Composition and Structure of Arabidopsis thaliana Extrachromosomal Circular DNAs Revealed by Nanopore Sequencing
Abstract
1. Introduction
2. Results
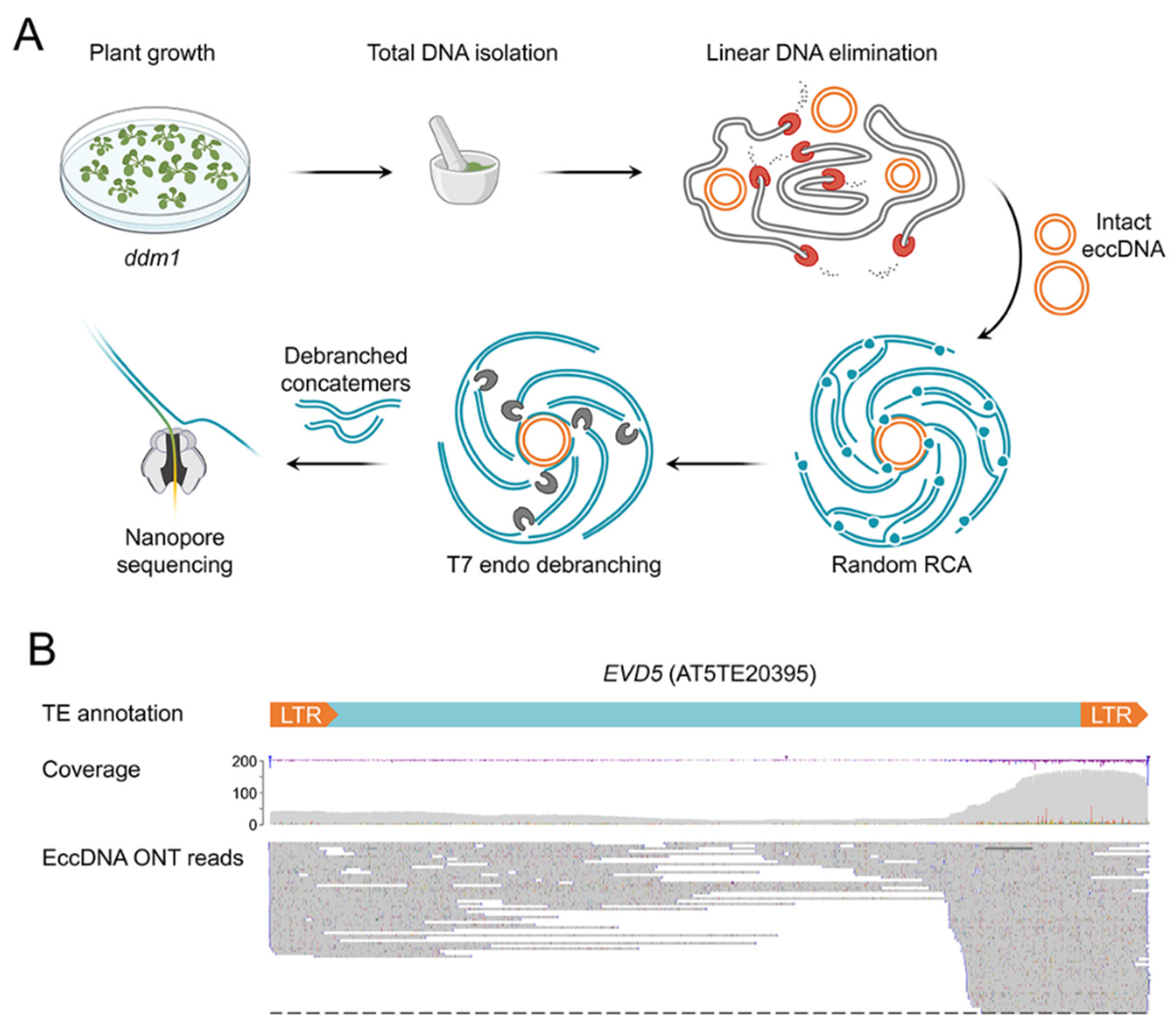
2.1. Extrachromosomal Circular DNA (eccDNA) Sequencing of ddm1 Plants
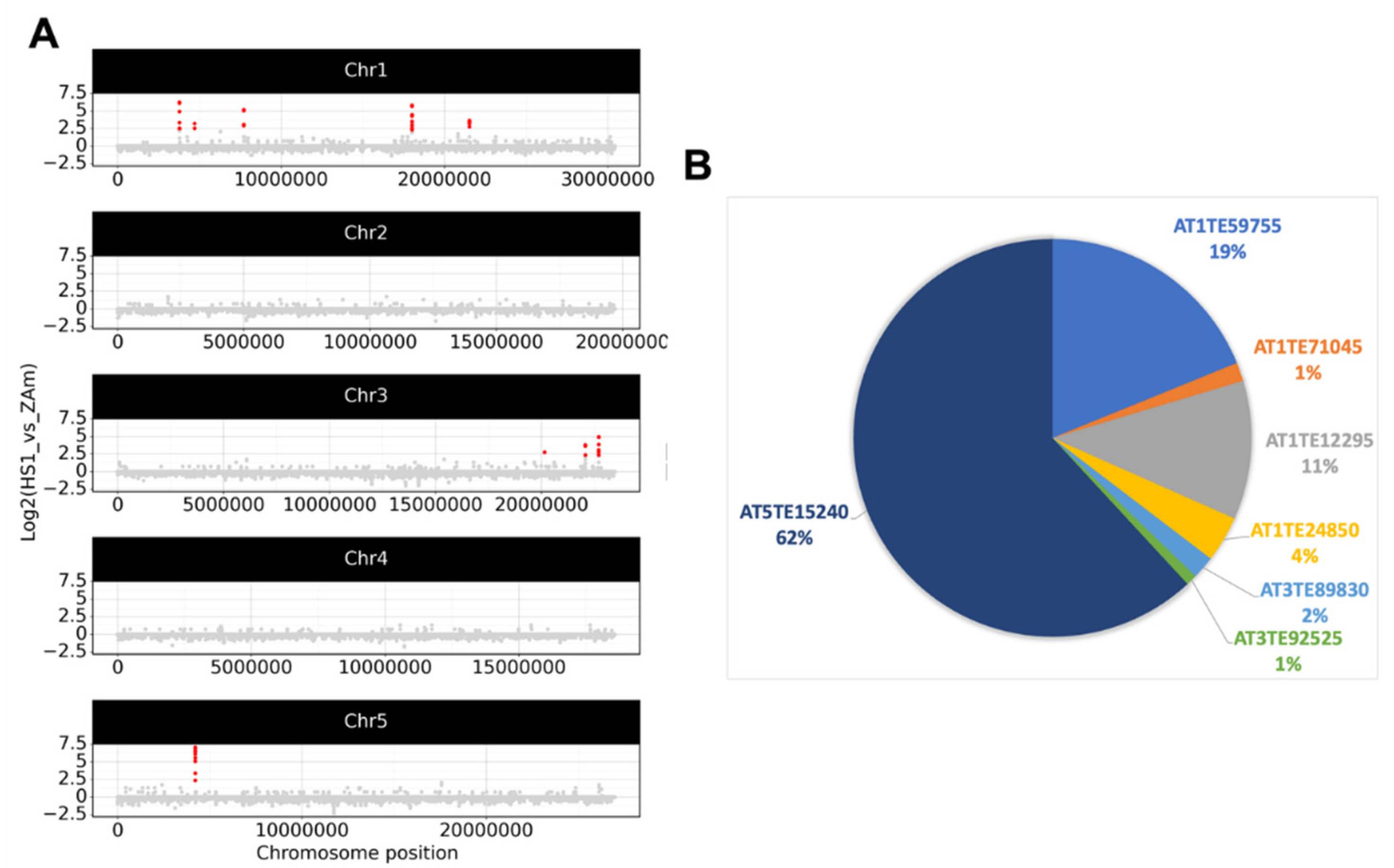
2.2. Epigenetic Stress Does Not Shift eccDNA Composition in A. thaliana
2.3. Detection of eccDNAs under Abiotic, Hormone, and Flagellin Treatment of Epigenetically Stressed Plants
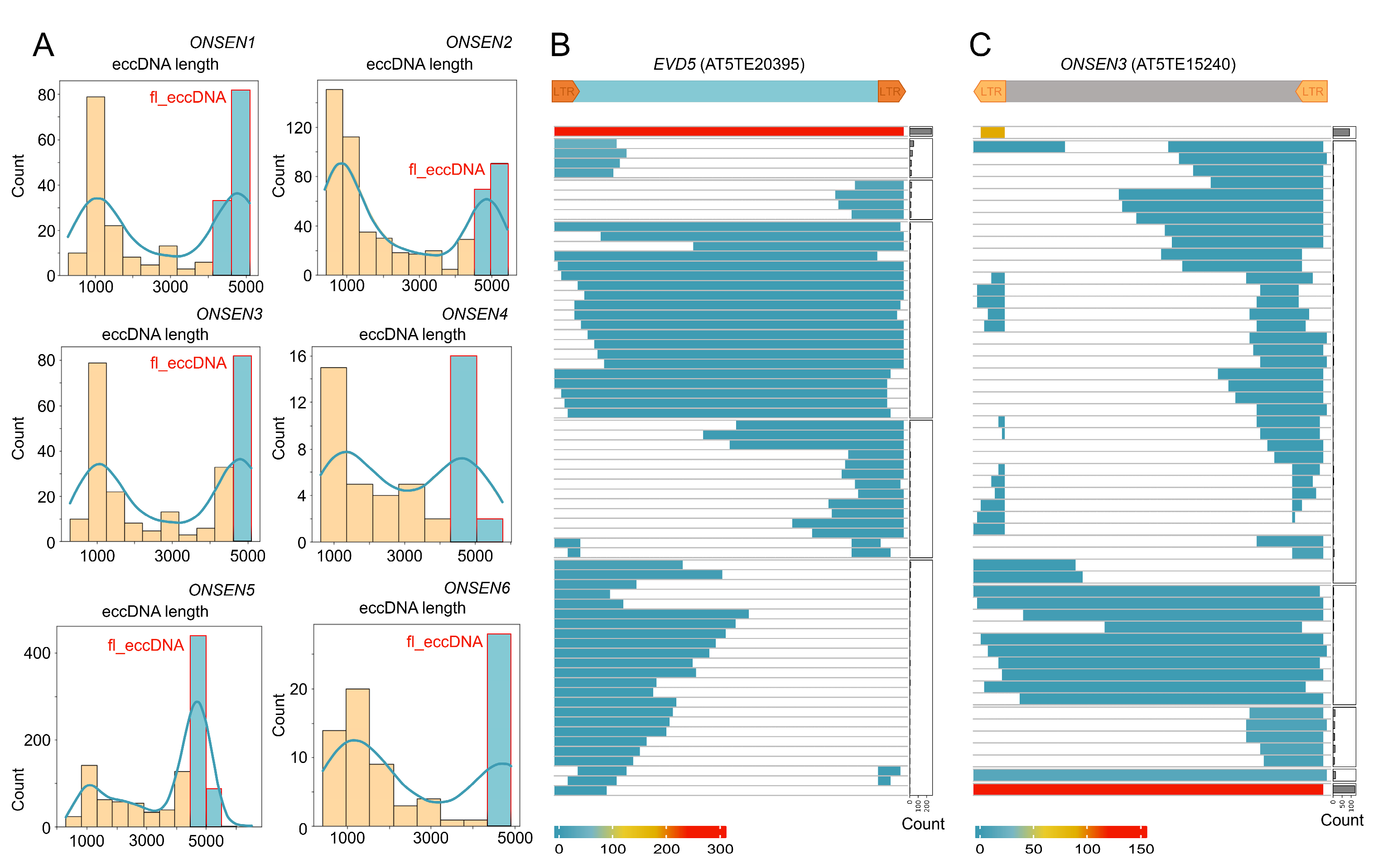
