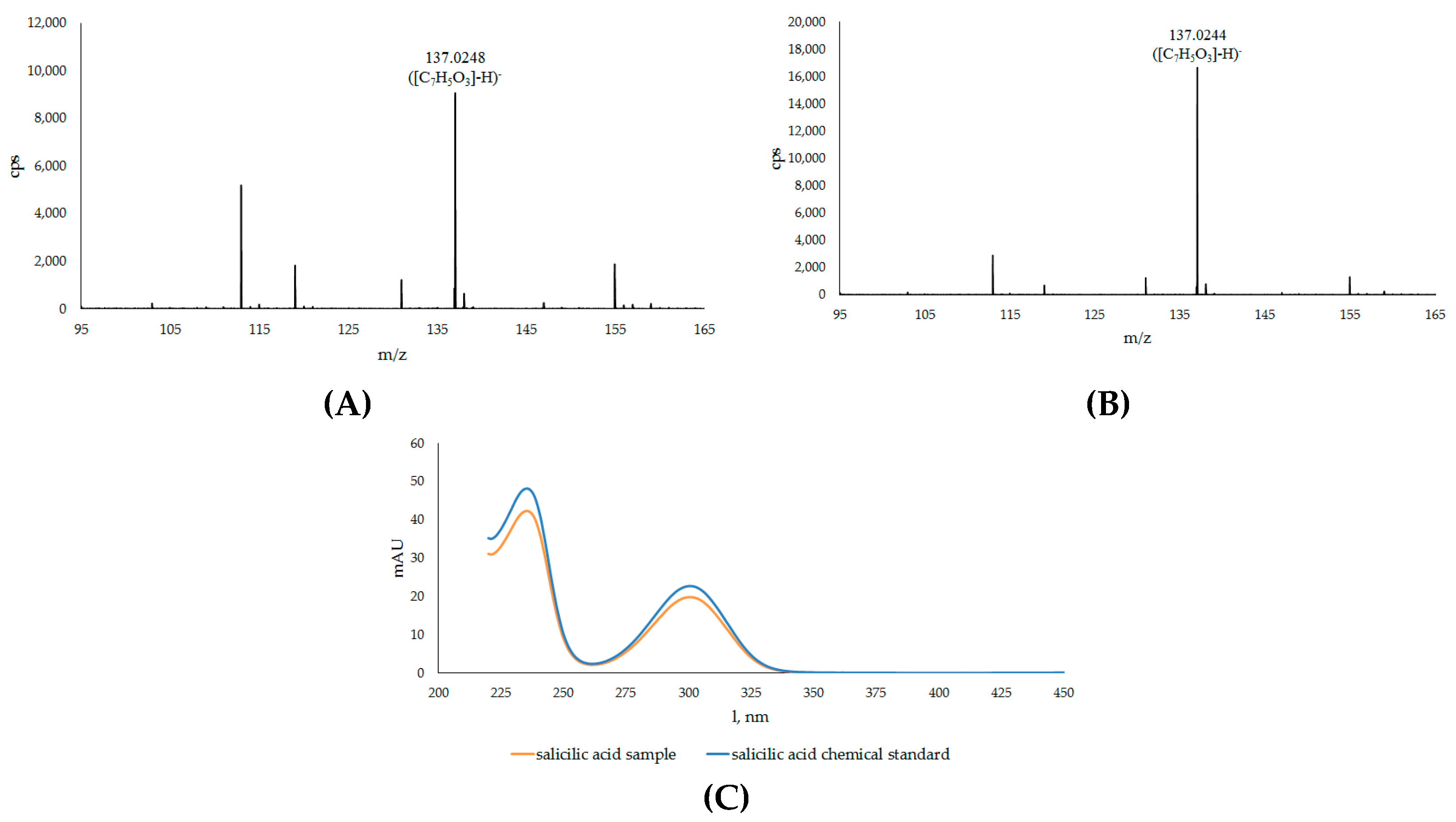




| No. | Compound | RT a (min) | m/z exp. b | m/z calc. c | (M-H)− | Error (ppm) |
|---|---|---|---|---|---|---|
| 1B | 2,5 Dihydroxybenzoic acid (Gentisic acid) | 3.711 | 153.0194 | 153.0193 | C7H5O4 | −0.65 |
| 2B | 2,3 Dihydroxybenzoic acid | 5.102 | 153.0196 | 153.0193 | C7H5O4 | −1.55 |
| 3B | Catechin | 5.881 | 289.0728 | 289.0718 | C15H13O6 | −3.45 |
| 4B | Methyl benzoate | 5.997 | 135.0468 | 135.0452 | C8H7O2 | −11.84 |
| 5B | Epicatechin | 6.874 | 289.0723 | 289.0718 | C15H13O6 | −1.72 |
| 6B | p-Coumaric acid | 7.785 | 163.0410 | 163.0401 | C9H7O3 | −5.52 |
| 7B | Ferulic acid | 8.845 | 193.0514 | 193.0506 | C10H9O4 | −4.14 |
| 8B | o-hydroxybenzoic acid (Salicylic acid) | 9.756 | 137.0259 | 137.0255 | C7H5O3 | −2.91 |
| 9B | Quercetin 3-O-glucuronide | 9.952 | 477.0653 | 477.0675 | C21H17O13 | 4.61 |
| 10B | Myricetin | 10.869 | 317.0316 | 317.0303 | C15H9O8 | −4.10 |
| 11B | Quercetin | 13.231 | 301.0355 | 301.0354 | C15H9O7 | −0.33 |
| 12B | Kaempferol | 15.023 | 285.0395 | 285.0404 | C15H9O6 | 3.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nutricati, E.; De Pascali, M.; Negro, C.; Bianco, P.A.; Quaglino, F.; Passera, A.; Pierro, R.; Marcone, C.; Panattoni, A.; Sabella, E.; et al. Signaling Cross-Talk between Salicylic and Gentisic Acid in the ‘Candidatus Phytoplasma Solani’ Interaction with Sangiovese Vines. Plants 2023, 12, 2695. https://doi.org/10.3390/plants12142695
Nutricati E, De Pascali M, Negro C, Bianco PA, Quaglino F, Passera A, Pierro R, Marcone C, Panattoni A, Sabella E, et al. Signaling Cross-Talk between Salicylic and Gentisic Acid in the ‘Candidatus Phytoplasma Solani’ Interaction with Sangiovese Vines. Plants. 2023; 12(14):2695. https://doi.org/10.3390/plants12142695
Chicago/Turabian StyleNutricati, Eliana, Mariarosaria De Pascali, Carmine Negro, Piero Attilio Bianco, Fabio Quaglino, Alessandro Passera, Roberto Pierro, Carmine Marcone, Alessandra Panattoni, Erika Sabella, and et al. 2023. "Signaling Cross-Talk between Salicylic and Gentisic Acid in the ‘Candidatus Phytoplasma Solani’ Interaction with Sangiovese Vines" Plants 12, no. 14: 2695. https://doi.org/10.3390/plants12142695
APA StyleNutricati, E., De Pascali, M., Negro, C., Bianco, P. A., Quaglino, F., Passera, A., Pierro, R., Marcone, C., Panattoni, A., Sabella, E., De Bellis, L., & Luvisi, A. (2023). Signaling Cross-Talk between Salicylic and Gentisic Acid in the ‘Candidatus Phytoplasma Solani’ Interaction with Sangiovese Vines. Plants, 12(14), 2695. https://doi.org/10.3390/plants12142695
















