A Study of GUS Expression in Arabidopsis as a Tool for the Evaluation of Gene Evolution, Function and the Role of Expression Derived from Gene Duplication
Abstract
1. Introduction
2. Results
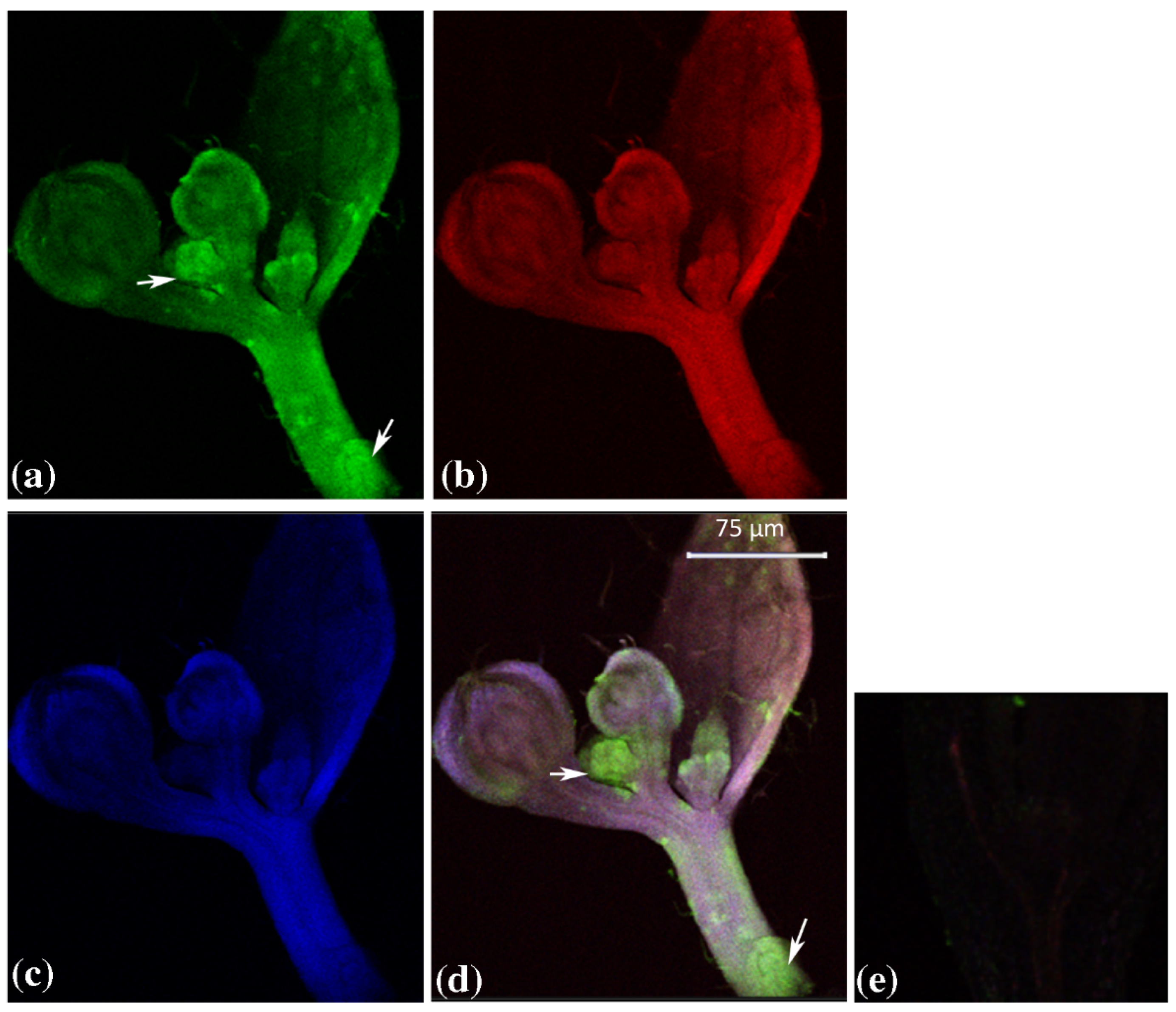
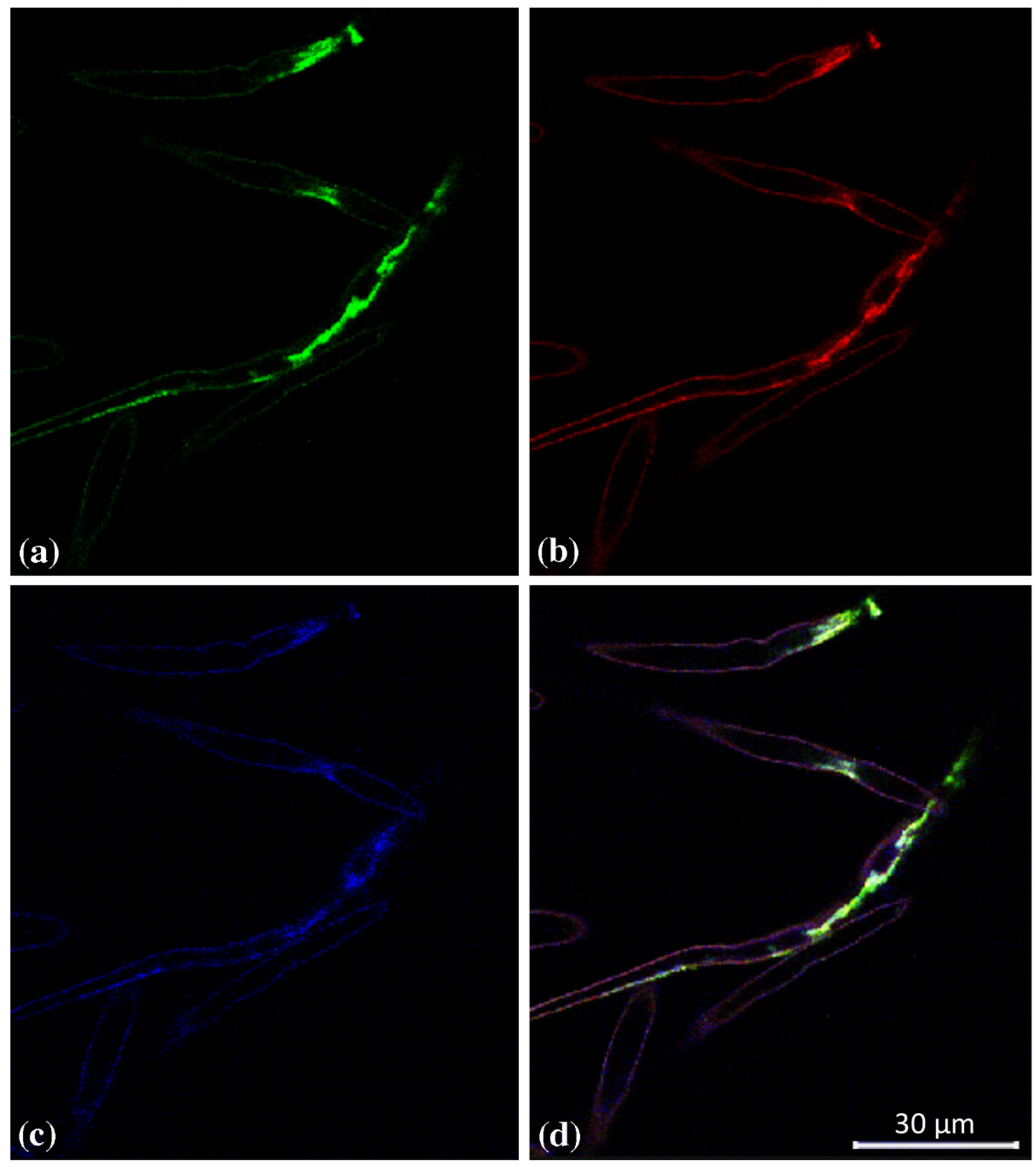
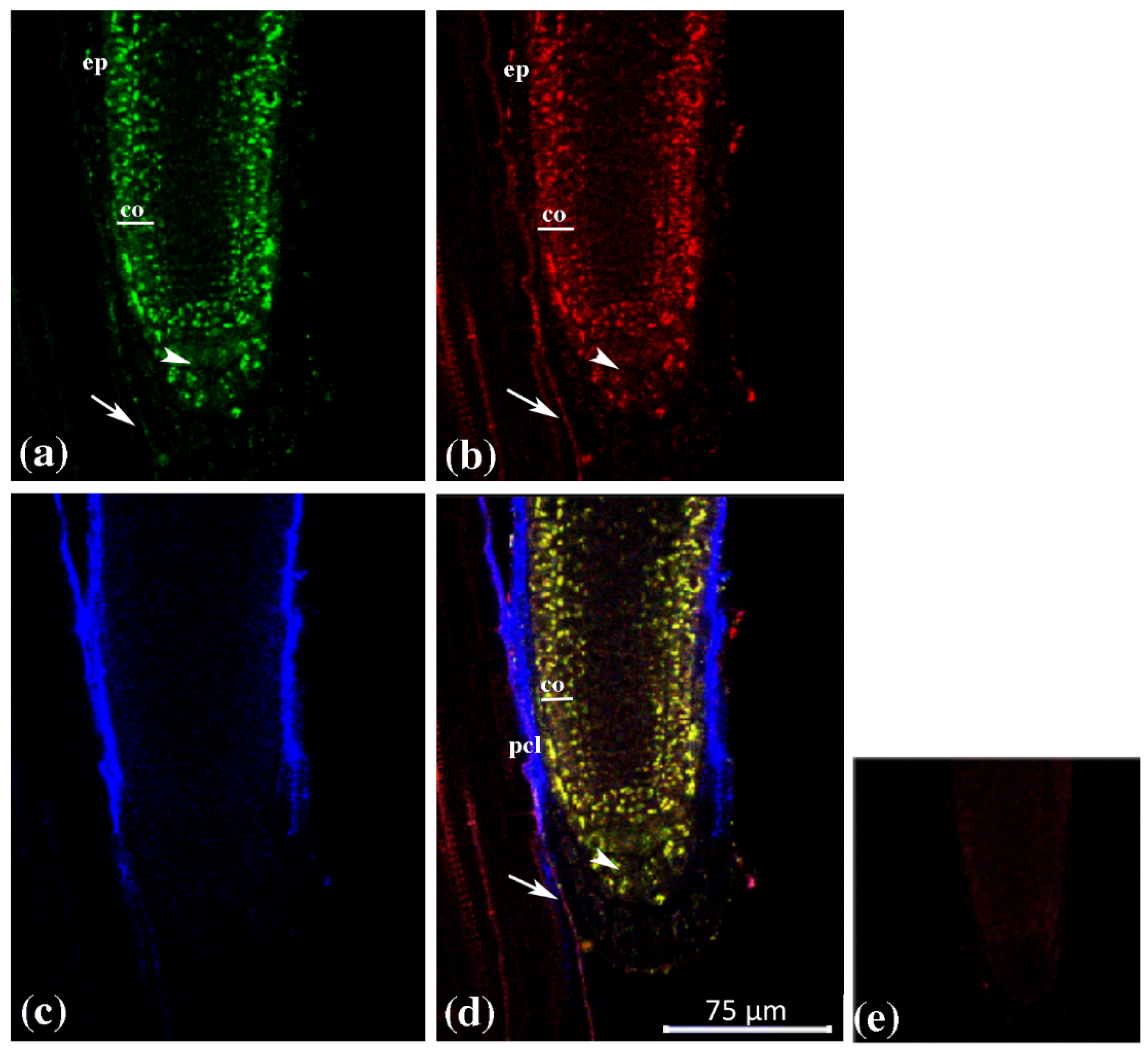
2.1. Tissue Specific Expression of GUS Genes in Arabidopsis thaliana
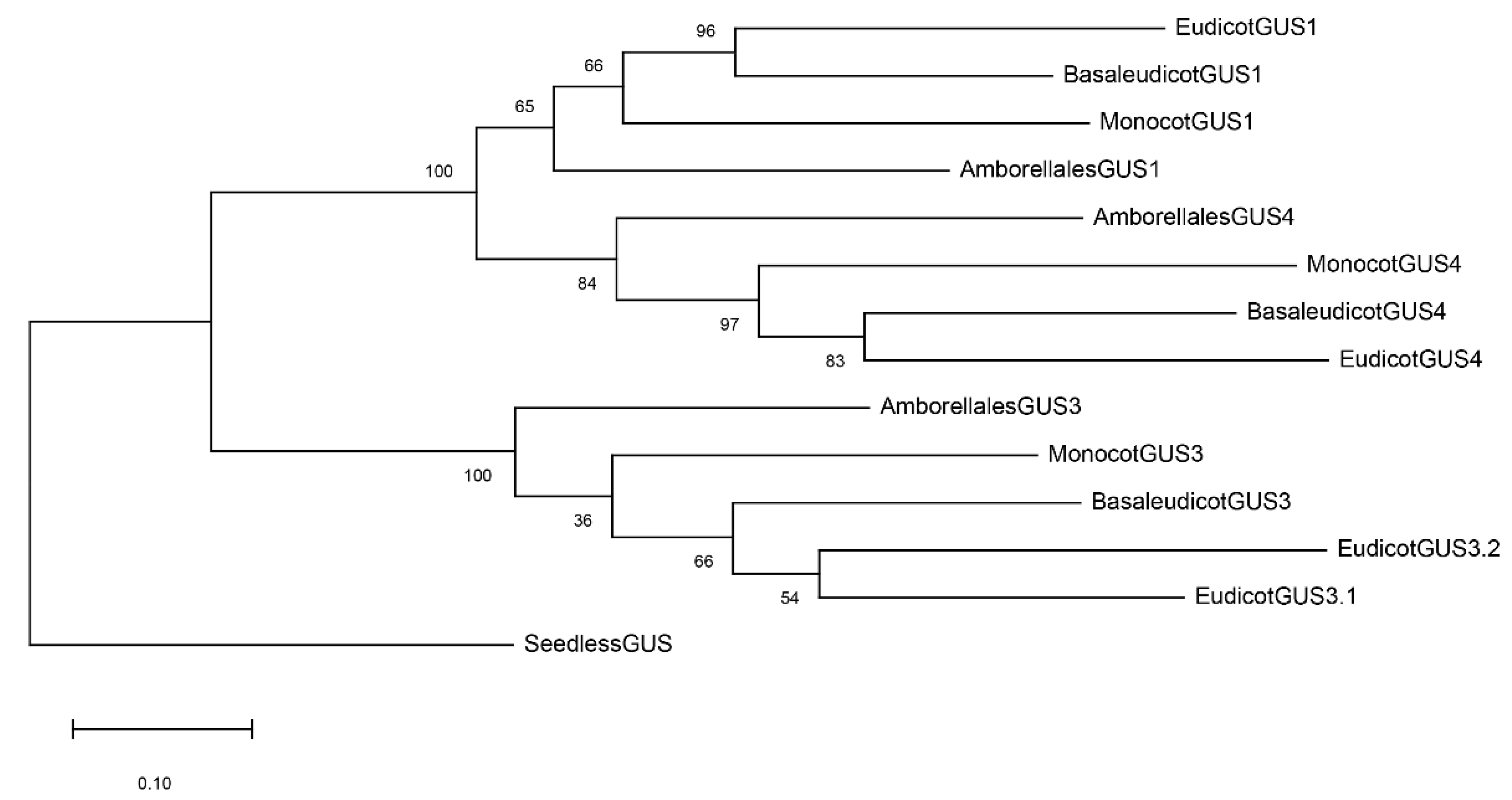
2.2. GUS Duplications and Its Evolutionary Tree
3. Discussion
Conclusions and Future Perspectives
4. Materials and Methods
4.1. Plant Material: Growth and Fixation
4.2. Multiprobe In Situ Hybridization
4.3. Confocal Visualization
4.4. Sequence Retrieval from Databases
4.5. Evolutionary Tree Construction
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Maere, S.; Bodt, S.D.; Raes, J.; Casneuf, T.; Montagu, M.V.; Kuiper, M.; de Peer, Y.V. Modeling Gene and Genome Duplications in Eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Flagel, L.E.; Wendel, J.F. Gene Duplication and Evolutionary Novelty in Plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Salse, J.; Abrouk, M.; Bolot, S.; Guilhot, N.; Courcelle, E.; Faraut, T.; Waugh, R.; Close, T.J.; Messing, J.; Feuillet, C. Reconstruction of Monocotelydoneous Proto-Chromosomes Reveals Faster Evolution in Plants than in Animals. Proc. Natl. Acad. Sci. USA 2009, 106, 14908–14913. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murat, F.; Van de Peer, Y.; Salse, J. Decoding Plant and Animal Genome Plasticity from Differential Paleo-Evolutionary Patterns and Processes. Genome Biol. Evol. 2012, 4, 917–928. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J. Evolution by Gene Duplication: An Update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef][Green Version]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral Polyploidy in Seed Plants and Angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Doyle, J.J.; Luckow, M.A. The Rest of the Iceberg. Legume Diversity and Evolution in a Phylogenetic Context. Plant Physiol. 2003, 131, 900–910. [Google Scholar] [CrossRef][Green Version]
- De Bodt, S.; Maere, S.; Van de Peer, Y. Genome Duplication and the Origin of Angiosperms. Trends Ecol. Evol. 2005, 20, 591–597. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The Grapevine Genome Sequence Suggests Ancestral Hexaploidization in Major Angiosperm Phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef][Green Version]
