The Levels of DAHP Synthase, the First Enzyme of the Shikimate Pathway, Are Related to Free Aromatic Amino Acids and Glutamine Content in Nicotiana plumbaginifolia Cell Cultures
Abstract
1. Introduction
2. Results
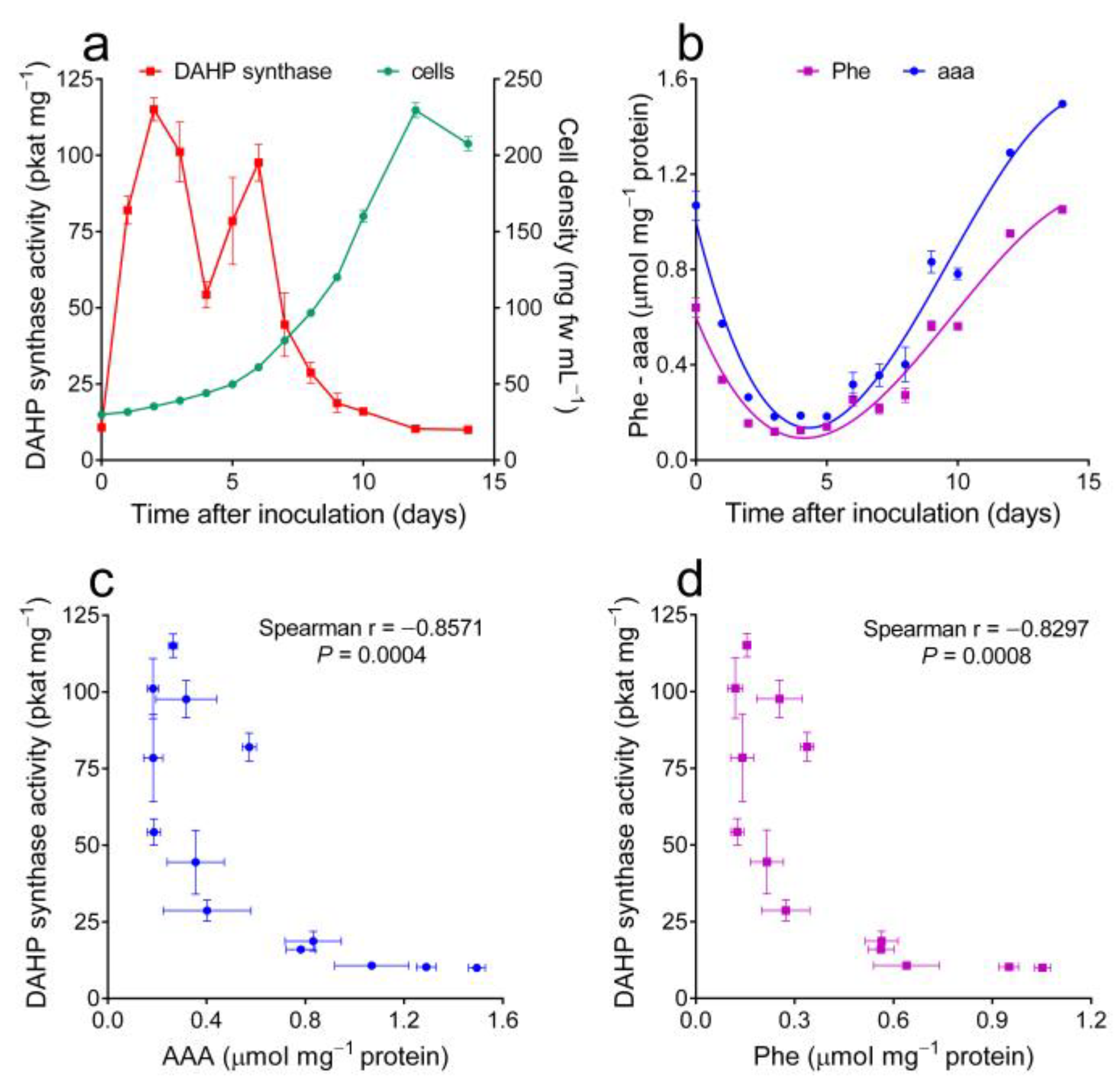
2.1. Concentration of Free Amino Acids in Wild Tobacco Cells Greatly Varies with the Culture Growth Stage, and Aromatic Amino Acid Contents Are Negatively Correlatde with DAHP Synthase Specific Activity Levels
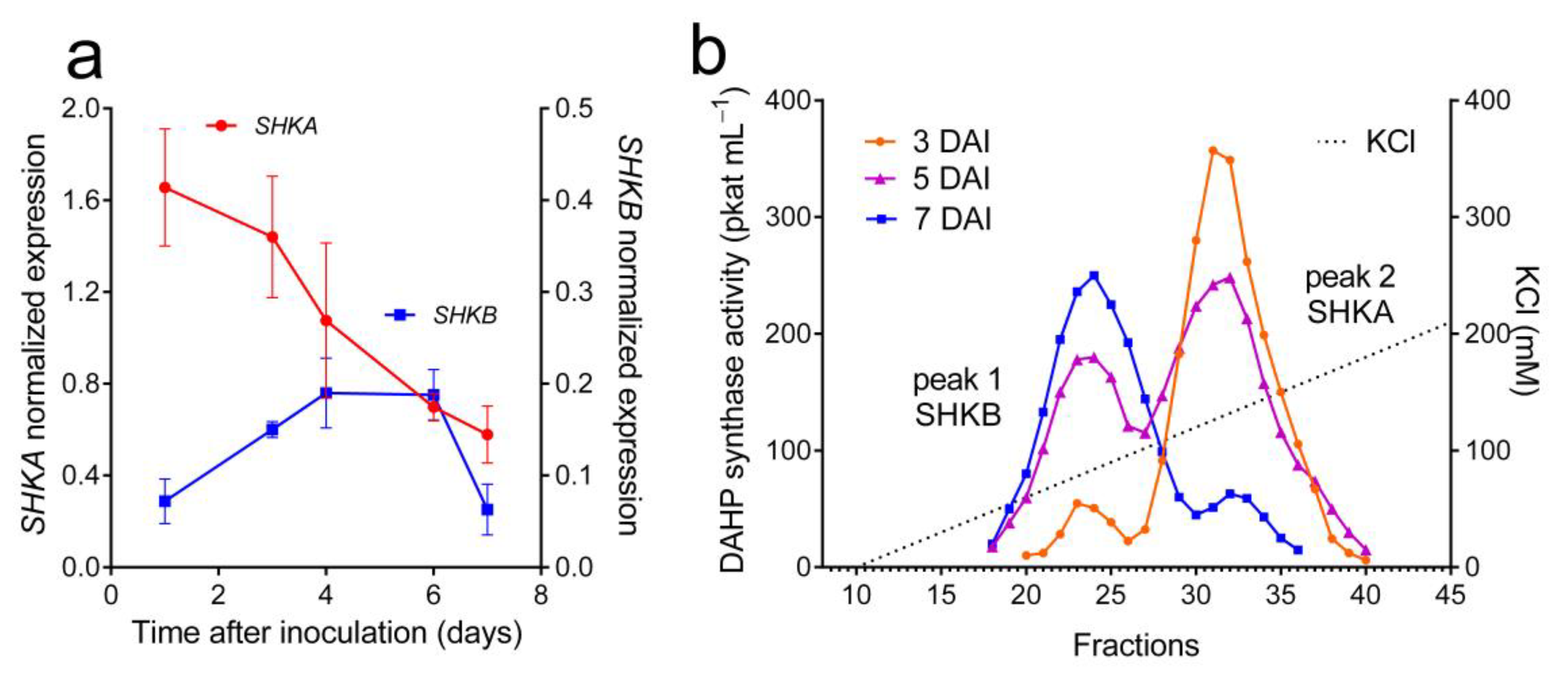
2.2. Two DAHP Synthase Isogenes Are Expressed in N. plumbaginifolia Cells, and the Corresponding Proteins Can Be Resolved through Anion-Exchange Liquid Chromatography
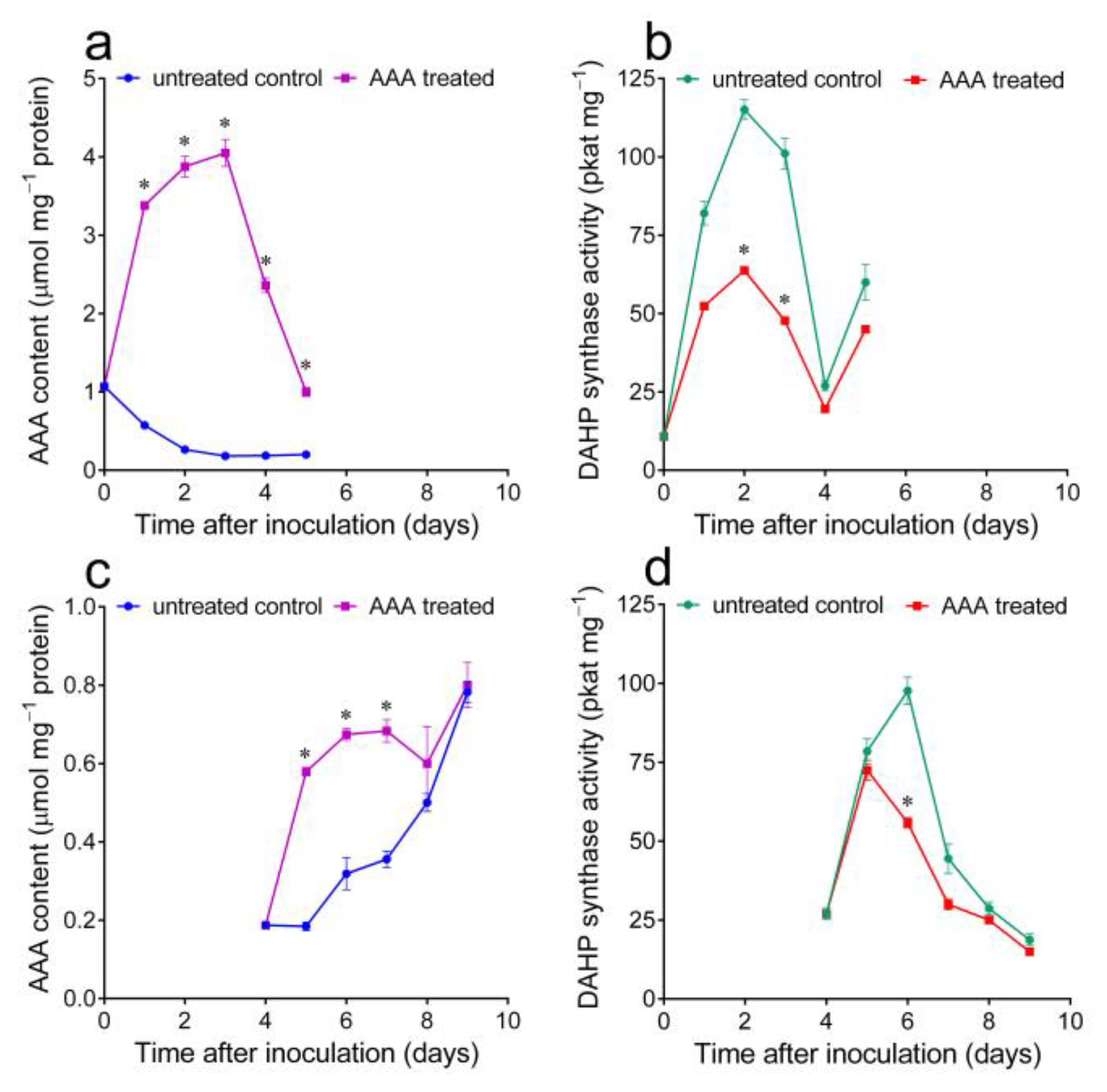
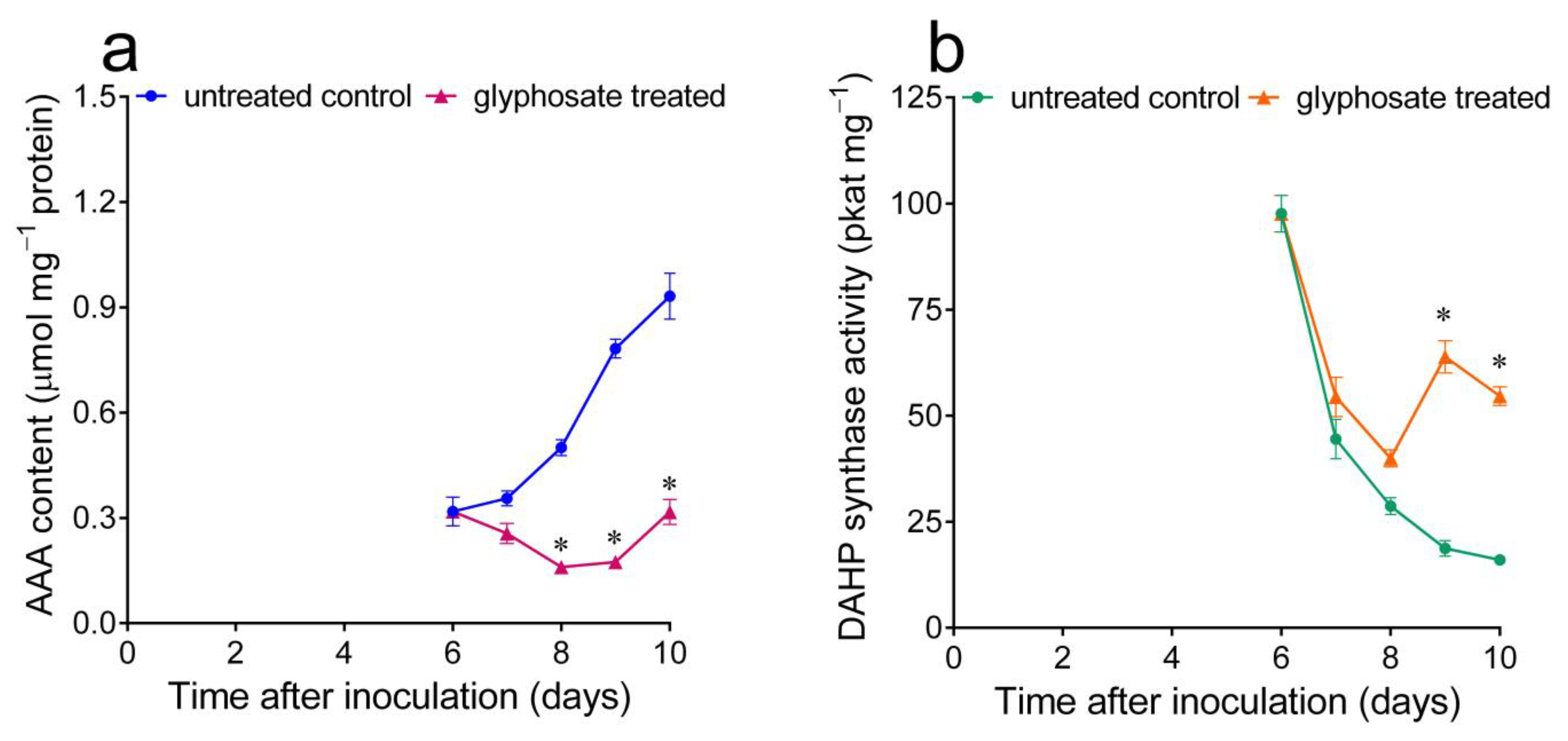
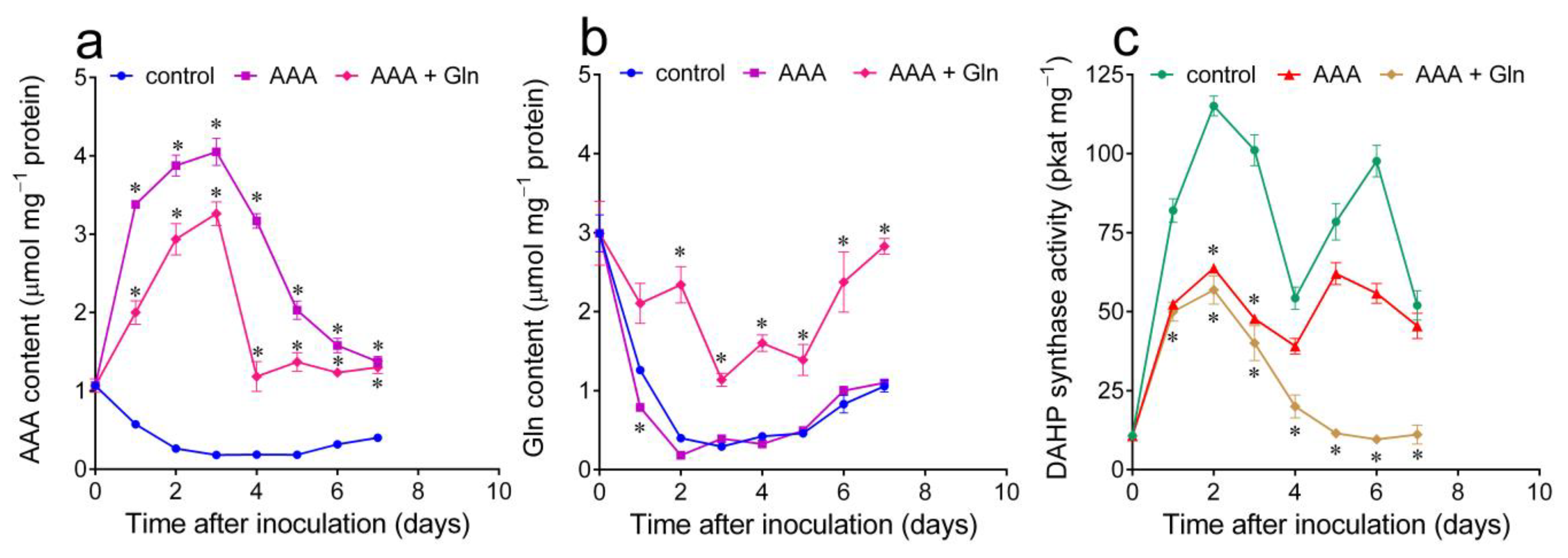
2.3. Variations in Intracellular Free Aromatic Amino Acid Pools, Induced by Treatment with Either Exogenous AAA or the Aromatic Synthesis Inhibitor Glyphosate, Cause the Consistent and Opposite Fluctuation of DAHP Synthase Specific Activity
2.4. The Intracellular Concentration of Free Glutamine Also Influences DAHP Synthase Levels in Actively Proliferating Cells
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Amino Acid Treatments
4.2. DAHP Synthase Extraction
4.3. DAHP Synthase Assay
4.4. Chromatographic Separation of DAHP Synthase Isoforms
4.5. Amino Acid Extraction, Separation and Quantification
4.6. PCR Cloning and RT-PCR Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bate, N.J.; Orr, J.; Ni, W.; Meromi, A.; Nadler-Hassar, T.; Doerner, P.W.; Dixon, R.A.; Lamb, C.J.; Elkind, Y. Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA 1994, 91, 7608–7612. [Google Scholar] [CrossRef] [PubMed]
- Gonzali, S.; Mazzucato, A.; Perata, P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci. 2009, 14, 237–241. [Google Scholar] [CrossRef]
- Henstrand, J.M.; McCue, K.F.; Brink, K.; Handa, A.K.; Herrmann, K.M.; Conn, E.E. Light and fungal elicitor induce 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase mRNA in suspension cultured cells of parsley (Petroselinum crispum L.). Plant Physiol. 1992, 98, 761–763. [Google Scholar] [CrossRef]
