Proteomic Analysis of Bt cry1Ac Transgenic Oilseed Rape (Brassica napus L.)
Abstract
1. Introduction
2. Results
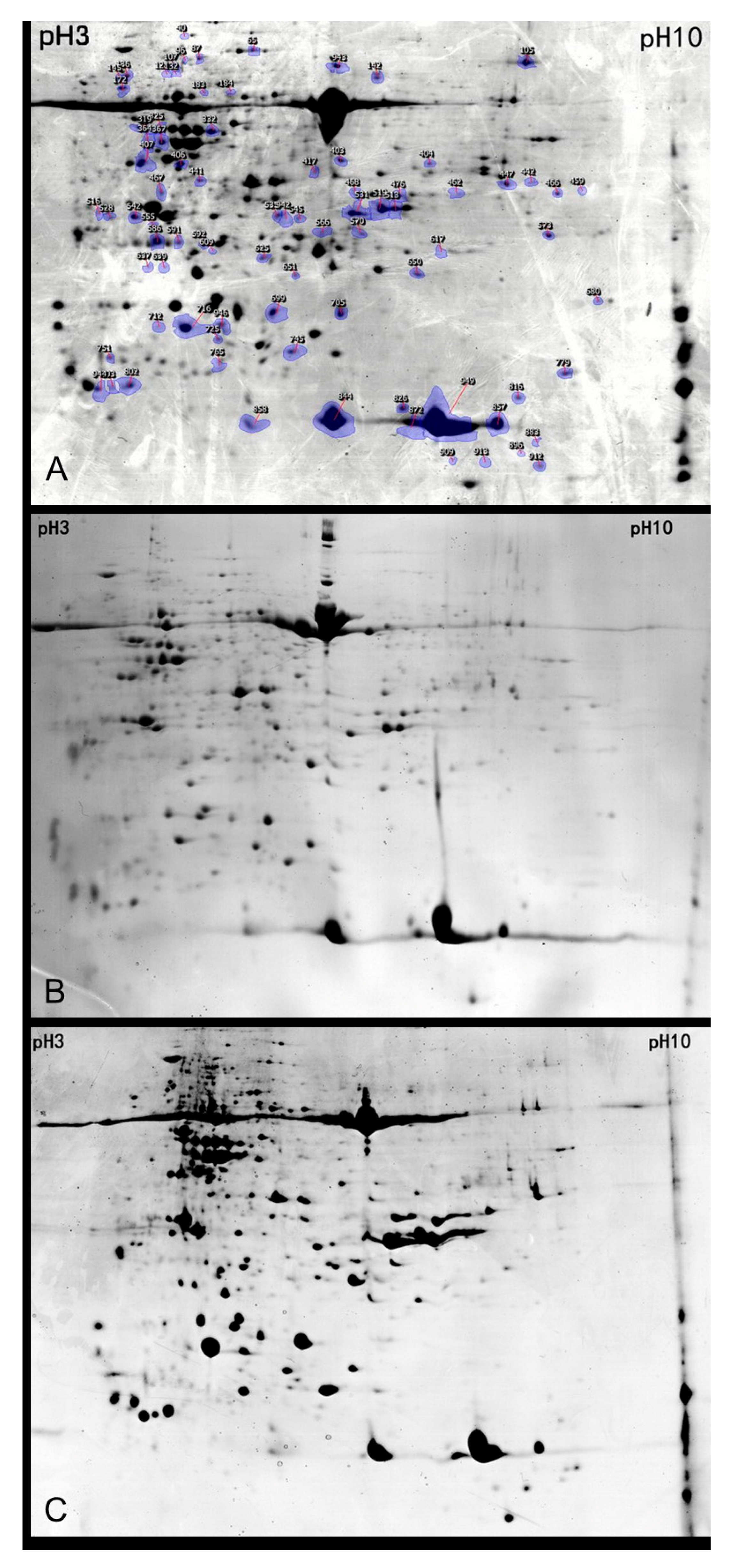
2.1. Comparative Proteomic Analysis of Transgenic and Non-Transgenic Bt Oilseed Rape Leaves
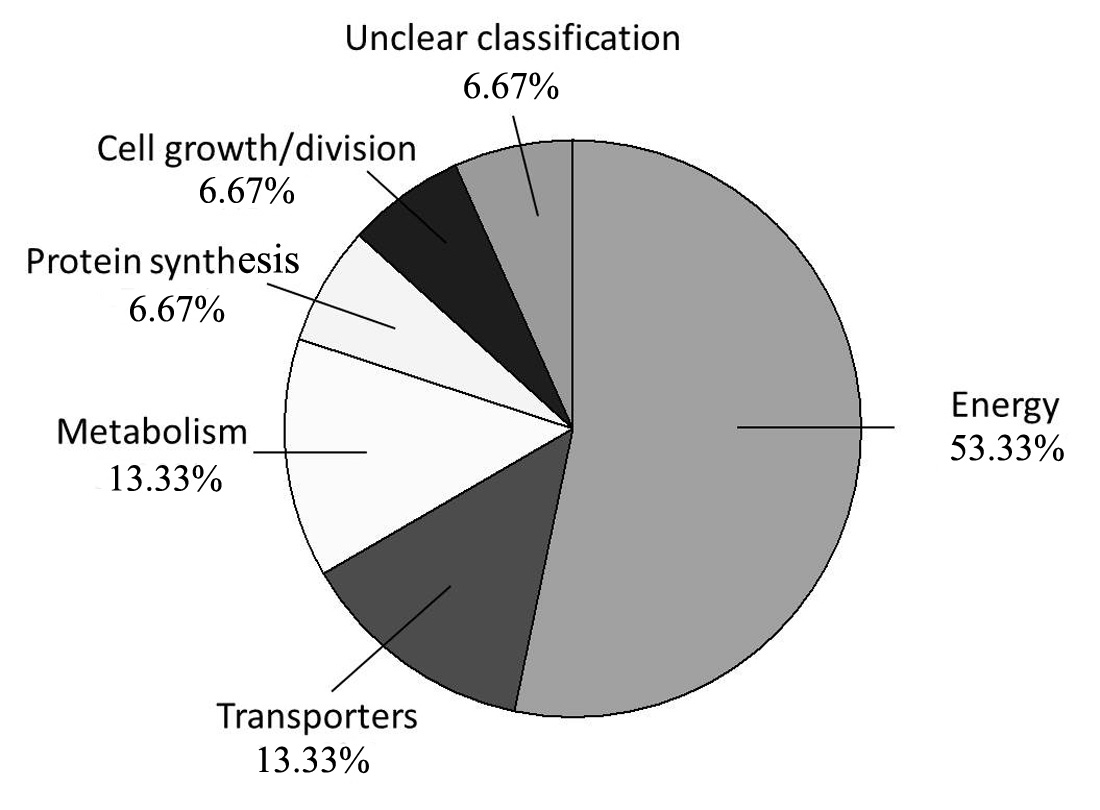
2.2. Identification and Functional Evaluation of the Differentially Expressed Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Planting
4.2. Protein Extraction and Quantification
4.3. 2D Gel Electrophoresis
4.4. Image and MS Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, X.; Ying, Y. Food safety evaluation based on near infrared spectroscopy and imaging: A Review. Crit. Rev. Food Sci. 2016, 56, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Boqvist, S.; Söderqvist, K.; Vågsholm, I. Food safety challenges and one health within Europe. Acta Vet. Scand. 2018, 60, 1. [Google Scholar] [CrossRef] [PubMed]
- Satoh, R.; Nakamura, R.; Komatsu, A.; Oshima, M.; Teshima, R. Proteomic analysis of known and candidate rice allergens between non-transgenic and transgenic plants. Regul. Toxicol. Pharm. 2011, 59, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Oboa, B.; Doicd, F.; Jop, E. Transgenic plant-mediated phytoremediation: Applications, challenges, and prospects. In Assisted Phytoremediation; Vimal, P., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2021; pp. 179–202. [Google Scholar]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91–118. [Google Scholar] [CrossRef]
- Sanchis, V. From microbial sprays to insect-resistant transgenic plants: History of the biospesticide Bacillus thuringiensis. A review. Agron. Sustain. Dev. 2010, 1, 217–231. [Google Scholar] [CrossRef]
- Ricroch, A.E.; Bergé, J.B.; Kuntz, M. Evaluation of genetically engineered crops using transcriptomic, proteomic, and metabolomic profiling techniques. Plant Physiol. 2011, 155, 1752–1761. [Google Scholar] [CrossRef]
- Herman, R.A. Unintended compositional changes in genetically modified (GM) crops, 20 years of research. J. Agric. Food Chem. 2013, 61, 11695–11701. [Google Scholar] [CrossRef]
- Kim, J.-K.; Park, S.-Y.; Lee, S.-M.; Lim, S.-H.; Kim, H.-J.; Oh, S.-D.; Yeo, Y.; Cho, H.-S.; Ha, S.-H. Unintended polar metabolite profiling of carotenoid-biofortified transgenic rice reveals substantial equivalence to its non-transgenic counterpart. Plant Biotechnol. Rep. 2013, 7, 121–128. [Google Scholar] [CrossRef]
- De Schrijver, A.; De Clercq, P.; Maagd, R.A.; van Frankenhuyzen, K. Relevance of Bt toxin interaction studies for environmental risk assessment of genetically modified crops. Plant Biotechnol. J. 2016, 13, 1221–1223. [Google Scholar] [CrossRef]