- Fawcett, J.A.; Maere, S.; de Peer, Y.V. Plants with Double Genomes Might Have Had a Better Chance to Survive the Cretaceous–Tertiary Extinction Event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef][Green Version]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; Depamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and Angiosperm Diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef][Green Version]
- Van de Peer, Y.; Maere, S.; Meyer, A. The Evolutionary Significance of Ancient Genome Duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef][Green Version]
- Jiao, Y.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J.; et al. A Genome Triplication Associated with Early Diversification of the Core Eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eric Schranz, M.; Mohammadin, S.; Edger, P.P. Ancient Whole Genome Duplications, Novelty and Diversification: The WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 2012, 15, 147–153. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The Draft Genome of a Diploid Cotton Gossypium Raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Project, A.G. The Amborella Genome and the Evolution of Flowering Plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Robinson, S.J.; Nixon, J.; Xiao, R.; Huebert, T.; Condie, J.; Kessler, D.; Clarke, W.E.; Edger, P.P.; Links, M.G.; et al. Polyploid Evolution of the Brassicaceae during the Cenozoic Era. Plant Cell 2014, 26, 2777–2791. [Google Scholar] [CrossRef][Green Version]
- Vanneste, K.; Baele, G.; Maere, S.; Van de Peer, Y. Analysis of 41 Plant Genomes Supports a Wave of Successful Genome Duplications in Association with the Cretaceous-Paleogene Boundary. Genome Res. 2014, 24, 1334–1347. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mabry, M.E.; Brose, J.M.; Blischak, P.D.; Sutherland, B.; Dismukes, W.T.; Bottoms, C.A.; Edger, P.P.; Washburn, J.D.; An, H.; Hall, J.C.; et al. Phylogeny and Multiple Independent Whole-Genome Duplication Events in the Brassicales. Am. J. Bot. 2020, 107, 1148–1164. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; Mason, A. Plant Speciation through Chromosome Instability and Ploidy Change: Cellular Mechanisms, Molecular Factors and Evolutionary Relevance. Curr. Plant Biol. 2014, 1, 10–33. [Google Scholar] [CrossRef][Green Version]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef][Green Version]