- Weaver, L.M.; Herrmann, K.M. Dynamics of the shikimate pathway in plants. Trends Plant Sci. 1997, 2, 346–351. [Google Scholar] [CrossRef]
- Herrmann, K.M.; Weaver, L.M. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Laitinen, R.A.; Ainasoja, M.; Broholm, S.K.; Teeri, T.H.; Elomaa, P. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. J. Exp. Bot. 2008, 59, 3691–3703. [Google Scholar] [CrossRef]
- Ahmad, S.; Rightmire, B.; Jensen, R.A. Evolution of the regulatory isozymes of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase present in the Escherichia coli genealogy. J. Bacteriol. 1986, 165, 146–154. [Google Scholar] [CrossRef]
- Jensen, R.A.; Ahmad, S. Evolution and phylogenetic distribution of the specialized isozymes of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase in superfamily-B prokaryotes. Microbiol. Sci. 1988, 5, 316–319. [Google Scholar]
- Hartmann, M.; Schneider, T.R.; Pfeil, A.; Heinrich, G.; Lipscomb, W.N.; Braus, G.H. Evolution of feedback-inhibited beta /alpha barrel isoenzymes by gene duplication and a single mutation. Proc. Natl. Acad. Sci. USA 2003, 100, 862–867. [Google Scholar] [CrossRef]
- Doong, R.L.; Jensen, R.A. Synonymy of the three apparent isoenzymes of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase in Pisum sativum L. with 3-deoxy-d-manno-octulosonate 8-phosphate synthase and the DS-Co/DS-Mn isoenzyme pair. New Phytol. 1992, 121, 165–171. [Google Scholar] [CrossRef]
- Doong, R.L.; Ganson, R.J.; Jensen, R.A. Plastid-localized 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DS-Mn): The early-pathway target of sequential feedback inhibition in higher plants. Plant Cell Environ. 1993, 16, 393–402. [Google Scholar] [CrossRef]
- Ganson, R.J.; D’Amato, T.A.; Jensen, R.A. The two-isozyme system of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase in Nicotiana silvestris and other higher plants. Plant Physiol. 1986, 82, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Doong, R.L.; Gander, J.E.; Ganson, R.J.; Jensen, R.A. The cytosolic isoenzyme of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase in Spinacia oleracea and other higher plants: Extreme substrate ambiguity and other properties. Physiol. Plant. 1992, 84, 351–360. [Google Scholar] [CrossRef]
