Jujube Fruit Metabolomic Profiles Reveal Cultivar Differences and Function as Cultivar Fingerprints
Abstract
:1. Introduction
2. Results
2.1. Jujube Cultivar and Fruit Characteristics
2.2. Total Detected Compounds and Their Categories
2.3. Location Effect
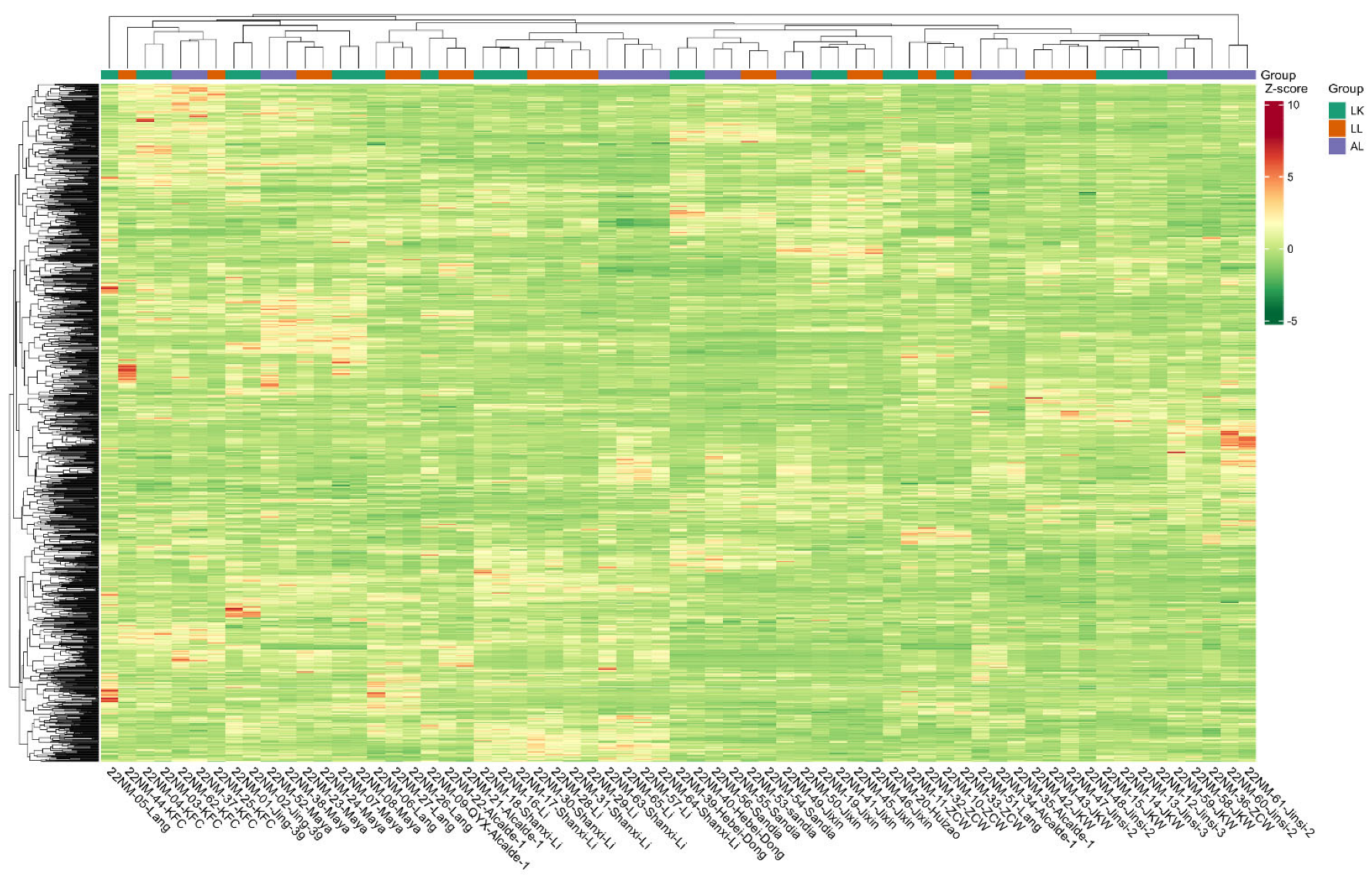
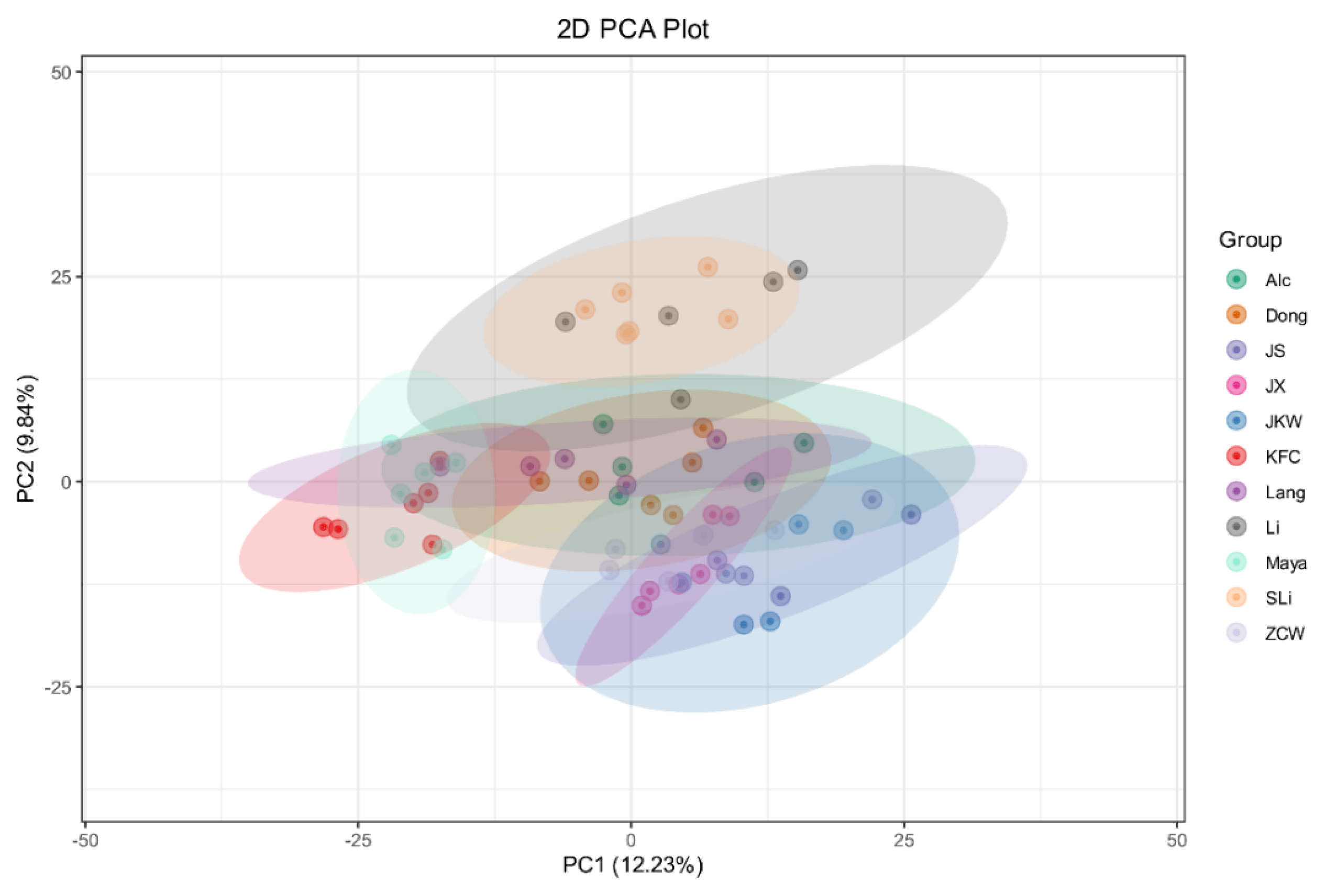
2.4. Global Cultivar Metabolic Profiling
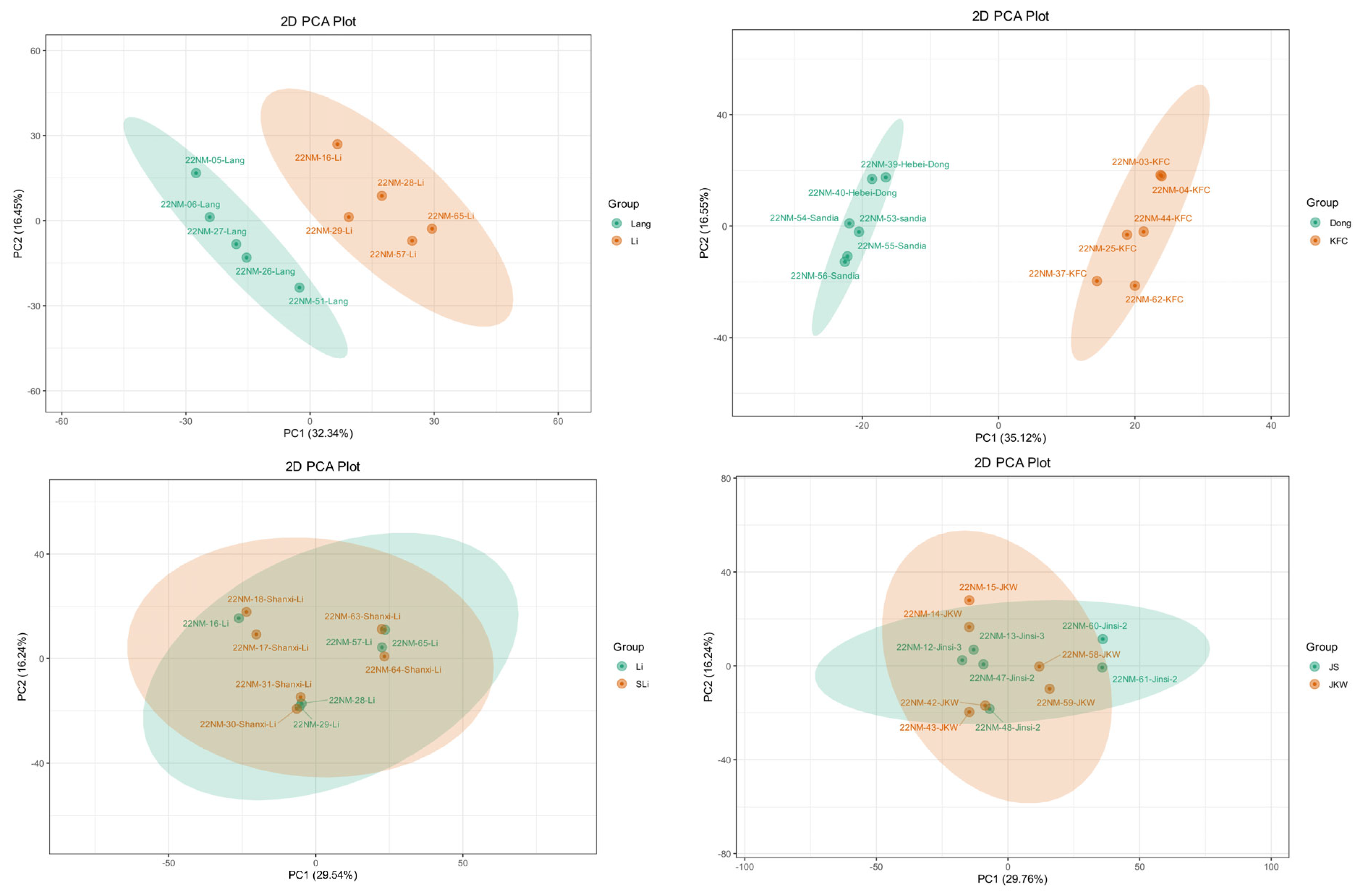
2.5. Cultivar-Specific Pairwise Metabolic Comparison
3. Discussion
3.1. Cultivar Defines Metabolomics Profile and Location Plays a Minor Role
3.2. Cultivar Metabolomic Profile Functions as Cultivar Metabolomic Fingerprint
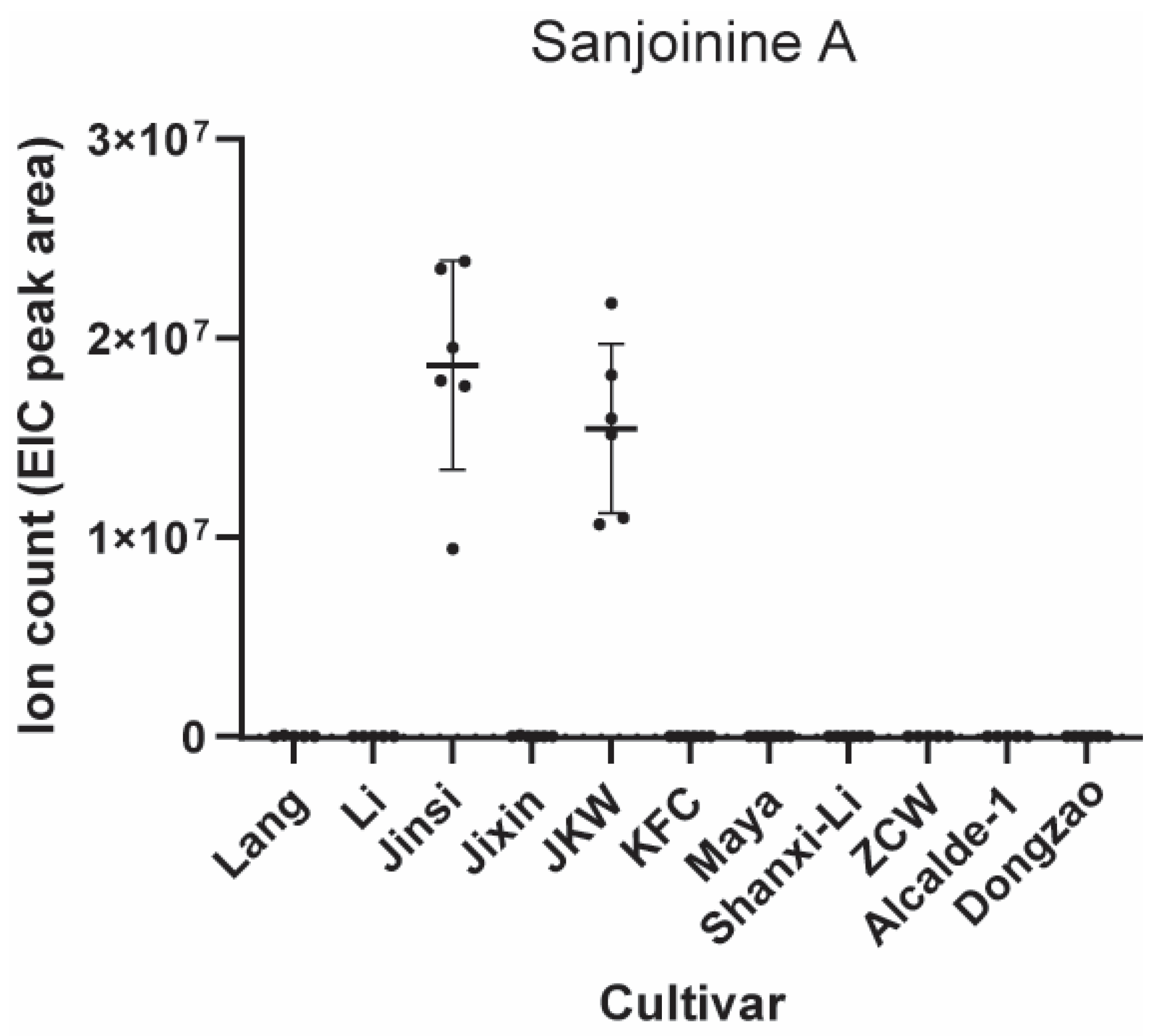
3.3. Important Nutritional and Medicinal Components in Jujube Fruits
4. Materials and Methods
4.1. Jujube Sampling
4.2. Metabolomics Sample Preparation
4.3. UPLC–MS Analysis
4.4. Qualitative and Quantitative Analysis of Metabolite
4.5. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomas, C. Chinese Jujube in Southwestern United States. In USDA Yearbook of Agriculture 1926; United States Department of Agriculture: Washington, DC, USA, 1927; pp. 212–215. Available online: https://naldc.nal.usda.gov/download/IND43842740/pdf (accessed on 15 February 2023).
- Ackerman, W.L. Flowering Pollination, Self-Sterility and Seed Development of Chinese Jujube. Proc. Am. Soc. Hoticultural Sci. 1961, 77, 265. [Google Scholar]
- Bonner, F.T. Rudolf. Ziziphus Mill-jujube. In Seeds of Wood Plants in the United States. Agricultural Handbook No. 450; USDA Forest Service: Washington, DC, USA, 1974; pp. 862–863. [Google Scholar]
- Kader, A.A.; Li, Y.; Chordas, A. Postharvest respiration, ethylene production, and compositional changes in Chinese jujube fruits [Zizyphus jujuba, Chinese date, maturation, ascorbic acid, chilling injury]. HortScience 1982, 17, 678–679. [Google Scholar] [CrossRef]