- Veitia, R.A.; Bottani, S.; Birchler, J.A. Cellular Reactions to Gene Dosage Imbalance: Genomic, Transcriptomic and Proteomic Effects. Trends Genet. 2008, 24, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Veitia, R.A. The Gene Balance Hypothesis: Implications for Gene Regulation, Quantitative Traits and Evolution. New Phytol. 2010, 186, 54–62. [Google Scholar] [CrossRef][Green Version]
- Edger, P.P.; Pires, J.C. Gene and Genome Duplications: The Impact of Dosage-Sensitivity on the Fate of Nuclear Genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef][Green Version]
- Blanc, G.; Wolfe, K.H. Functional Divergence of Duplicated Genes Formed by Polyploidy during Arabidopsis Evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seoighe, C.; Gehring, C. Genome Duplication Led to Highly Selective Expansion of the Arabidopsis Thaliana Proteome. Trends Genet. 2004, 20, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M.; Rapaka, L.; Lyons, E.; Pedersen, B.; Thomas, B.C. G-Boxes, Bigfoot Genes, and Environmental Response: Characterization of Intragenomic Conserved Noncoding Sequences in Arabidopsis. Plant Cell 2007, 19, 1441–1457. [Google Scholar] [CrossRef][Green Version]
- Schnable, J.C.; Springer, N.M.; Freeling, M. Differentiation of the Maize Subgenomes by Genome Dominance and Both Ancient and Ongoing Gene Loss. Proc. Natl. Acad. Sci. USA 2011, 108, 4069–4074. [Google Scholar] [CrossRef][Green Version]
- Schnable, J.C.; Freeling, M.; Lyons, E. Genome-Wide Analysis of Syntenic Gene Deletion in the Grasses. Genome Biol. Evol. 2012, 4, 265–277. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Freeling, M.; Thomas, B.C. Gene-Balanced Duplications, like Tetraploidy, Provide Predictable Drive to Increase Morphological Complexity. Genome Res. 2006, 16, 805–814. [Google Scholar] [CrossRef][Green Version]
- Duarte, J.M.; Cui, L.; Wall, P.K.; Zhang, Q.; Zhang, X.; Leebens-Mack, J.; Ma, H.; Altman, N.; de Pamphilis, C.W. Expression Pattern Shifts Following Duplication Indicative of Subfunctionalization and Neofunctionalization in Regulatory Genes of Arabidopsis. Mol. Biol. Evol. 2006, 23, 469–478. [Google Scholar] [CrossRef] [PubMed]