- Sato, K.; Mase, K.; Nakano, Y.; Nishikubo, N.; Sugita, R.; Tsuboi, Y.; Kajita, S.; Zhou, J.; Kitano, H.; Katayama, Y. 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase is regulated for the accumulation of polysaccharide-linked hydroxycinnamoyl esters in rice (Oryza sativa L.) internode cell walls. Plant Cell Rep. 2006, 25, 676–688. [Google Scholar] [CrossRef]
- Janzik, I.; Preiskowski, S.; Kneifel, H. Ozone has dramatic effects on the regulation of the prechorismate pathway in tobacco (Nicotiana tabacum L. cv. Bel W3). Planta 2005, 223, 20–27. [Google Scholar] [CrossRef]
- Zhao, J.; Weaver, L.M.; Herrmann, K.M. Translocation of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase precursor into isolated chloroplasts. Planta 2002, 216, 180–186. [Google Scholar] [CrossRef]
- Entus, R.; Poling, M.; Herrmann, K.M. Redox regulation of Arabidopsis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Plant Physiol. 2002, 129, 1866–1871. [Google Scholar] [CrossRef]
- Keith, B.; Dong, X.N.; Ausubel, F.M.; Fink, G.R. Differential induction of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase genes in Arabidopsis thaliana by wounding and pathogenic attack. Proc. Natl. Acad. Sci. USA 1991, 88, 8821–8825. [Google Scholar] [CrossRef]
- Görlach, J.; Raesecke, H.R.; Rentsch, D.; Regenass, M.; Roy, P.; Zala, M.; Keel, C.; Boller, T.; Amrhein, N.; Schmid, J. Temporally distinct accumulation of transcripts encoding enzymes of the prechorismate pathway in elicitor-treated, cultured tomato cells. Proc. Natl. Acad. Sci. USA 1995, 92, 3166–3170. [Google Scholar] [CrossRef]
- McCue, K.F.; Conn, E.E. Induction of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase activity by fungal elicitor in cultures of Petroselinum crispum. Proc. Natl. Acad. Sci. USA 1989, 86, 7374–7377. [Google Scholar] [CrossRef]