- Narva, K.E.; Wang, N.X.; Herman, R. Safety considerations derived from Cry34Ab1/Cry35Ab1 structure and function. J. Invertebr Pathol. 2017, 142, 27–33. [Google Scholar] [CrossRef]
- ISAAA. Global Status of Commercialized Biotech/GM Crops in 2019: Biotech Crops Drive Socio-Economic Development and Sustainable Environment in the New Frontier; ISAAA Brief No. 55; ISAAA: Ithaca, NY, USA, 2019. [Google Scholar]
- Cao, D.; Stewart, C.N., Jr.; Zheng, M.; Guan, Z.-J.; Tang, Z.-X.; Wei, W.; Ma, K.-P. Stable Bacillus thuringiensis transgene introgression from Brassica napus to wild mustard B. Juncea. Plant Sci. 2014, 227, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-B.; Stewart, C.N., Jr.; Li, J.-S.; Wei, W. One species to another: Sympatric Bt transgene gene flow from Brassica napus alters the reproductive strategy of wild relative Brassica juncea under herbivore treatment. Ann. Bot. 2018, 122, 617. [Google Scholar] [CrossRef] [PubMed]
- Coll, A.; Nadal, A.; Rossignol, M.; Puigdomènech, P.; Pla, M. Proteomic analysis of MON810 and comparable non-GM maize varieties grown in agricultural fields. Transgenic Res. 2011, 20, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Zhao, W.; Hao, J.; Luo, Y.; Tang, X.; Zhang, Y.; Huang, K. Comparative analysis of the proteomic and nutritional composition of transgenic rice seeds with Cry1ab/ac genes and their non-transgenic counterparts. J. Cereal Sci. 2012, 55, 226–233. [Google Scholar] [CrossRef]
- Gong, C.-Y.; Wang, T. Proteomic evaluation of genetically modified crops: Current status and challenges. Front. Plant Sci. 2013, 4, 41. [Google Scholar] [CrossRef]
- Agapito-Tenfen, S.Z.; Vilperte, V.; Benevenuto, R.F.; Rover, C.M.; Traavik, T.I.; Nodari, R.O. Effect of stacking insecticidal cry and herbicide tolerance epsps transgenes on transgenic maize proteome. BMC Plant Biol. 2014, 14, 346–352. [Google Scholar] [CrossRef]
- Shukla, P.; Gautam, R.; Singh, N.K. A proteomic study of cysteine protease induced cell death in anthers of male sterile tobacco transgenic plants. Physiol. Mol. Biol. Plants 2019, 25, 1073–1082. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, X.; Niu, F.; Sun, X.; Hu, Z.; Zhang, H. iTRAQ-based quantitative proteomic analysis of wheat roots in response to salt stress. Proteomics 2017, 17, 1600265. [Google Scholar] [CrossRef]
- Xue, K.; Yang, J.; Liu, B.; Xue, D. The integrated risk assessment of transgenic rice Oryza sativa: A comparative proteomics approach. Food Chem. 2012, 135, 314–318. [Google Scholar] [CrossRef]