- De Smet, R.; Van de Peer, Y. Redundancy and Rewiring of Genetic Networks Following Genome-Wide Duplication Events. Curr. Opin. Plant Biol. 2012, 15, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.A.; Van de Peer, Y.; Maere, S. Significance and Biological Consequences of Polyploidization in Land Plant Evolution. In Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Vienna, Austria, 2013; pp. 277–293. ISBN 978-3-7091-1160-4. [Google Scholar]
- Bruno, L.; Ronchini, M.; Gagliardi, O.; Corinti, T.; Chiappetta, A.; Gerola, P.; Bitonti, M.B. Analysis of AtGUS1 and AtGUS2 in Arabidopsis Root Apex by a Highly Sensitive TSA-MISH Method. Int. J. Dev. Biol. 2015, 59, 221–228. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arul, L.; Benita, G.; Sudhakar, D.; Thayumanavan, B.; Balasubramanian, P. Β-glucuronidase of Family-2 Glycosyl Hydrolase: A Missing Member in Plants. Bioinformation 2008, 3, 194–197. [Google Scholar] [CrossRef][Green Version]
- Henrissat, B.; Coutinho, P.M.; Davies, G.J. A Census of Carbohydrate-Active Enzymes in the Genome of Arabidopsis Thaliana. Plant Mol. Biol. 2001, 47, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B. A Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1991, 280 Pt 2, 309–316. [Google Scholar] [CrossRef]
- Henrissat, B.; Bairoch, A. New Families in the Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1993, 293, 781–788. [Google Scholar] [CrossRef]
- Henrissat, B.; Bairoch, A. Updating the Sequence-Based Classification of Glycosyl Hydrolases. Biochem. J. 1996, 316, 695–696. [Google Scholar] [CrossRef][Green Version]
- Sudan, C.; Prakash, S.; Bhomkar, P.; Jain, S.; Bhalla-Sarin, N. Ubiquitous Presence of β-Glucuronidase (GUS) in Plants and Its Regulation in Some Model Plants. Planta 2006, 224, 853–864. [Google Scholar] [CrossRef]
- Sasaki, K.; Taura, F.; Shoyama, Y.; Morimoto, S. Molecular Characterization of a Novel Beta-Glucuronidase from Scutellaria Baicalensis Georgi. J. Biol. Chem. 2000, 275, 27466–27472. [Google Scholar] [CrossRef]
- Woo, H.-H.; Jeong, B.R.; Hirsch, A.M.; Hawes, M.C. Characterization of Arabidopsis AtUGT85A and AtGUS Gene Families and Their Expression in Rapidly Dividing Tissues. Genomics 2007, 90, 143–153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Honys, D.; Twell, D. Transcriptome Analysis of Haploid Male Gametophyte Development in Arabidopsis. Genome Biol. 2004, 5, R85. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pina, C.; Pinto, F.; Feijó, J.A.; Becker, J.D. Gene Family Analysis of the Arabidopsis Pollen Transcriptome Reveals Biological Implications for Cell Growth, Division Control, and Gene Expression Regulation. Plant Physiol. 2005, 138, 744–756. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Konishi, T.; Kotake, T.; Soraya, D.; Matsuoka, K.; Koyama, T.; Kaneko, S.; Igarashi, K.; Samejima, M.; Tsumuraya, Y. Properties of Family 79 β-Glucuronidases That Hydrolyze β-Glucuronosyl and 4-O-Methyl-β-Glucuronosyl Residues of Arabinogalactan-Protein. Carbohydr. Res. 2008, 343, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- Matas, A.J.; Yeats, T.H.; Buda, G.J.; Zheng, Y.; Chatterjee, S.; Tohge, T.; Ponnala, L.; Adato, A.; Aharoni, A.; Stark, R.; et al. Tissue- and Cell-Type Specific Transcriptome Profiling of Expanding Tomato Fruit Provides Insights into Metabolic and Regulatory Specialization and Cuticle Formation. Plant Cell 2011, 23, 3893–3910. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hafidh, S.; Breznenová, K.; Růžička, P.; Feciková, J.; Čapková, V.; Honys, D. Comprehensive Analysis of Tobacco Pollen Transcriptome Unveils Common Pathways in Polar Cell Expansion and Underlying Heterochronic Shift during Spermatogenesis. BMC Plant Biol. 2012, 12, 24. [Google Scholar] [CrossRef][Green Version]
- Sood, P.P. Histoenzymological Compartmentation of β-Glucuronidase in the Germinating Pollen Grains OfPortulaca Grandiflora. Biol. Plant 1980, 22, 124–127. [Google Scholar] [CrossRef]
- Schulz, M.; Weissenböck, G. Partial Purification and Characterization of a Luteolin-Triglucuronide-Specific β-Glucuronidase from Rye Primary Leaves (Secale Cereale). Phytochemistry 1987, 26, 933–937. [Google Scholar] [CrossRef]
- Plegt, L.; Bino, R.J. β-Glucuronidase Activity during Development of the Male Gametophyte from Transgenic and Non-Transgenic Plants. Mol. Gen. Genet. 1989, 216, 321–327. [Google Scholar] [CrossRef]