- McCue, K.F.; Conn, E.E. Induction of shikimic acid pathway enzymes by light in suspension cultured cells of parsley (Petroselinum crispum). Plant Physiol. 1990, 94, 507–510. [Google Scholar] [CrossRef]
- Pinto, J.E.; Dyer, W.E.; Weller, S.C.; Herrmann, K.M. glyphosate induces 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase in potato (Solanum tuberosum L.) cells grown in suspension culture. Plant Physiol. 1988, 87, 891–893. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Lejczak, B.; Kafarski, P. The herbicidally active compound N-2-(6-methyl-pyridyl)-aminomethylene bisphosphonic acid inhibits in vivo aromatic biosynthesis. J. Plant Growth Regul. 1999, 18, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Lejczak, B.; Kafarski, P. The herbicidally active compound N-2-(5-chloro-pyridyl) aminomethylene bisphosphonic acid acts by inhibiting both glutamine and aromatic amino acid biosynthesis. Aust. J. Plant Physiol. 2000, 27, 677–683. [Google Scholar] [CrossRef]
- Bongue-Bartelsman, M.; Phillips, D.A. Nitrogen stress regulates gene expression of enzymes in the flavonoid biosynthetic pathway of tomato. Plant Physiol. Biochem. 1995, 33, 539–546. [Google Scholar]
- Løvdal, T.; Olsen, K.M.; Slimestad, R.; Verheul, M.; Lillo, C. Synergetic effects of nitrogen depletion, temperature, and light on the content of phenolic compounds and gene expression in leaves of tomato. Phytochemistry 2010, 71, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Feyissa, D.N.; Løvdal, T.; Olsen, K.M.; Slimestad, R.; Lillo, C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta 2009, 230, 747–754. [Google Scholar] [CrossRef]
- Umeda, M.; Hara, C.; Matsubayashi, Y.; Li, H.H.; Liu, Q.; Tadokoro, F.; Aotsuka, S.; Uchimiya, H. Expressed sequence tags from cultured cells of rice (Oryza sativa L.) under stressed conditions: Analysis of transcripts of genes engaged in ATP-generating pathways. Plant Mol. Biol. 1994, 25, 469–478. [Google Scholar] [CrossRef]
- van Heerden, P.S.; Towers, G.H.; Lewis, N.G. Nitrogen metabolism in lignifying Pinus taeda cell cultures. J. Biol. Chem. 1996, 271, 12350–12355. [Google Scholar] [CrossRef]