- Sestili, F.; Paoletti, F.; Botticella, E.; Masci, S.; Saletti, R.; Muccilli, V.; Lafiandra, D. Comparative proteomic analysis of kernel proteins of two high amylose transgenic durum wheat lines obtained by biolistic and Agrobacterium-mediated transformations. J. Cereal Sci. 2013, 58, 15–22. [Google Scholar] [CrossRef]
- Liu, Y.-B.; Zhang, Y.-X.; Song, S.-Q.; Li, J.-S.; Stewart, C.N., Jr.; Wei, W.; Zhao, Y.-J.; Wang, W.-Q. A proteomic analysis of seeds from Bt-transgenic Brassica napus and hybrids with wild B. Juncea. Sci. Rep.-UK 2015, 5, 15480. [Google Scholar] [CrossRef]
- Vidal, N.; Barbosa, H.; Jacob, S.; Arruda, M. Comparative study of transgenic and non-transgenic maize (Zea mays) flours commercialized in Brazil, focusing on proteomic analyses. Food Chem. 2015, 180, 288–294. [Google Scholar] [CrossRef] [PubMed]
- García-Molina, M.D.; Muccilli, V.; Saletti, R.; Foti, S.; Masci, S.; Barro, F. Comparative proteomic analysis of two transgenic low-gliadin wheat lines and non-transgenic wheat control. J. Proteom. 2017, 165, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liang, L.; Zhang, Z.; Dong, M.; Jin, W. iTRAQ-based quantitative proteomic analysis of transgenic and non-transgenic maize seeds. J. Food Compos. Anal. 2020, 92, 103564. [Google Scholar] [CrossRef]
- Yang, Y.; Dai, L.; Zhu, K.; Xia, H.; Chen, L.; Liu, H.; Chen, K. Foreign protein detection in transgenic rice revealed by comparative proteomic analysis. Crop. Sci. 2015, 552, 2225–2233. [Google Scholar] [CrossRef]
- Bathellier, C.; Tcherkez, G.; Lorimer, G.H.; Farquhar, G.D. Rubisco is not really so bad. Plant Cell Environ. 2018, 41, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Gulfishan, M.; Jahan, A.; Bhat, T.A.; Sahab, D. Plant senescence and organ abscission. In Senescence Signalling and Control in Plants; Sarwat, M., Narendra Tuteja, N., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 255–272. [Google Scholar]
- Tommasi, I.C. The mechanism of Rubisco catalyzed carboxylation reaction: Chemical aspects involving acid-base chemistry and functioning of the molecular machine. Catalysts 2021, 11, 813. [Google Scholar] [CrossRef]
- Salesse-Smith, C.E.; Sharwood, R.E.; Busch, F.A.; Kromdijk, J.; Bardal, V.; Stern, D.B. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 2018, 4, 802–810. [Google Scholar] [CrossRef]
- Li, W.; Qiang, X.-J.; Han, X.-R.; Jiang, L.-L.; Zhang, S.H.; Han, J.; He, R.; Cheng, X.G. Ectopic Expression of a Thellungiella salsuginea aquaporin gene, TsPIP1;1, increased the salt tolerance of rice. Int. J. Mol. Sci. 2018, 19, 2229–2235. [Google Scholar] [CrossRef]
- Zhao, B.; Huo, J.; Liu, N.; Zhang, J.; Dong, J. Transketolase is identified as a target of herbicidal substance α-terthienyl by proteomics. Toxins 2018, 10, 41–43. [Google Scholar] [CrossRef]