- Locke, L.F. The chinese jujube: A promising fruit tree for the southwest. In Proceedings of the Second Annual Oklahoma Crops and Soils Conference; Oklahoma Agricultural Experiment Station: Stillwater, OK, USA, 1948; pp. 78–81. [Google Scholar]
- Lyrene, P. Flowering and fruiting of Chinese jujubes in Florida. HortScience 1983, 18, 208–209. [Google Scholar] [CrossRef]
- Yao, S. Past, Present, and Future of Jujubes—Chinese Dates in the United States. HortScience 2013, 48, 672–680. [Google Scholar] [CrossRef]
- Yao, S.; Heyduck, R.; Guldan, S. Early Performance of Jujube Fresh Eating Cultivars in the Southwestern United States. HortScience 2019, 54, 1941–1946. [Google Scholar] [CrossRef] [Green Version]
- Yao, S.; Heyduck, R.; Guldan, S.; Sapkota, G. Early performance of jujube drying and multipurpose cultivars in the southwestern United States. HortScience 2020, 55, 1804–1810. [Google Scholar] [CrossRef]
- Liu, M.; Wang, J.; Wang, L.; Liu, P.; Zhao, J.; Zhao, Z.; Yao, S.; Stănică, F.; Liu, Z.; Wang, L.; et al. The historical and current research progress on jujube–a superfruit for the future. Hortic. Res. 2020, 7, 119. [Google Scholar] [CrossRef]
- Gao, Q.-H.; Wu, C.-S.; Wang, M. The jujube (Ziziphus jujuba Mill.) fruit: A review of current knowledge of fruit composition and health benefits. J. Agric. Food Chem. 2013, 61, 3351–3363. [Google Scholar] [CrossRef]
- Cyong, J.-C.; Hanabusa, K. Cyclic adenosine monophosphate in fruits of Zizyphus jujuba. Phytochemistry 1980, 19, 2747–2748. [Google Scholar] [CrossRef]
- Cyong, J.-C.; Takahashi, M. Identification of guanosine 3′: 5′-monophosphate in the fruit of Zizyphus jujuba. Phytochemistry 1982, 21, 1871–1874. [Google Scholar] [CrossRef]
