Apple-Net: A Model Based on Improved YOLOv5 to Detect the Apple Leaf Diseases
Abstract
1. Introduction
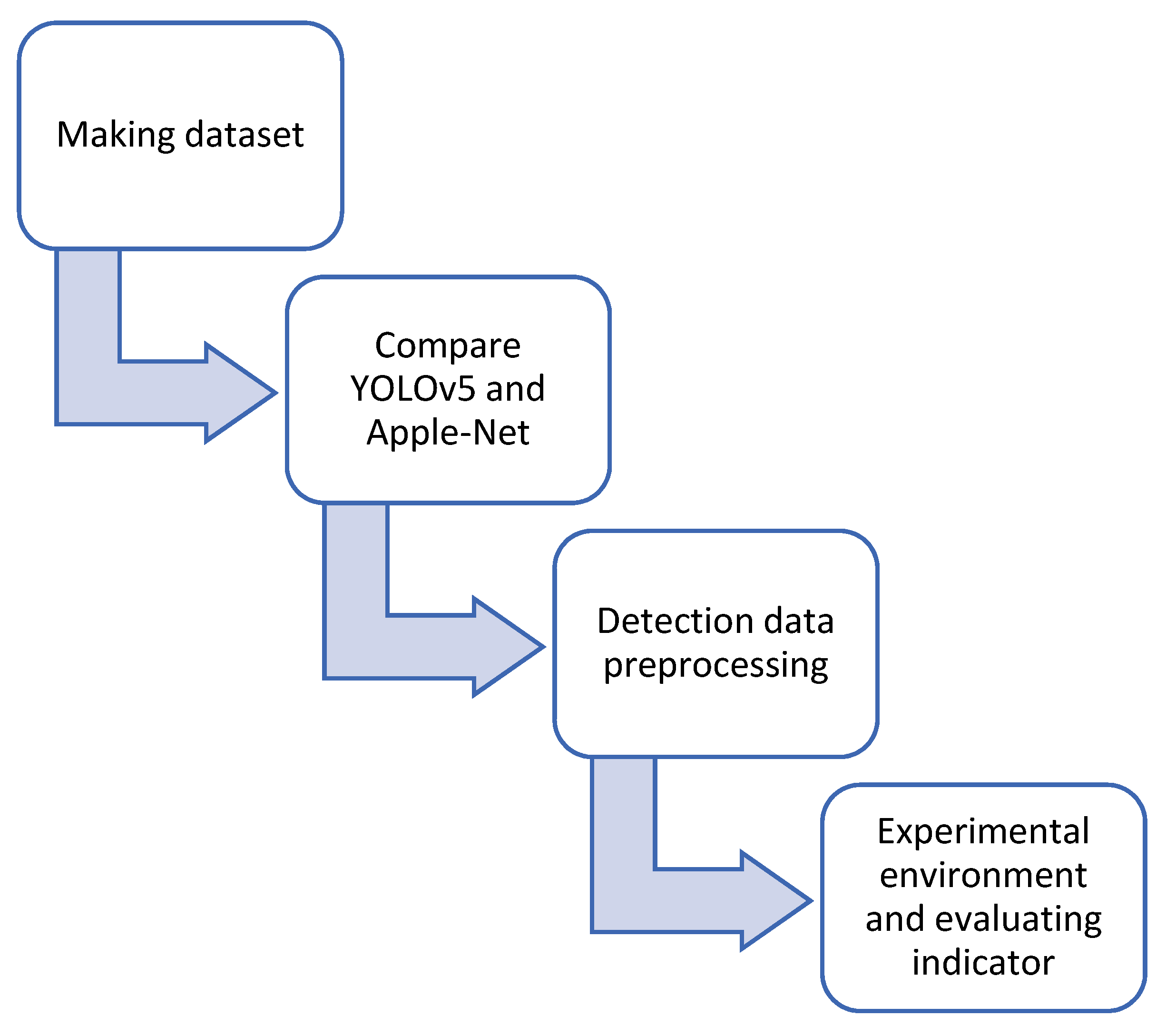