- Bi, H.; Li, F.; Wang, H.; Ai, X. Overexpression of transketolase gene promotes chilling tolerance by increasing the activities of photosynthetic enzymes, alleviating oxidative damage and stabilizing cell structure in Cucumis sativus L. Physiol. Plant. 2018, 167, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, W.; Lin, Y.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Insight into transketolase of Pyropia haitanensis under desiccation stress based on integrative analysis of omics and transformation. BMC Plant Biol. 2019, 19, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, X.; Jia, X.; Huang, Q.; Tan, Y.; Guo, A. Comparative proteomics of Bt-transgenic and non-transgenic cotton leaves. Proteome Sci. 2015, 13, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Hisabori, T. Regulation machineries of ATP synthase from phototroph. In Advances in Botanical Research; Hisabori, T., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 96, pp. 1–26. [Google Scholar]
- Wang, F.-W.; Wang, C.; Sun, Y.; Wang, N.; Li, X.-W.; Dong, Y.-Y.; Yao, N.; Liu, X.M.; Chen, H.; Chen, X.-F.; et al. Overexpression of vacuolar proton pump ATPase (V-H+-ATPase) subunits B, C and H confers tolerance to salt and saline-alkali stresses in transgenic alfalfa (Medicago sativa L.). J. Integr. Agric. 2016, 15, 2279–2289. [Google Scholar] [CrossRef]
- Jaarsma, R.; de Boer, A.H. Salinity tolerance of two potato cultivars (Solanum tuberosum) correlates with differences in vacuolar transport activity. Front. Plant Sci. 2018, 9, 737–741. [Google Scholar] [CrossRef]
- Costa, A.; Luoni, L.; Marrano, C.A.; Hashimoto, K.; Köster, P.; Giacometti, S.; De Michelis, M.I.; Kudla, J.; Bonza, M.C. Ca2+-dependent phosphoregulation of the plasma membrane Ca2+-ATPase ACA8 modulates stimulus-induced calcium signatures. J. Exp. Bot. 2017, 68, 3215–3230. [Google Scholar] [CrossRef]
- Yadav, A.K. Role of plant Ca2+-ATPase in calcium homeostasis during development and stresses. In Calcium Transport Elements in Plants; Upadhyay, S.K., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 103–128. [Google Scholar]
- Zhong, M.; Liu, X.; Liu, F.; Ren, Y.; Wang, Y.; Zhu, J.; Teng, X.; Duan, E.; Wang, F.; Zhang, H.; et al. FLOURY ENDOSPERM12 encoding alanine aminotransferase 1 regulates carbon and nitrogen metabolism in rice. J. Plant Biol. 2019, 62, 61–73. [Google Scholar] [CrossRef]
- Wang, X.; Wei, Y.; Shi, L.; Ma, X.; Theg, S.-M. New isoforms and assembly of glutamine synthetase in the leaf of wheat (Triticum aestivum L.). J. Exp. Bot. 2015, 66, 6827–6834. [Google Scholar] [CrossRef]