- Huang, J.; Heyduck, R.; Richins, R.D.; VanLeeuwen, D.; O’Connell, M.A.; Yao, S. Jujube Cultivar Vitamin C Profile and Nutrient Dynamics during Maturation. HortScience 2017, 52, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Sapkota, G.; Delgado, E.; VanLeeuwen, D.; Holguin, F.O.; Flores, N.; Heyduck, R.; Yao, S. Dynamics of Nutrients in Jujube (Ziziphus jujuba Mill.) at Different Maturity Stages, Cultivars, and Locations in the Southwest United States. HortScience 2023, 58, 155–163. [Google Scholar] [CrossRef]
- Chigumba, D.N.; Mydy, L.S.; de Waal, F.; Li, W.; Shafiq, K.; Wotring, J.W.; Mohamed, O.G.; Mladenovic, T.; Tripathi, A.; Sexton, J.Z. Discovery and biosynthesis of cyclic plant peptides via autocatalytic cyclases. Nat. Chem. Biol. 2022, 18, 18–28. [Google Scholar] [CrossRef]
- Lima, S.T.; Ampolini, B.G.; Underwood, E.B.; Graf, T.N.; Earp, C.E.; Khedi, I.C.; Pasquale, M.A.; Chekan, J.R. A Widely Distributed Biosynthetic Cassette Is Responsible for Diverse Plant Side Chain Cross-Linked Cyclopeptides. Angew. Chem. 2023, 135, e202218082. [Google Scholar] [CrossRef]
- Kang, K.B.; Ernst, M.; van der Hooft, J.J.; da Silva, R.R.; Park, J.; Medema, M.H.; Sung, S.H.; Dorrestein, P.C. Comprehensive mass spectrometry-guided phenotyping of plant specialized metabolites reveals metabolic diversity in the cosmopolitan plant family Rhamnaceae. Plant J. 2019, 98, 1134–1144. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.B.; Ming, G.; Kim, G.J.; Choi, H.; Oh, W.K.; Sung, S.H. Jubanines F–J, cyclopeptide alkaloids from the roots of Ziziphus jujuba. Phytochemistry 2015, 119, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Han, H.; Eun, J.S.; Kim, H.-C.; Hong, J.-T.; Oh, K.-W. Sanjoinine A isolated from Zizyphi Spinosi Semen augments pentobarbital-induced sleeping behaviors through the modification of GABA-ergic systems. Biol. Pharm. Bull. 2007, 30, 1748–1753. [Google Scholar] [CrossRef] [Green Version]