- Hu, C.Y.; Chee, P.P.; Chesney, R.H.; Zhou, J.H.; Miller, P.D.; O’Brien, W.T. Intrinsic GUS-like Activities in Seed Plants. Plant Cell Rep. 1990, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Alwen, A.; Moreno, R.M.B.; Vicente, O.; Heberle-Bors, E. Plant Endogenous β-Glucuronidase Activity: How to Avoid Interference with the Use of TheE. Coli β-Glucuronidase as a Reporter Gene in Transgenic Plants. Transgenic Res. 1992, 1, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Anhalt, S.; Weissenböck, G. Subcellular Localization of Luteolin Glucuronides and Related Enzymes in Rye Mesophyll. Planta 1992, 187, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, C.A.; Owens, L.D. Native β-Glucuronidase Activity in Sugarbeet (Beta Vulgaris). Physiol. Plant. 1994, 90, 763–771. [Google Scholar] [CrossRef]
- Morimoto, S.; Harioka, T.; Shoyama, Y. Purification and Characterization of Flavone-Specific β-Glucuronidase from Callus Cultures of Scutellaria Baicalensis Georgi. Planta 1995, 195, 535–540. [Google Scholar] [CrossRef]
- Morimoto, S.; Tateishi, N.; Matsuda, T.; Tanaka, H.; Taura, F.; Furuya, N.; Matsuyama, N.; Shoyama, Y. Novel Hydrogen Peroxide Metabolism in Suspension Cells of Scutellaria Baicalensis Georgi. Int. J. STD AIDS 1998, 273, 12606–12611. [Google Scholar] [CrossRef][Green Version]
- Schoenbeck, M.A.; Swanson, G.A.; Brommer, S.J. β-Glucuronidase Activity in Seedlings of the Parasitic Angiosperm Cusctua Pentagona: Developmental Impact of the β-Glucuronidase Inhibitor Saccharic Acid 1,4-Lactone. Funct. Plant Biol. 2007, 34, 811–821. [Google Scholar] [CrossRef][Green Version]
- Fior, S.; Gerola, P.D. Impact of Ubiquitous Inhibitors on the GUS Gene Reporter System: Evidence from the Model Plants Arabidopsis, Tobacco and Rice and Correction Methods for Quantitative Assays of Transgenic and Endogenous GUS. Plant Methods 2009, 5, 19. [Google Scholar] [CrossRef][Green Version]
- Eudes, A.; Mouille, G.; Thévenin, J.; Goyallon, A.; Minic, Z.; Jouanin, L. Purification, Cloning and Functional Characterization of an Endogenous Beta-Glucuronidase in Arabidopsis Thaliana. Plant Cell Physiol. 2008, 49, 1331–1341. [Google Scholar] [CrossRef][Green Version]
- Wen, F.; Woo, H.-H.; Hirsch, A.M.; Hawes, M.C. Lethality of Inducible, Meristem-Localized Ectopic β-Glucuronidase Expression in Plants. Plant Mol. Biol. Rep. 2004, 22, 7–14. [Google Scholar] [CrossRef]
- Woo, H.-H.; Jeong, B.R.; Hawes, M.C. Flavonoids: From Cell Cycle Regulation to Biotechnology. Biotechnol. Lett. 2005, 27, 365–374. [Google Scholar] [CrossRef]