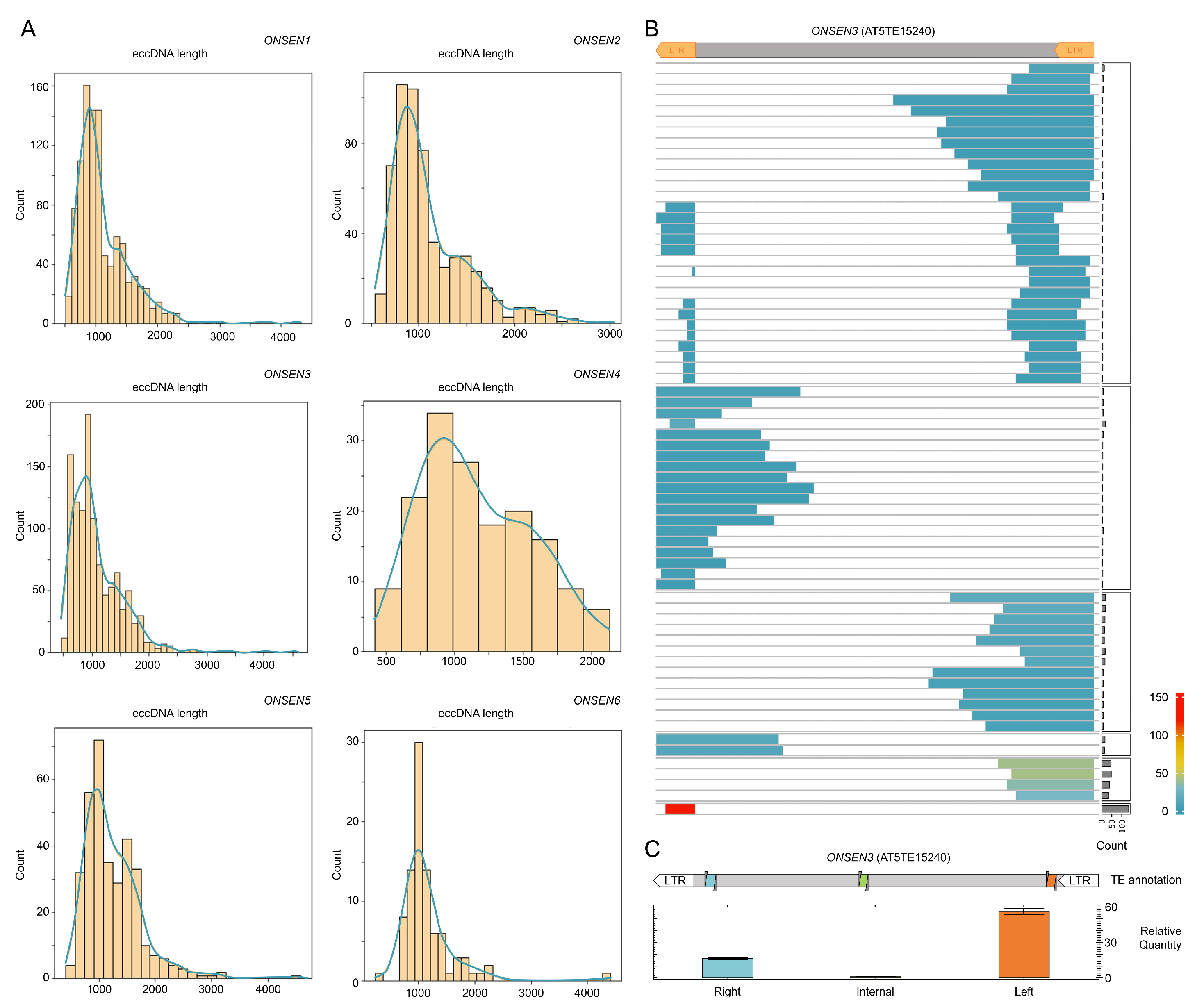
2.4. ONT Reads Revealed Structure of eccDNA Molecules
3. Discussion
4. Materials and Methods
4.1. Plant Material and In Vitro Growth Conditions
4.2. Stress Conditions
4.3. DNA Isolation
4.4. Extrachromosomal Circular DNA (eccDNA) Enrichment
4.5. RT-qPCR Analysis of fl_eccDNA and tr_eccDNAs of ONSEN
4.6. Nanopore Library Preparation and Sequencing
4.7. Bioinformatic Analysis of eccDNA Sequencing and Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hotta, Y.; Bassel, A. Molecular Size And Circularity