- Tan, N.-H.; Zhou, J. Plant cyclopeptides. Chem. Rev. 2006, 106, 840–895. [Google Scholar] [CrossRef] [PubMed]
- Tschesche, R.; Last, H. Alkaloide aus rhamnaceen, V franganin und frangufolin, zwei weitere peptid-alkaloide aus Rhamnus frangula L. Tetrahedron Lett. 1968, 9, 2993–2998. [Google Scholar] [CrossRef] [PubMed]
- Daneshmand, F.; Zare-Zardini, H.; Ebrahimi, L. Investigation of the antimicrobial activities of Snakin-Z, a new cationic peptide derived from Zizyphus jujuba fruits. Nat. Prod. Res. 2013, 27, 2292–2296. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Zhao, X.; Liu, F.; Luo, Z.; Chen, S.; Liu, Z.; Zhao, Z.; Liu, M.; Wang, L. Triterpenoids in Jujube: A Review of Composition, Content Diversity, Pharmacological Effects, Synthetic Pathway, and Variation during Domestication. Plants 2023, 12, 1501. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Kanu, A.B. Recent developments in sample preparation techniques combined with high-performance liquid chromatography: A critical review. J. Chromatogr. A 2021, 1654, 462444. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.J.; Ren, J.L.; Zhang, A.H.; Sun, H.; Yan, G.L.; Han, Y.; Liu, L. Novel applications of mass spectrometry-based metabolomics in herbal medicines and its active ingredients: Current evidence. Mass Spectrom. Rev. 2019, 38, 380–402. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Mu, X.; Liu, J.; Li, B.; Liu, H.; Zhang, B.; Xiao, P. Plant metabolomics: A new strategy and tool for quality evaluation of Chinese medicinal materials. Chin. Med. 2022, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Han, G.; Liu, Y.; Jiang, J.; Jia, Y.; Li, X. Nutrient composition and quality traits of dried jujube fruits in seven producing areas based on metabolomics analysis. Food Chem. 2022, 385, 132627. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Gao, Z.; Jiao, X.; Shi, J.; Wang, R. Widely targeted metabolomic analysis of active compounds at different maturity stages of ‘Hupingzao’jujube. J. Food Compos. Anal. 2020, 88, 103417. [Google Scholar] [CrossRef]
- Lu, D.; Zhang, L.; Wu, Y.; Pan, Q.; Zhang, Y.; Liu, P. An integrated metabolome and transcriptome approach reveals the fruit flavor and regulatory network during jujube fruit development. Front. Plant Sci. 2022, 13, 952698. [Google Scholar] [CrossRef]
- Shen, B.; Zhang, Z.; Shi, Q.; Du, J.; Xue, Q.; Li, X. Active compound analysis of Ziziphus jujuba cv. Jinsixiaozao in different developmental stages using metabolomic and transcriptomic approaches. Plant Physiol. Biochem. 2022, 189, 14–23. [Google Scholar] [CrossRef]
- Xue, X.; Zhao, A.; Wang, Y.; Ren, H.; Li, Y.; Li, D.; Du, J. Metabolomics-based analysis of flavonoid metabolites in Chinese jujube and sour jujube fruits from different harvest periods. J. Food Sci. 2022, 87, 3752–3765. [Google Scholar] [CrossRef]
- Wang, L.; Gu, S.; Wu, Y. Analysis of flavonoids in fruit of jujube (Ziziphus jujuba) by UPLC-MS-based metabonomics. Shipin Kexue/Food Sci. 2020, 41, 155–161. [Google Scholar]
- Lu, D.; Wu, Y.; Pan, Q.; Zhang, Y.; Qi, Y.; Bao, W. Identification of key genes controlling L-ascorbic acid during jujube (Ziziphus jujuba mill.) fruit development by integrating transcriptome and metabolome analysis. Front. Plant Sci. 2022, 13, 950103. [Google Scholar] [CrossRef]