- Zulet-González, A.; Barco-Antoñanzas, M.; Gil-Monreal, M.; Royuela, M.; Zabalza, A. Increased glyphosate-induced gene expression in the shikimate pathway is abolished in the presence of aromatic amino acids and mimicked by shikimate. Front. Plant Sci. 2020, 11, 459. [Google Scholar] [CrossRef]
- Herrmann, K.M. The shikimate pathway: Early steps in the biosynthesis of aromatic compounds. Plant Cell 1995, 7, 907–919. [Google Scholar] [CrossRef]
- Maeda, H.; Shasany, A.K.; Schnepp, J.; Orlova, I.; Taguchi, G.; Cooper, B.R.; Rhodes, D.; Pichersky, E.; Dudareva, N. RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell 2010, 22, 832–849. [Google Scholar] [CrossRef]
- Zhu, J.; Patzoldt, W.L.; Shealy, R.T.; Vodkin, L.O.; Clough, S.J.; Tranel, P.J. Transcriptome response to glyphosate in sensitive and resistant soybean. J. Agric. Food Chem. 2008, 56, 6355–6363. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Hofius, D.; Hajirezaei, M.R.; Fernie, A.R.; Börnke, F.; Sonnewald, U. Functional analysis of the essential bifunctional tobacco enzyme 3-dehydroquinate dehydratase/shikimate dehydrogenase in transgenic tobacco plants. J. Exp. Bot. 2007, 58, 2053–2067. [Google Scholar] [CrossRef]
- Batz, O.; Logemann, E.; Reinold, S.; Hahlbrock, K. Extensive reprogramming of primary and secondary metabolism by fungal elicitor or infection in parsley cells. Biol. Chem. 1998, 379, 1127–1135. [Google Scholar] [CrossRef]
- Forlani, G.; Kafarski, P.; Lejczak, B.; Wieczorek, P. Mode of action of herbicidal derivatives of aminomethylenebisphosphonic acid.2. Reversal of herbicidal action by aromatic amino acids. J. Plant Growth Regul. 1997, 16, 147–152. [Google Scholar] [CrossRef]
- Bonner, C.A.; Rodrigues, A.M.; Miller, J.A.; Jensen, R.A. Amino acids are general growth inhibitors of Nicotiana silvestris in tissue culture. Physiol. Plant. 1992, 84, 319–328. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Scheible, W.R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef]
- Lam, H.-M.; Chiao, Y.A.; Li, M.-W.; Yung, Y.-K.; Ji, S. Putative nitrogen sensing systems in higher plants. J. Integr. Plant Biol. 2006, 48, 873–888. [Google Scholar] [CrossRef]