Of DNA In Cells Of Mammals And Higher Plants*. Proc. Natl. Acad. Sci. USA 1965, 53, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Billheimer, F.E.; Avers, C.J. Nuclear And Mitochondrial DNA From Wild-Type And Petite Yeast: Circularity, Length, And Buoyant Density*. Proc. Natl. Acad. Sci. USA 1969, 64, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Clark-Walker, G.D. Size Distribution of Circular DNA from Petite-Mutant Yeast Lacking ρDNA. Eur. J. Biochem. 1973, 32, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Mirouze, M.; Bucher, E. Extrachromosomal Circular DNA: A Neglected Nucleic Acid Molecule in Plants. Curr. Opin. Plant Biol. 2022, 69, 102263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Peng, H.; Llauro, C.; Bucher, E.; Mirouze, M. Ecc_finder: A Robust and Accurate Tool for Detecting Extrachromosomal Circular DNA From Sequencing Data. Front. Plant Sci. 2021, 12, 743742. [Google Scholar] [CrossRef]
- Lanciano, S.; Carpentier, M.-C.; Llauro, C.; Jobet, E.; Robakowska-Hyzorek, D.; Lasserre, E.; Ghesquière, A.; Panaud, O.; Mirouze, M. Sequencing the Extrachromosomal Circular Mobilome Reveals Retrotransposon Activity in Plants. PLoS Genet. 2017, 13, e1006630. [Google Scholar] [CrossRef]
- Vanderschuren, H.; Zaidi, S.S.-A.; Shakir, S.; Mehta, D.; Nguyen, V.; Gutzat, R. Profiling of EccDNAs in Arabidopsis Indicates Shift in Population of TE-Derived EccDNAs in Response to Stress, Cellular State, and Epigenetic Processes. 2022. Available online: https://assets.researchsquare.com/files/rs-2256837/v3_covered.pdf?c=1680543096 (accessed on 1 March 2023).