- Ouerghi, Z.; Lachaâl, M.; Chebbi, M.; Ben Abdallah, S.; Amdouni, T.; Msilini, N. Does the source of nitrogen affect the response of fenugreek plants to saline stress? In Agrochimica: International Journal of Plant Chemistry, Soil Science and Plant Nutrition of the University of Pisa; Pisa University Press: Pisa, Italy, 2017; Volume 61, pp. 2283–5431. [Google Scholar]
- Bao, A.; Zhao, Z.; Ding, G.; Shi, L.; Xu, F.; Cai, F. The stable level of glutamine synthetase 2 plays an important role in rice growth and in carbon-nitrogen metabolic balance. Int. J. Mol Sci. 2015, 16, 12713–12736. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Q.; Huan, Z.; Zhong, M.; Chen, W.; Du, H. Identification and characterization of glutamine synthetase isozymes in Gracilaria lemaneiformis. Aquat. Bot. 2018, 146, 23–30. [Google Scholar] [CrossRef]
- Hinck, A.P.; Markus, M.A.; Huang, S.R.; Grzesiek, S.; Kustonovich, I.; Draper, D.E.; Torchia, D.A. The RNA binding domain of ribosomal protein L11: Three- dimensional structure of the RNA-bound form of the protein and its interaction with 23 S rRNA. J. Mol. Biol. 2015, 274, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.-L.; Xie, R.-R.; Tian, H.; Wang, Q.-L.; Guo, F.-Q. Putative zeatin O-glucosyltransferase OscZOG1 regulates root and shoot development and formation of agronomic traits in rice. J. Integr. Plant Biol. 2016, 58, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Halfhill, M.D.; Richards, H.A.; Mabon, S.A.; Stewart, C.N., Jr. Expression of GFP and Bt transgenes in Brassica napus and hybridization with Brassica rapa. Theor. Appl. Genet. 2001, 103, 659–667. [Google Scholar] [CrossRef]
- Joosen, R.; Cordewener, J.; Supena, E.D.J.; Vorst, O.; Lammers, M.; Maliepaard, C.; Zeilmaker, T.; Miki, B.; America, T.; Custers, J.; et al. Combined transcriptome and proteome analysis identifies pathways and markers associated with the establishment of rapeseed microspore-derived embryo development. Plant Physiol. 2007, 144, 155–172. [Google Scholar] [CrossRef]
- Ramagli, L.S. Quantifying protein in 2-D PAGE solubilization buffers. In 2-D Proteome Analysis Protocols. Methods in Molecular Biology; Link, A.J., Ed.; Humana Press: Totowa, NJ, USA, 1999; Volume 112, pp. 99–103. [Google Scholar]
- Li, X.-H.; Wu, X.-F.; Yue, W.-F.; Liu, J.-M.; Li, G.-L.; Miao, Y.-G. Proteomic analysis of the silkworm (Bombyx mori L.) hemolymph during developmental stage. J. Proteome Res. 2006, 5, 2809–2814. [Google Scholar] [CrossRef] [PubMed]
| Spot Number a | Protein Name | Accession No. b | Score c | Theor. pI d | Exp. pI d | Theor. MW e | Exp. MW e | Ratio f |