2. Materials and Methods
2.1. Data
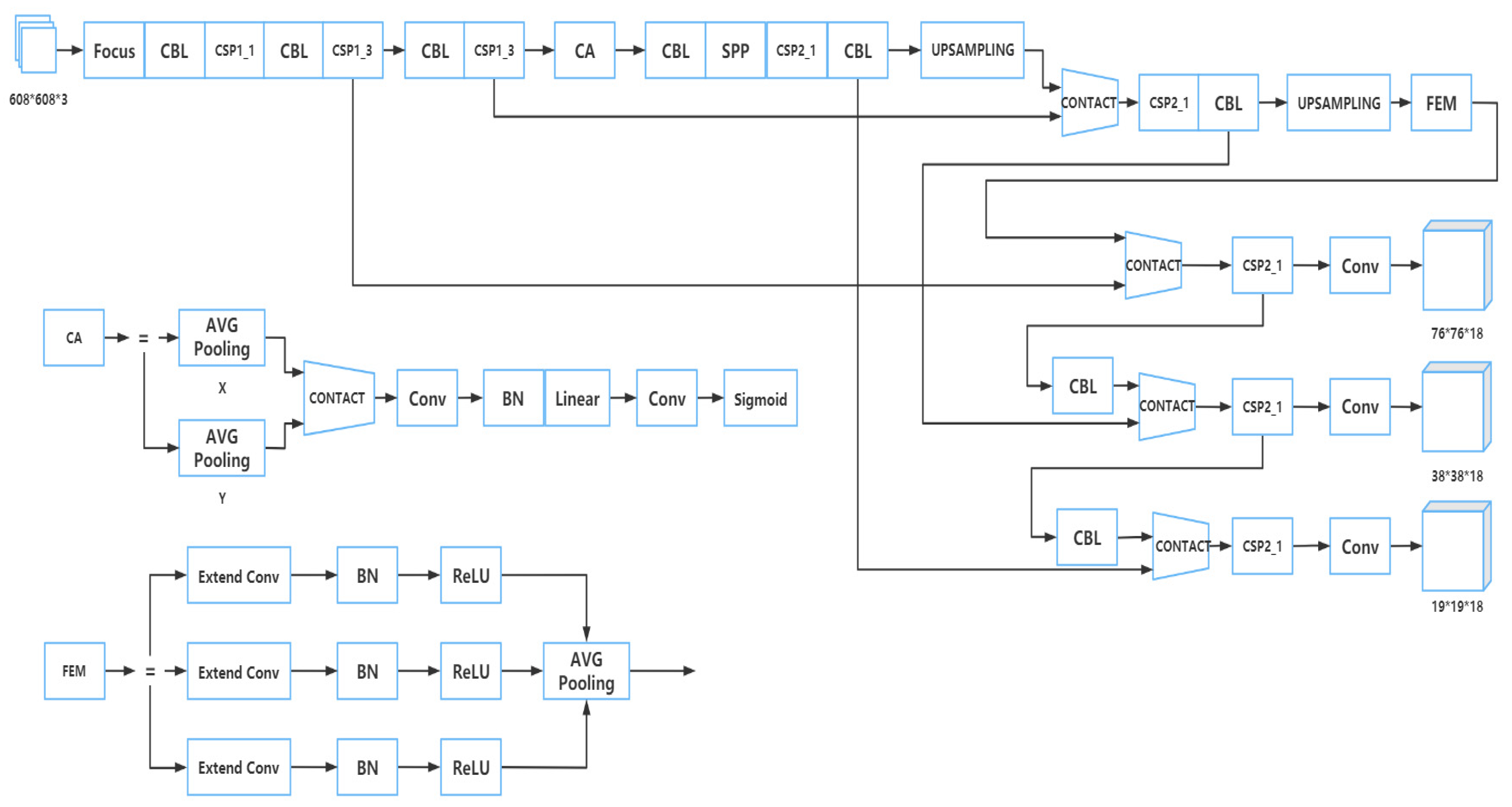
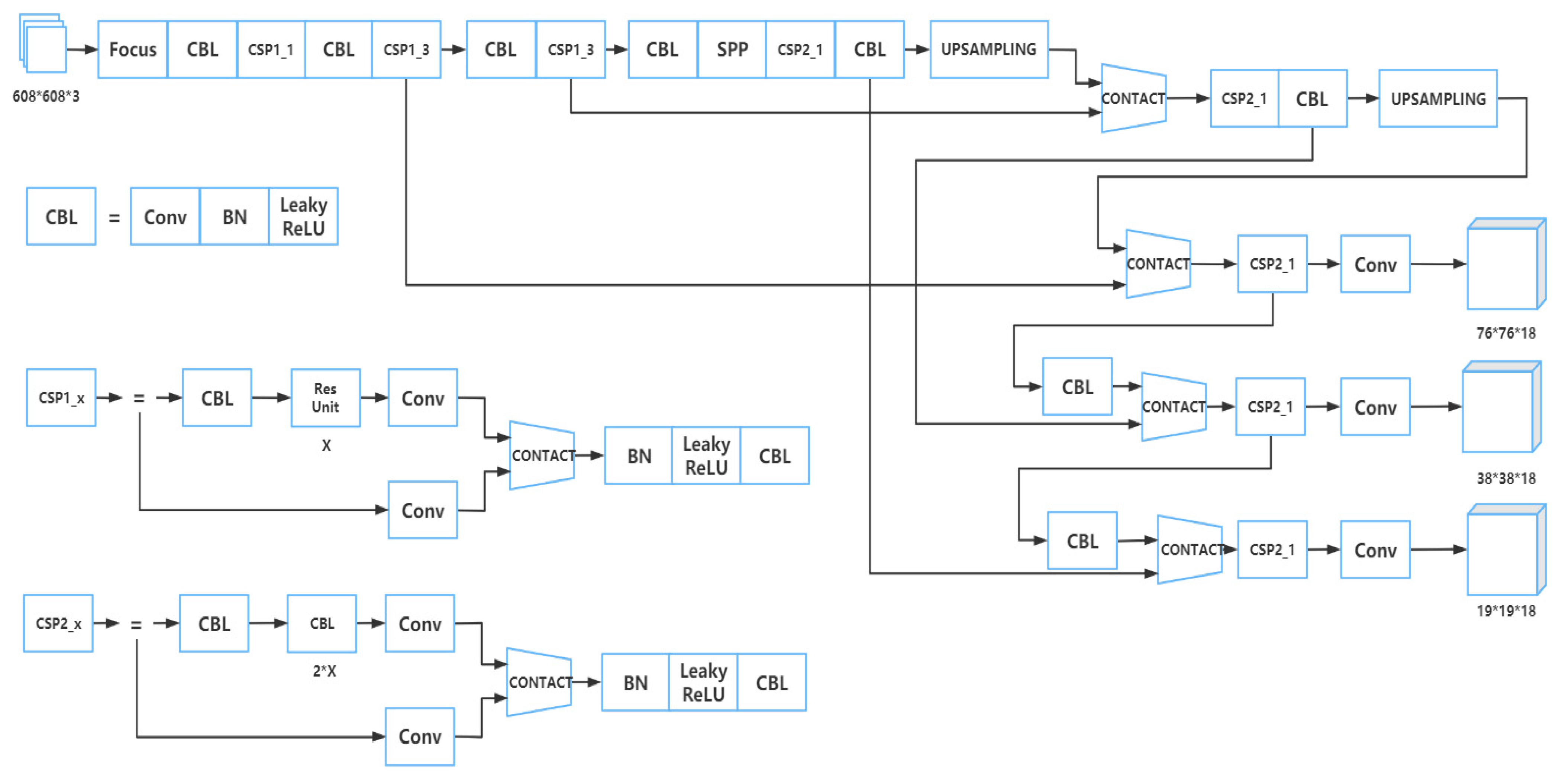
2.2. Training Model