- Badur, R. Molecular and Functional Analysis of Plant Isozymes Considering as Example the Fructose-1,6-bisphosphate Aldolase, the Phosphoglucoisomerase and the 3-deoxy-D-arabino-heptulosonate-7-phosphate Synthase. Ph.D. Thesis, University of Göttingen, Göttingen, Germany, 1998. [Google Scholar]
- Görlach, J.; Beck, A.; Henstrand, J.M.; Handa, A.K.; Herrmann, K.M.; Schmid, J.; Amrhein, N. Differential expression of tomato (Lycopersicon esculentum L.) genes encoding shikimate pathway isoenzymes. I. 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Plant Mol. Biol. 1993, 23, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Hagendoorn, M.J.M.; Zethof, J.L.M.; van Hunnik, E.; van der Plas, L.H.W. Regulation of anthocyanin and lignin synthesis in Petunia hybrida cell suspensions. Plant Cell Tissue Organ Cult. 1991, 27, 141–147. [Google Scholar] [CrossRef]
- Forlani, G. Differential in vitro responses of rice cultivars to Italian lineages of the blast pathogen Pyricularia grisea. 2. Aromatic biosynthesis. J. Plant Physiol. 2010, 167, 928–932. [Google Scholar] [CrossRef]
- Gent, L.; Forde, B.G. How do plants sense their nitrogen status? J. Exp. Bot. 2017, 68, 2531–2539. [Google Scholar] [CrossRef]
- Liu, Y.; Duan, X.; Zhao, X.; Ding, W.; Wang, Y.; Xiong, Y. Diverse nitrogen signals activate convergent ROP2-TOR signaling in Arabidopsis. Dev. Cell 2021, 56, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.M.; Oh, G.G.K.; Lee, C.P.; Millar, A.H. Metabolite regulatory interactions control plant respiratory metabolism via Target of Rapamycin (TOR) kinase activation. Plant Cell 2019, 32, 666–682. [Google Scholar] [CrossRef] [PubMed]
- Lillo, C.; Lea, U.S.; Ruoff, P. Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 2008, 31, 587–601. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Forlani, G.; Lejczak, B.; Kafarski, P. N-pyridyl-aminomethylene-bisphosphonic acids inhibit the first enzyme in the shikimate pathway, 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase. Pestic. Biochem. Physiol. 1996, 55, 180–188. [Google Scholar] [CrossRef]