- Hirunuma, M.; Shoyama, Y.; Sasaki, K.; Sakamoto, S.; Taura, F.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Flavone-Catalyzed Apoptosis in Scutellaria Baicalensis. Phytochemistry 2011, 72, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Doseff, A.I.; Grotewold, E. Flavones: From Biosynthesis to Health Benefits. Plants 2016, 5, 27. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iwai, H.; Masaoka, N.; Ishii, T.; Satoh, S. A Pectin Glucuronyltransferase Gene Is Essential for Intercellular Attachment in the Plant Meristem. Proc. Natl. Acad. Sci. USA 2002, 99, 16319–16324. [Google Scholar] [CrossRef][Green Version]
- Zhong, R.; Peña, M.J.; Zhou, G.-K.; Nairn, C.J.; Wood-Jones, A.; Richardson, E.A.; Morrison, W.H.; Darvill, A.G.; York, W.S.; Ye, Z.-H. Arabidopsis Fragile Fiber8, Which Encodes a Putative Glucuronyltransferase, Is Essential for Normal Secondary Wall Synthesis. Plant Cell 2005, 17, 3390–3408. [Google Scholar] [CrossRef][Green Version]
- Blair, M.W.; Cortés, A.J.; Farmer, A.D.; Huang, W.; Ambachew, D.; Penmetsa, R.V.; Carrasquilla-Garcia, N.; Assefa, T.; Cannon, S.B. Uneven Recombination Rate and Linkage Disequilibrium across a Reference SNP Map for Common Bean (Phaseolus vulgaris L.). PLoS ONE 2018, 13, e0189597. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burleigh, J.G.; Bansal, M.S.; Eulenstein, O.; Hartmann, S.; Wehe, A.; Vision, T.J. Genome-Scale Phylogenetics: Inferring the Plant Tree of Life from 18,896 Gene Trees. Syst. Biol. 2011, 60, 117–125. [Google Scholar] [CrossRef][Green Version]
- Ruhfel, B.R.; Gitzendanner, M.A.; Soltis, P.S.; Soltis, D.E.; Burleigh, J.G. From Algae to Angiosperms–Inferring the Phylogeny of Green Plants (Viridiplantae) from 360 Plastid Genomes. BMC Evol. Biol. 2014, 14, 23. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murad, L.; Lim, K.Y.; Christopodulou, V.; Matyasek, R.; Lichtenstein, C.P.; Kovarik, A.; Leitch, A.R. The Origin of Tobacco’s T Genome Is Traced to a Particular Lineage within Nicotiana Tomentosiformis (Solanaceae). Am. J. Bot. 2002, 89, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Levvy, G.A. Baicalinase, a Plant β-Glucuronidase. Biochem. J. 1954, 58, 462–469. [Google Scholar] [CrossRef]
- Miwa, T. Baicalinase, a Flavone Glucuronide-Splitting Enzyme (I). Acta Phytochim. 1932, 6, 155–171. [Google Scholar]
- β-Glucuronidases in Plants. Available online: https://irinsubria.uninsubria.it/handle/11383/2090374 (accessed on 26 April 2023).