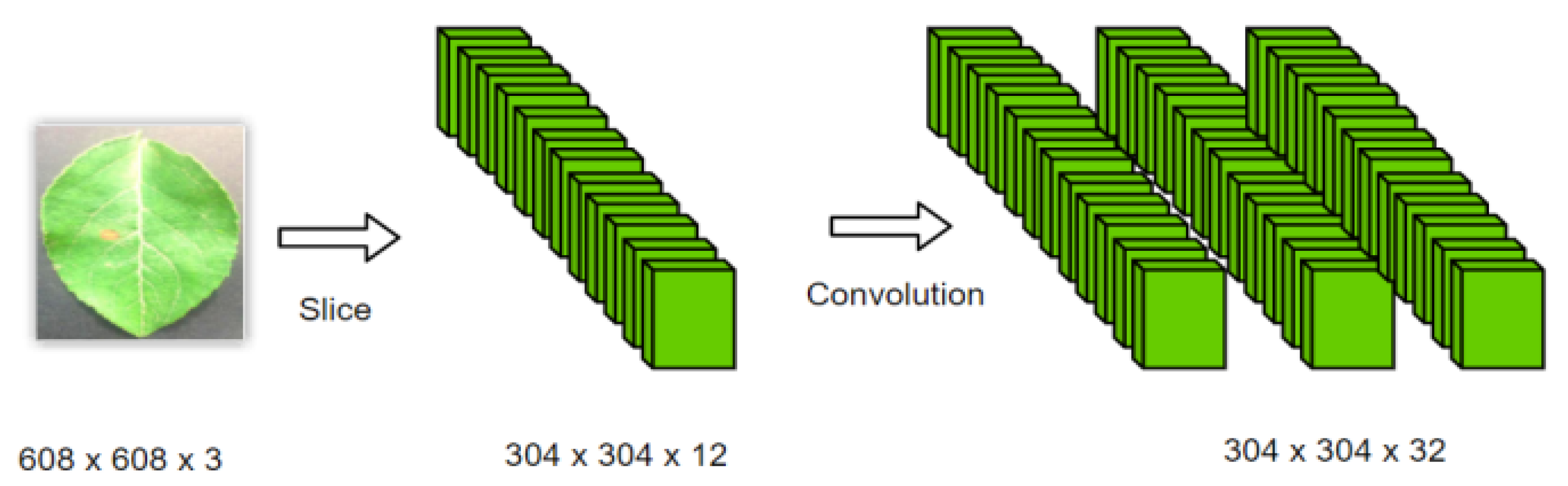
2.2.1. Part of Input
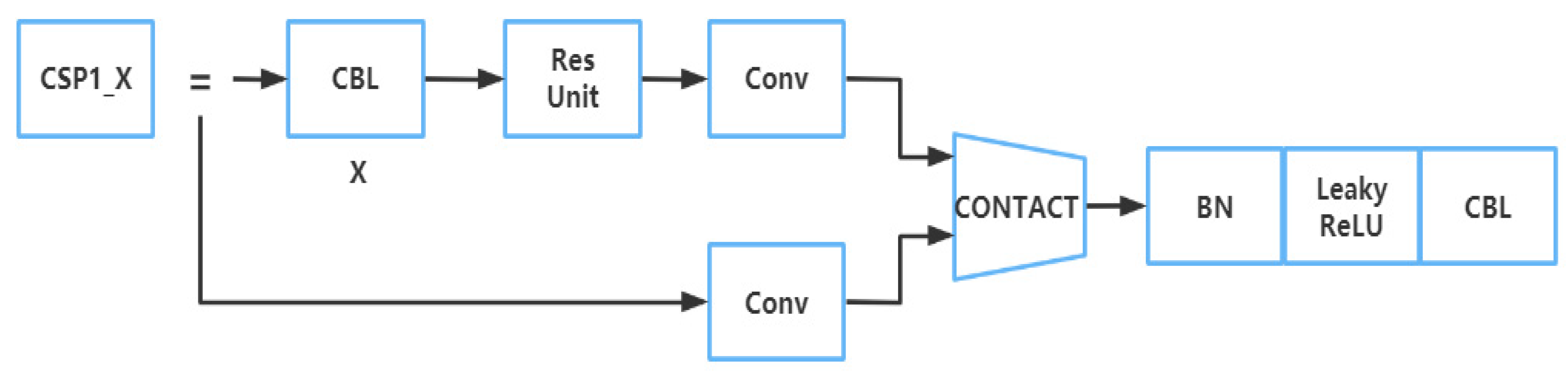
2.2.2. Part of Backbone