- Elaloui, M.; Laamouri, A.; Fabre, J.; Mathieu, C.; Vilarem, G.; Hasnaoui, B. Distribution of free amino acids, polyphenols and sugars of Ziziphus jujuba pulps harvested from plants grown in Tunisia. Nat. Prod. Res. 2015, 29, 94–97. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wang, L.; Xu, M.; Deng, B.; Liu, H.; Zhao, X. Phytochemical analysis of Ziziphus jujube leaf at different foliar ages based on widely targeted metabolomics. Open Chem. 2022, 20, 1485–1493. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, Q.; Wang, B.; Ma, A.; Wang, Y.; Xue, Q.; Shen, B.; Hamaila, H.; Tang, T.; Qi, X. Jujube metabolome selection determined the edible properties acquired during domestication. Plant J. 2022, 109, 1116–1133. [Google Scholar] [CrossRef]
- Corrêa dos Santos, C.H.; Geraldo de Carvalho, M.; Laub, A.; Franke, K.; Wessjohann, L. UHPLC-ESI-Orbitrap-HR-MS Analysis of Cyclopeptide Alkaloids From Ziziphus joazeiro. Nat. Prod. Commun. 2021, 16, 1934578X211054955. [Google Scholar] [CrossRef]
- Sapkota, D.; Zhang, D.; Meinhardt, L.; Yao, S. Preliminary Report of Jujube Cultivar Genotyping in the United States. Available online: https://www.researchgate.net/profile/Dikshya-Sapkota/publication/366411885_Preliminary_Report_of_Jujube_Cultivar_Genotyping_in_the_United_States/links/63a0b5b740358f78eb018913/Preliminary-Report-of-Jujube-Cultivar-Genotyping-in-the-United-States.pdf (accessed on 29 May 2023).
- Jiang, J.-G.; Huang, X.-J.; Chen, J.; Lin, Q.-S. Comparison of the sedative and hypnotic effects of flavonoids, saponins, and polysaccharides extracted from Semen Ziziphus jujube. Nat. Prod. Res. 2007, 21, 310–320. [Google Scholar] [CrossRef]
- Western Regional Climate Center. New Mexico Climate Summaries. 2023. Available online: https://wrcc.dri.edu/summary/climsmnm.html (accessed on 15 February 2023).