- Kwolek, K.; Kędzierska, P.; Hankiewicz, M.; Mirouze, M.; Panaud, O.; Grzebelus, D.; Macko-Podgórni, A. Diverse and Mobile: EccDNA-based Identification of Carrot Low-copy-number LTR Retrotransposons Active in Callus Cultures. Plant J. 2022, 110, 1811–1828. [Google Scholar] [CrossRef]
- Mann, L.; Seibt, K.M.; Weber, B.; Heitkam, T. ECCsplorer: A Pipeline to Detect Extrachromosomal Circular DNA (EccDNA) from next-Generation Sequencing Data. BMC Bioinform. 2022, 23, 40. [Google Scholar] [CrossRef]
- Thieme, M.; Lanciano, S.; Balzergue, S.; Daccord, N.; Mirouze, M.; Bucher, E. Inhibition of RNA Polymerase II Allows Controlled Mobilisation of Retrotransposons for Plant Breeding. Genome Biol. 2017, 18, 134. [Google Scholar] [CrossRef]
- Wang, K.; Tian, H.; Wang, L.; Wang, L.; Tan, Y.; Zhang, Z.; Sun, K.; Yin, M.; Wei, Q.; Guo, B.; et al. Deciphering Extrachromosomal Circular DNA in Arabidopsis. Comput. Struct. Biotechnol. J. 2021, 19, 1176–1183. [Google Scholar] [CrossRef]
- Hirochika, H.; Otsuki, H. Extrachromosomal Circular Forms of the Tobacco Retrotransposon Ttol. Gene 1995, 165, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Barteri, F.; Casacuberta, J.; Mirouze, M.; Carputo, D.; Aversano, R. LTR-TEs Abundance, Timing and Mobility in Solanum commersonii and S. tuberosum Genomes Following Cold-Stress Conditions. Planta 2019, 250, 1781–1787. [Google Scholar] [CrossRef]
- Mehta, D.; Hirsch-Hoffmann, M.; Were, M.; Patrignani, A.; Zaidi, S.S.-A.; Were, H.; Gruissem, W.; Vanderschuren, H. A New Full-Length Circular DNA Sequencing Method for Viral-Sized Genomes Reveals That RNAi Transgenic Plants Provoke a Shift in Geminivirus Populations in the Field. Nucleic Acids Res. 2018, 47, e9. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Cornet, L.; Hirsch-Hoffmann, M.; Zaidi, S.S.-A.; Vanderschuren, H. Full-Length Sequencing of Circular DNA Viruses and Extrachromosomal Circular DNA Using CIDER-Seq. Nat. Protoc. 2020, 15, 1673–1689. [Google Scholar] [CrossRef] [PubMed]
- Kirov, I.; Merkulov, P.; Dudnikov, M.; Polkhovskaya, E.; Komakhin, R.A.; Konstantinov, Z.; Gvaramiya, S.; Ermolaev, A.; Kudryavtseva, N.; Gilyok, M.; et al. Transposons Hidden in Arabidopsis thaliana Genome Assembly Gaps and Mobilization of Non-Autonomous LTR Retrotransposons Unravelled by Nanotei Pipeline. Plants 2021, 10, 2681. [Google Scholar] [CrossRef] [PubMed]
- Kirov, I.; Merkulov, P.; Polkhovskaya, E.; Konstantinov, Z.; Kazancev, M.; Saenko, K.; Polkhovskiy, A.; Dudnikov, M.; Garibyan, T.; Demurin, Y.; et al. Epigenetic Stress and Long-Read cDNA Sequencing of Sunflower (Helianthus annuus L.) Revealed the Origin of the Plant Retrotranscriptome. Plants 2022, 11, 3579. [Google Scholar] [CrossRef] [PubMed]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Scheid, O.M. How a Retrotransposon Exploits the Plant’s Heat Stress Response for Its Activation. PLoS Genet. 2014, 10, e1004115. [Google Scholar] [CrossRef]
- Baduel, P.; Leduque, B.; Ignace, A.; Gy, I.; Gil, J.; Loudet, O.; Colot, V.; Quadrana, L. Genetic and Environmental Modulation of Transposition Shapes the Evolutionary Potential of Arabidopsis thaliana. Genome Biol. 2021, 22, 138. [Google Scholar] [CrossRef]
- Quadrana, L.; Silveira, A.B.; Mayhew, G.F.; LeBlanc, C.; Martienssen, R.A.; Jeddeloh, J.A.; Colot, V. The Arabidopsis thaliana Mobilome and Its Impact at the Species Level. eLife 2016, 5, e15716. [Google Scholar] [CrossRef]