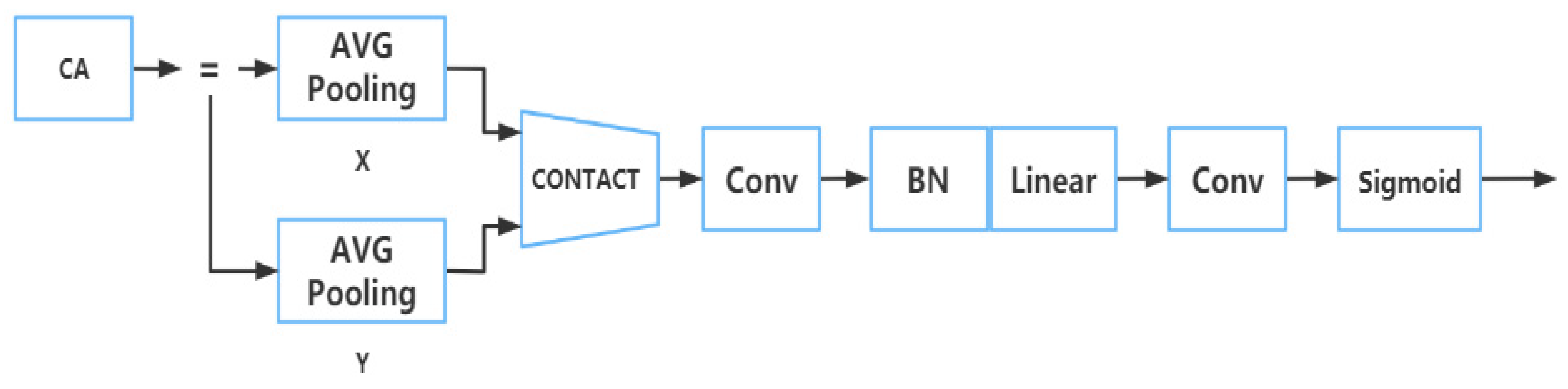
2.2.3. Part of Neck
2.2.4. Part of Prediction
2.3. Detection Model
2.3.1. Raindrop Image Recognition Network