- Cao, H.; Ji, Y.; Li, S.; Lu, L.; Tian, M.; Yang, W.; Li, H. Extensive metabolic profiles of leaves and stems from the medicinal plant Dendrobium officinale Kimura et Migo. Metabolites 2019, 9, 215. [Google Scholar] [CrossRef] [Green Version]
- Lozada, D.N.; Pulicherla, S.R.; Holguin, F.O. Widely Targeted Metabolomics Reveals Metabolite Diversity in Jalapeño and Serrano Chile Peppers (Capsicum annuum L.). Metabolites 2023, 13, 288. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tu, H.; Wan, J.; Chen, W.; Liu, X.; Luo, J.; Xu, J.; Zhang, H. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chem. 2016, 199, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bujak, R.; Daghir-Wojtkowiak, E.; Kaliszan, R.; Markuszewski, M.J. PLS-based and regularization-based methods for the selection of relevant variables in non-targeted metabolomics data. Front. Mol. Biosci. 2016, 3, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Li, Q.; Zhao, Y.; Wei, H.; Wang, J.; Baker, C.J.; Liu, Q.; Wei, W. Integration of metabolomics and existing omics data reveals new insights into phytoplasma-induced metabolic reprogramming in host plants. PLoS ONE 2021, 16, e0246203. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]





| Cultivar Groups (Fruit Types) | All Significant Differential Metabolites | Up-Regulated Metabolites | Down-Regulated Metabolites | Up-Regulated Metabolic Pathways | Down-Regulated Metabolic Pathways | Differential Secondary Metabolites [%] |
|---|---|---|---|---|---|---|
| Alcalde1/ Dongzao (fresh/fresh) | 475 | 246 | 229 | Stilbenoid, Flavone and Flavonol, Glutathione, Sulfur | Arginine, Linoleic acid, Alanine/ Aspartate /Glutamate, Proline, Porphyrin, Nitrogen, Cyanoamino acid, Valine/Leucine /Isoleucine, D-amino acid, Pyrimidine, Glyoxylate and decarboxylate, Butanoate, Propanoate | 41.4 |
| Alcalde1/Jinsi (fresh/MP) | 375 | 192 | 183 | Arginine and Proline, Purine alkaloids, Phenylpropanoid, Pyrimidine, Nucleotides, Tropane/piperidine/pyridine alkaloids, Betalain, Fatty acid, alpha-linoleic acid, glutathione, Purine, Stilbenoid | Sphingolipid, Flavonoid | 40.5 |
| Alcalde1/KFC (fresh/MP) | 483 | 365 | 118 | Flavonoid, Flavone and Flavonol, Phenylpropanoid, Stilbenoid | Tryptophan | 53.7 |
| Alcalde1/Lang (fresh/dry) | 330 | 212 | 118 | Flavonoid, Phenylpropanoid, Tryptophan, Phenylalanine, Glutathione | None | 38.5 |
| Alcalde1/Li (fresh/fresh) | 336 | 206 | 130 | Flavonoid, Sphingolipid, Quinone | Tyrosine, Isoquinoline alkaloids, Nucleotide | 42.6 |
| Alcalde1/Maya (fresh/fresh) | 489 | 368 | 121 | Phenylalanine, Phenylpropanoid, Flavonoid, Arginine and Proline | Flavone and Flavonol | 42.5 |
| Dongzao/JKW (fresh/MP) | 564 | 316 | 248 | Linoleic acid, Nucleotide, Purine, Arginine and Proline, Phenylalanine, D-amino acid, | Flavone and Flavonol | 39.4 |
| Dongzao/KFC (fresh/MP) | 531 | 403 | 128 | Arginine and Proline, Flavonoid, Isoquinoline alkaloid, Linoleic acid, Flavone and Flavonol, Tyrosine, Phenylalanine | Glutathione | 42.4 |
| Dongzao/Li (fresh/fresh) | 487 | 260 | 227 | Amino acid, Flavonoid, Linoleic acid, Sphingolipid | Phenylpropanoid, Flavone and Flavonol, Sulfur | 50 |
| Dongzao/Maya (fresh/fresh) | 575 | 400 | 175 | Linoleic acid, Quinone, Tyrosine, Arginine and proline, Tropane/piperidine/pyridine alkaloid, Isoquinoline alkaloid, Cyanoamino acid, Purine, Pyrimidine, Alanine/Aspartate/Glutamate, Nucleotide | None | 42.4 |
| JKW/Li (MP/fresh) | 452 | 207 | 245 | Flavonoid, Flavone and Flavonol | Linoleic acid | 44.3 |
| JKW/Maya (MP/fresh) | 561 | 392 | 169 | Phenylalanine, Sphingolipid, Tyrosine, Tryptophan, Arginine and proline | Purine, Phenylpropanoid | 40 |