- Phylogenetic Analysis of B-Glucoronidasi Genes in Angiosperm. Available online: https://iris.unical.it/handle/20.500.11770/172841 (accessed on 26 April 2023).
- Cortés, A.J.; López-Hernández, F.; Osorio-Rodriguez, D. Predicting Thermal Adaptation by Looking Into Populations’ Genomic Past. Front. Genet. 2020, 11, 564515. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.A.; Chitikineni, A.; Lam, H.-M.; Hickey, L.T.; Croser, J.S.; Bayer, P.E.; et al. Fast-Forward Breeding for a Food-Secure World. Trends Genet. 2021, 37, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- Valvekens, D.; Montagu, M.V.; Lijsebettens, M.V. Agrobacterium Tumefaciens-Mediated Transformation of Arabidopsis Thaliana Root Explants by Using Kanamycin Selection. Proc. Natl. Acad. Sci. USA 1988, 85, 5536–5540. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bruno, L.; Muto, A.; Spadafora, N.D.; Iaria, D.; Chiappetta, A.; Lijsebettens, M.V.; Bitonti, M.B. Multi-Probe in Situ Hybridization to Whole Mount Arabidopsis Seedlings. Int. J. Dev. Biol. 2011, 55, 197–203. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef][Green Version]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. GUIDANCE2: Accurate Detection of Unreliable Alignment Regions Accounting for the Uncertainty of Multiple Parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef][Green Version]
- Penn, O.; Privman, E.; Ashkenazy, H.; Landan, G.; Graur, D.; Pupko, T. GUIDANCE: A Web Server for Assessing Alignment Confidence Scores. Nucleic Acids Res. 2010, 38, W23–W28. [Google Scholar] [CrossRef][Green Version]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis Software for Microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef][Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruno, L.; Ronchini, M.; Binelli, G.; Muto, A.; Chiappetta, A.; Bitonti, M.B.; Gerola, P. A Study of GUS Expression in Arabidopsis as a Tool for the Evaluation of Gene Evolution, Function and the Role of Expression Derived from Gene Duplication. Plants 2023, 12, 2051. https://doi.org/10.3390/plants12102051
Bruno L, Ronchini M, Binelli G, Muto A, Chiappetta A, Bitonti MB, Gerola P. A Study of GUS Expression in Arabidopsis as a Tool for the Evaluation of Gene Evolution, Function and the Role of Expression Derived from Gene Duplication. Plants. 2023; 12(10):2051. https://doi.org/10.3390/plants12102051
Chicago/Turabian StyleBruno, Leonardo, Matteo Ronchini, Giorgio Binelli, Antonella Muto, Adriana Chiappetta, Maria Beatrice Bitonti, and Paolo Gerola. 2023. "A Study of GUS Expression in Arabidopsis as a Tool for the Evaluation of Gene Evolution, Function and the Role of Expression Derived from Gene Duplication" Plants 12, no. 10: 2051. https://doi.org/10.3390/plants12102051
APA StyleBruno, L., Ronchini, M., Binelli, G., Muto, A., Chiappetta, A., Bitonti, M. B., & Gerola, P. (2023). A Study of GUS Expression in Arabidopsis as a Tool for the Evaluation of Gene Evolution, Function and the Role of Expression Derived from Gene Duplication. Plants, 12(10), 2051. https://doi.org/10.3390/plants12102051