- Srinivasan, P.R.; Sprinson, D.B. 2-Keto-3-deoxy-D-arabo-heptonic acid 7-phosphate synthetase. J. Biol. Chem. 1959, 234, 716–722. [Google Scholar] [CrossRef]
- Mazzucotelli, E.; Tartari, A.; Cattivelli, L.; Forlani, G. Metabolism of gamma-aminobutyric acid during cold acclimation and freezing and its relationship to frost tolerance in barley and wheat. J. Exp. Bot. 2006, 57, 3755–3766. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Funck, D. A specific and sensitive enzymatic assay for the quantitation of L-proline. Front. Plant Sci. 2020, 11, 582026. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
| AA | 1 DAI | 4 DAI | 7 DAI | 10 DAI | ||||
|---|---|---|---|---|---|---|---|---|
| µmol (mg prot)−1 | % | µmol (mg prot)−1 | % | µmol (mg prot)−1 | % | µmol (mg prot)−1 | % | |
| Asp | 0.19 ± 0.01 | 3.5 | 0.07 ± 0.01 | 3.6 | 0.11 ± 0.00 | 2.7 | 0.19 ± 0.02 | 2.0 |
| Glu | 0.57 ± 0.01 | 10.7 | 0.39 ± 0.06 | 19.2 | 0.60 ± 0.05 | 13.9 | 0.91 ± 0.13 | 9.5 |
| Asn | 0.21 ± 0.01 | 4.0 | 0.03 ± 0.00 | 1.3 | 0.09 ± 0.01 | 2.1 | 0.23 ± 0.05 | 2.4 |
| Ser | 0.11 ± 0.01 | 2.1 | 0.15 ± 0.01 | 7.5 | 0.14 ± 0.01 | 3.3 | 0.12 ± 0.01 | 1.3 |
| Gln + His | 1.26 ± 0.06 | 23.8 | 0.42 ± 0.06 | 20.6 | 1.16 ± 0.06 | 27.0 | 3.06 ± 0.62 | 31.7 |
| Arg | 0.49 ± 0.01 | 9.3 | 0.05 ± 0.01 | 2.3 | 0.39 ± 0.03 | 9.0 | 0.84 ± 0.20 | 8.7 |
| Gly | 0.22 ± 0.07 | 4.1 | 0.03 ± 0.00 | 1.6 | 0.11 ± 0.01 | 2.5 | 0.28 ± 0.08 | 2.9 |
| Thr | 0.19 ± 0.04 | 3.7 | 0.06 ± 0.01 | 2.8 | 0.15 ± 0.01 | 3.4 | 0.34 ± 0.08 | 3.6 |
| Ala | 0.69 ± 0.01 | 13.0 | 0.29 ± 0.02 | 14.3 | 0.68 ± 0.05 | 15.8 | 1.89 ± 0.54 | 19.6 |
| Tyr | 0.18 ± 0.01 | 3.3 | 0.05 ± 0.00 | 2.2 | 0.16 ± 0.02 | 3.8 | 0.23 ± 0.03 | 2.4 |
| Trp | 0.06 ± 0.00 | 1.1 | 0.02 ± 0.00 | 0.8 | 0.03 ± 0.01 | 0.7 | 0.04 ± 0.01 | 0.4 |
| Met | 0.02 ± 0.00 | 0.3 | 0.01 ± 0.00 | 0.3 | 0.02 ± 0.01 | 0.6 | 0.03 ± 0.00 | 0.3 |
| Val | 0.11 ± 0.00 | 2.2 | 0.05 ± 0.01 | 2.5 | 0.07 ± 0.00 | 1.6 | 0.14 ± 0.03 | 1.5 |
| Phe | 0.34 ± 0.01 | 6.4 | 0.13 ± 0.01 | 6.1 | 0.22 ± 0.03 | 5.1 | 0.55 ± 0.01 | 5.7 |
| Ile | 0.06 ± 0.00 | 1.1 | 0.02 ± 0.00 | 1.2 | 0.04 ± 0.00 | 1.0 | 0.07 ± 0.01 | 0.8 |