| JKW/ZCW (MP/fresh) | 384 | 160 | 224 | Sphingolipid, Flavonoid, Flavone and Flavonol | Phenylalanine, Purine | 37 |
| Jinsi/JKW (dry/MP) | 47 | 36 | 11 | Amino acids, Tropane/piperidine/pyridine alkaloid, Glucosinolate, 2-oxocarboxylic acid | 62.5 | |
| Jinsi/KFC (MP/MP) | 494 | 379 | 115 | Sphingolipid, Flavonoid, Flavone and Flavonol | Cyanoamino acid, Thiamine, Amino acids, Nucleotide, Pantothenate and CoA, Purine | 46.8 |
| Jinsi/Lang (MP/dry) | 420 | 270 | 150 | Fructose and Mannose, Nucleotide | Linoleic acid, Phenylpropanoid, D-amino acid | 43.5 |
| Jinsi/Maya (MP/fresh) | 533 | 394 | 139 | Phenylalanine, Tyrosine, Arginine and proline | Purine, Nucleotide, Glutathione | 39.2 |
| Jixin/KFC (dry/MP) | 531 | 377 | 154 | Flavone and Flavonol, Flavonoid, Phenylpropanoid, Tropane/piperidine /pyridine alkaloid, Phenylalanine | Tryptophan, Purine | 56.7 |
| KFC/Lang (MP/dry) | 498 | 178 | 320 | Glutathione | Phenylpropanoid, Flavone and Flavonol, Tyrosine | 45.5 |
| KFC/Li (MP/fresh) | 492 | 154 | 338 | Glutathione, D-amino acid | Phenylpropanoid, Flavonoid, Tropane/piperidine/pyridine alkaloid, Flavone and Flavonol | 50.8 |
| KFC/Maya (MP/fresh) | 474 | 222 | 252 | Linoleic acid, Tryptophan | Purine, Nucleotide, Flavonoid, Flavone and Flavonol | 52.2 |
| Lang/Li (dry/fresh) | 355 | 149 | 206 | Purine, Nucleotide | Flavonoid, Isoquinoline alkaloid, Phenylpropanoid, Tyrosine, Phenylalanine | 50 |
| Lang/Maya (dry/fresh) | 437 | 265 | 172 | Tyrosine, Phenylpropanoid, Linoleic acid, Arginine and proline | Phenylalanine, Glutathione, Purine, Flavonoid, Nucleotide | 41.8 |
| Lang/ZCW (dry/fresh) | 445 | 181 | 264 | Arginine and proline, Tyrosine, Glutathione, Tropane/piperidine/pyridine alkaloid, phenylpropanoid | Nucleotide, Purine, D-amino acid, Flavonoid | 42.2 |
| Li/Maya (fresh/fresh) | 486 | 337 | 149 | Linoleic acid, Tryptophan, Phenylpropanoid | Purine, Glutathione, Nucleotide | 44.6 |
| Li/Shanxi-Li (fresh/fresh) | 42 | 36 | 6 | None | None | 75 |
| Li/ZCW (fresh/fresh) | 436 | 198 | 238 | None | Glucosinolate, Amino acids, 2-Oxocarboxylic acid | 42 |
| Maya/ZCW (fresh/fresh) | 519 | 130 | 389 | Tropane/piperidine/pyridine alkaloids | Phenylalanine, Tryptophan, Arginine and proline, Flavonoid | 53.1 |
| Cultivar | Code | Leyendecker (LK) | Los Lunas (LL) | Alcalde (AL) |
|---|---|---|---|---|
| Alcalde 1 (QYX) | Alc | 1 | 2 | 2 |
| Sandia/Hebei Dong | Dong | 2 | 2 | 2 |
| Jinsi 2 | JS | 2 | 2 | |
| Jinsi 3 | JS | 2 | ||
| Jixin | JX | 2 | 2 | 2 |
| Jinkuiwang | JKW | 2 | 2 | 2 |
| Kongfucui | KFC | 2 | 2 | 2 |
| Lang | Lang | 2 | 2 | 1 |
| Li | Li | 1 | 2 | 2 |
| Maya | Maya | 2 | 2 | 2 |
| Shanxi Li | SLi | 2 | 2 | 2 |
| Zaocuiwang | ZCW | 2 | 2 | 1 |
| Total | 20 | 22 | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, S.; Sapkota, D.; Hungerford, J.A.; Kersten, R.D. Jujube Fruit Metabolomic Profiles Reveal Cultivar Differences and Function as Cultivar Fingerprints. Plants 2023, 12, 2313. https://doi.org/10.3390/plants12122313
Yao S, Sapkota D, Hungerford JA, Kersten RD. Jujube Fruit Metabolomic Profiles Reveal Cultivar Differences and Function as Cultivar Fingerprints. Plants. 2023; 12(12):2313. https://doi.org/10.3390/plants12122313
Chicago/Turabian StyleYao, Shengrui, Dikshya Sapkota, Jordan A. Hungerford, and Roland D. Kersten. 2023. "Jujube Fruit Metabolomic Profiles Reveal Cultivar Differences and Function as Cultivar Fingerprints" Plants 12, no. 12: 2313. https://doi.org/10.3390/plants12122313
APA StyleYao, S., Sapkota, D., Hungerford, J. A., & Kersten, R. D. (2023). Jujube Fruit Metabolomic Profiles Reveal Cultivar Differences and Function as Cultivar Fingerprints. Plants, 12(12), 2313. https://doi.org/10.3390/plants12122313