- Arrey, G.; Keating, S.T.; Regenberg, B. A Unifying Model for Extrachromosomal Circular DNA Load in Eukaryotic Cells. Semin. Cell. Dev. Biol. 2022, 128, 40–50. [Google Scholar] [CrossRef]
- Møller, H.D.; Larsen, C.E.; Parsons, L.; Hansen, A.J.; Regenberg, B.; Mourier, T. Formation of Extrachromosomal Circular DNA from Long Terminal Repeats of Retrotransposons in Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2016, 6, 453–462. [Google Scholar] [CrossRef]
- Møller, H.D.; Parsons, L.; Jørgensen, T.S.; Botstein, D.; Regenberg, B. Extrachromosomal Circular DNA Is Common in Yeast. Proc. Natl. Acad. Sci. USA 2015, 112, E3114–E3122. [Google Scholar] [CrossRef] [PubMed]
- Deshayes, A.; Herrera-Estrella, L.; Caboche, M. Liposome-mediated Transformation of Tobacco Mesophyll Protoplasts by an Escherichia coli Plasmid. EMBO J. 1985, 4, 2731–2737. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Han, Y.; Meng, J.; Zhong, B.; Chen, J.; Zhang, H.; Qin, J.; Pang, J.; Liu, L. Small Extrachromosomal Circular DNA (EccDNA): Major Functions in Evolution and Cancer. Mol. Cancer 2021, 20, 113. [Google Scholar] [CrossRef] [PubMed]
- Molin, W.T.; Yaguchi, A.; Blenner, M.; Saski, C.A. The EccDNA Replicon: A Heritable, Extranuclear Vehicle That Enables Gene Amplification and Glyphosate Resistance in Amaranthus palmeri. Plant Cell 2020, 32, 2132–2140. [Google Scholar] [CrossRef]
- Pucker, B.; Kleinbölting, N.; Weisshaar, B. Large Scale Genomic Rearrangements in Selected Arabidopsis thaliana T-DNA Lines Are Caused by T-DNA Insertion Mutagenesis. BMC Genom. 2021, 22, 599. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, B.; Wang, Y.; Xing, Y. TideHunter: Efficient and Sensitive Tandem Repeat Detection from Noisy Long-Reads Using Seed-and-Chain. Bioinformatics 2019, 35, i200–i207. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Subgroup, 1000 Genome Project Data Processing The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Diesh, C.; Stevens, G.J.; Xie, P.; Martinez, T.D.J.; Hershberg, E.A.; Leung, A.; Guo, E.; Dider, S.; Zhang, J.; Bridge, C.; et al. JBrowse 2: A Modular Genome Browser with Views of Synteny and Structural Variation. Genome Biol. 2023, 24, 74. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z. Complex Heatmap Visualization. Imeta 2022, 1, e43. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merkulov, P.; Egorova, E.; Kirov, I. Composition and Structure of Arabidopsis thaliana Extrachromosomal Circular DNAs Revealed by Nanopore Sequencing. Plants 2023, 12, 2178. https://doi.org/10.3390/plants12112178
Merkulov P, Egorova E, Kirov I. Composition and Structure of Arabidopsis thaliana Extrachromosomal Circular DNAs Revealed by Nanopore Sequencing. Plants. 2023; 12(11):2178. https://doi.org/10.3390/plants12112178
Chicago/Turabian StyleMerkulov, Pavel, Ekaterina Egorova, and Ilya Kirov. 2023. "Composition and Structure of Arabidopsis thaliana Extrachromosomal Circular DNAs Revealed by Nanopore Sequencing" Plants 12, no. 11: 2178. https://doi.org/10.3390/plants12112178
APA StyleMerkulov, P., Egorova, E., & Kirov, I. (2023). Composition and Structure of Arabidopsis thaliana Extrachromosomal Circular DNAs Revealed by Nanopore Sequencing. Plants, 12(11), 2178. https://doi.org/10.3390/plants12112178