| Leu | 0.04 ± 0.00 | 0.8 | 0.03 ± 0.00 | 1.6 | 0.07 ± 0.01 | 1.7 | 0.15 ± 0.02 | 1.5 |
| Orn | 0.02 ± 0.00 | 0.4 | 0.01 ± 0.00 | 0.7 | 0.01 ± 0.00 | 0.3 | 0.03 ± 0.00 | 0.3 |
| Lys | 0.05 ± 0.00 | 0.9 | 0.01 ± 0.00 | 0.7 | 0.04 ± 0.01 | 1.0 | 0.07 ± 0.02 | 0.7 |
| Pro | 0.49 ± 0.01 | 9.2 | 0.22 ± 0.00 | 10.6 | 0.20 ± 0.01 | 4.6 | 0.47 ± 0.02 | 4.9 |
| All | 5.29 ± 0.08 | 100.0 | 2.05 ± 0.21 | 100.0 | 4.29 ± 0.25 | 100.0 | 9.65 ± 1.83 | 100.0 |
| Gene | Primers | cDNA Product Size |
|---|---|---|
| cDNA sequencing | ||
| SHKA | fwd AGCTGCCGAGTTACAGGGGAGACA rev ATGCACTCTGTGACGTTTTGGCCTG | 832 |
| SHKB | fwd GAAGCTTGGTGAGGCTGCATTGGG rev GCTCATGCACATCGAAGAAAGCTCG | 998 |
| qRT-PCR analysis | ||
| SHKA | fwd GCCAACCCCCTTGGGATAAA rev TTGCCCAGCTCTTCTCACTG | 180 |
| SHKB | fwd TTGACCACCCTATCATGGCG rev CATTTGCATGAGCACCGTCA | 177 |
| Actin 7 | fwd1 TGCTATTCAGGCTGTGCTCT rev1 CGAAGAATGGCATGGGGCAA | 129 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forlani, G.; Giberti, S.; Doria, E. The Levels of DAHP Synthase, the First Enzyme of the Shikimate Pathway, Are Related to Free Aromatic Amino Acids and Glutamine Content in Nicotiana plumbaginifolia Cell Cultures. Plants 2023, 12, 2524. https://doi.org/10.3390/plants12132524
Forlani G, Giberti S, Doria E. The Levels of DAHP Synthase, the First Enzyme of the Shikimate Pathway, Are Related to Free Aromatic Amino Acids and Glutamine Content in Nicotiana plumbaginifolia Cell Cultures. Plants. 2023; 12(13):2524. https://doi.org/10.3390/plants12132524
Chicago/Turabian StyleForlani, Giuseppe, Samuele Giberti, and Enrico Doria. 2023. "The Levels of DAHP Synthase, the First Enzyme of the Shikimate Pathway, Are Related to Free Aromatic Amino Acids and Glutamine Content in Nicotiana plumbaginifolia Cell Cultures" Plants 12, no. 13: 2524. https://doi.org/10.3390/plants12132524
APA StyleForlani, G., Giberti, S., & Doria, E. (2023). The Levels of DAHP Synthase, the First Enzyme of the Shikimate Pathway, Are Related to Free Aromatic Amino Acids and Glutamine Content in Nicotiana plumbaginifolia Cell Cultures. Plants, 12(13), 2524. https://doi.org/10.3390/plants12132524