|---|---|---|---|---|---|---|---|---|
| Energy | ||||||||
| 802 (↑) | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|11466371 | 86 | 5.06 | 6.14 | 52,405 | 53,186.5 | 1.07 |
| 65 (↑) | Transketolase | gi|18411711 | 97 | 5.81 | 5.94 | 79,837 | 80,374.4 | 1.06 |
| 858 (↑) | Ribulose bisphosphate carboxylase small chain 1B | gi|15240912 | 68 | 6.27/ | 7.59 | 20,155 | 20,558.2 | 1.06 |
| 367 (↑) | Ribulose bisphosphate carboxylase/oxygenase activase | gi|297612474 | 82 | 4.98 | 7.56 | 38,775 | 39,108.5 | 1.08 |
| 699 (↑) | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|11466371 | 134 | 5.06 | 6.14 | 52,405 | 53,186.5 | 1.12 |
| 844 (↑) | Ribulose bisphosphate carboxylase small chain F1 | gi|132091 | 390 | 6.27 | 8.23 | 14,358 | 20,455.2 | 1.06 |
| 476 (↑) | Ribulose bisphosphate carboxylase large chain | gi|2500677 | 78 | 6.28 | 6.39 | 48,953 | 49,069.4 | 1.08 |
| 872 (↑) | Ribulose bisphosphate carboxylase small chain F1 | gi|132091 | 103 | 6.27 | 8.23 | 14,358 | 20,455.2 | 1.06 |
| Transporters | ||||||||
| 586 (↓) | Calcium-transporting ATPase | gi|302756809 | 63 | 8.07 | 6 | 112,406 | 113,321.0 | 0.93 |
| 87 (↓) | V-type proton ATPase catalytic subunit A | gi|15219234 | 96 | 4.53 | 5.11 | 68,682 | 69,111.0 | 0.93 |
| Metabolism | ||||||||
| 651 (↓) | Alanine aminotransferase 2-like | gi|30698866 | 59 | 4.95 | 5.95 | 59,380 | 59,986.2 | 0.85 |
| 325 (↑) | Glutamine synthetase | gi|12643761 | 114 | 4.24 | 6.16 | 47,214 | 47,714.0 | 1.09 |
| Protein synthesis | ||||||||
| 513 (↑) | Ribosomal protein L11 | gi|56404772 | 66 | 9.60 | 9.27 | 14,973 | 15,151.0 | 1.10 |
| Cell growth/division | ||||||||
| 319 (↑) | Cis-zeatin O-glucosyltransferase | gi|242093988 | 63 | 4.79 | 6.06 | 50,372 | 50,813.9 | 1.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Z.-J.; Zheng, M.; Tang, Z.-X.; Wei, W.; Stewart, C.N., Jr. Proteomic Analysis of Bt cry1Ac Transgenic Oilseed Rape (Brassica napus L.). Plants 2023, 12, 2319. https://doi.org/10.3390/plants12122319
Guan Z-J, Zheng M, Tang Z-X, Wei W, Stewart CN Jr. Proteomic Analysis of Bt cry1Ac Transgenic Oilseed Rape (Brassica napus L.). Plants. 2023; 12(12):2319. https://doi.org/10.3390/plants12122319
Chicago/Turabian StyleGuan, Zheng-Jun, Min Zheng, Zhi-Xi Tang, Wei Wei, and C. Neal Stewart, Jr. 2023. "Proteomic Analysis of Bt cry1Ac Transgenic Oilseed Rape (Brassica napus L.)" Plants 12, no. 12: 2319. https://doi.org/10.3390/plants12122319
APA StyleGuan, Z.-J., Zheng, M., Tang, Z.-X., Wei, W., & Stewart, C. N., Jr. (2023). Proteomic Analysis of Bt cry1Ac Transgenic Oilseed Rape (Brassica napus L.). Plants, 12(12), 2319. https://doi.org/10.3390/plants12122319