2.3.2. Noise Reduction
2.4. Experimental Environment and Evaluating Indicators
3. Results
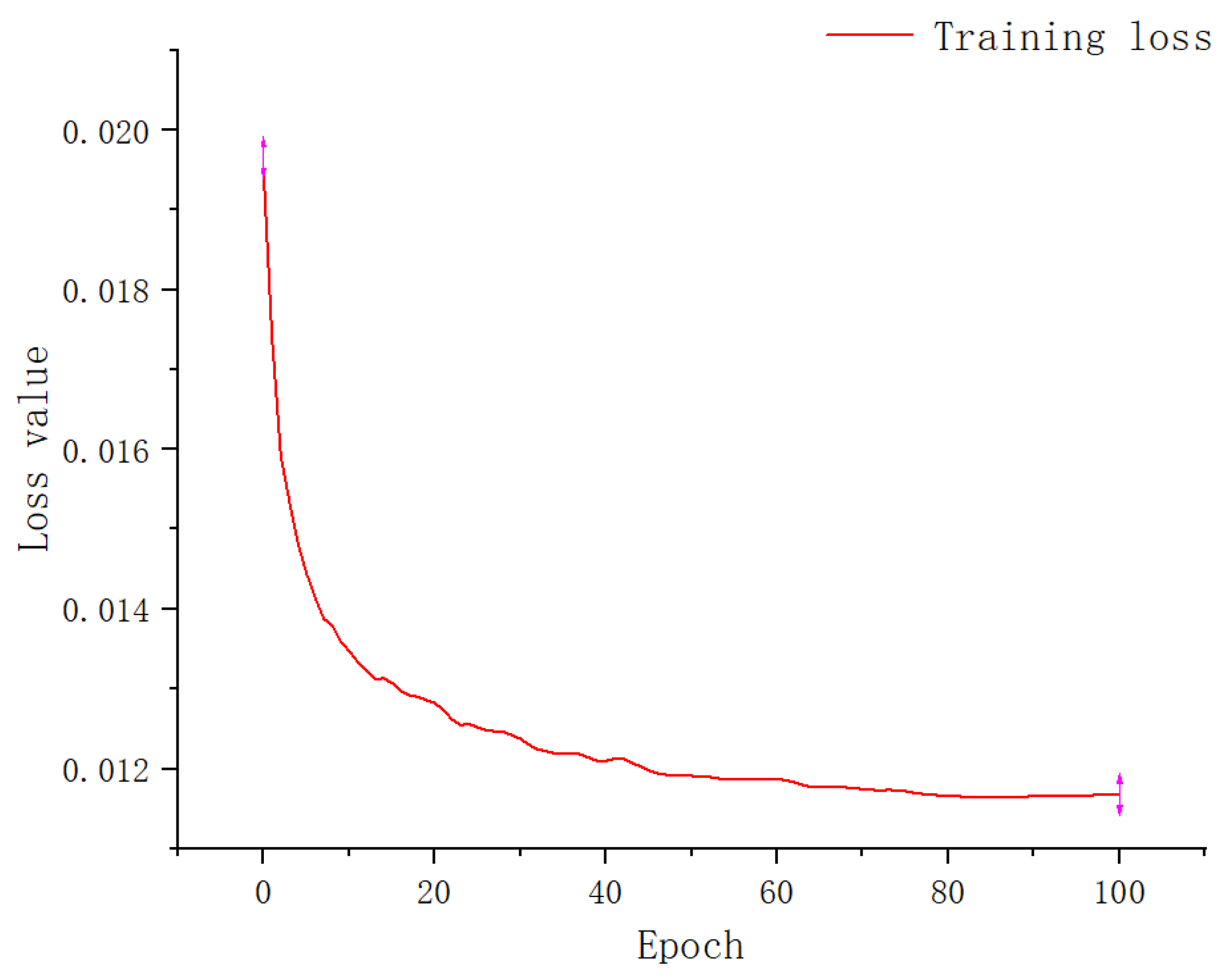
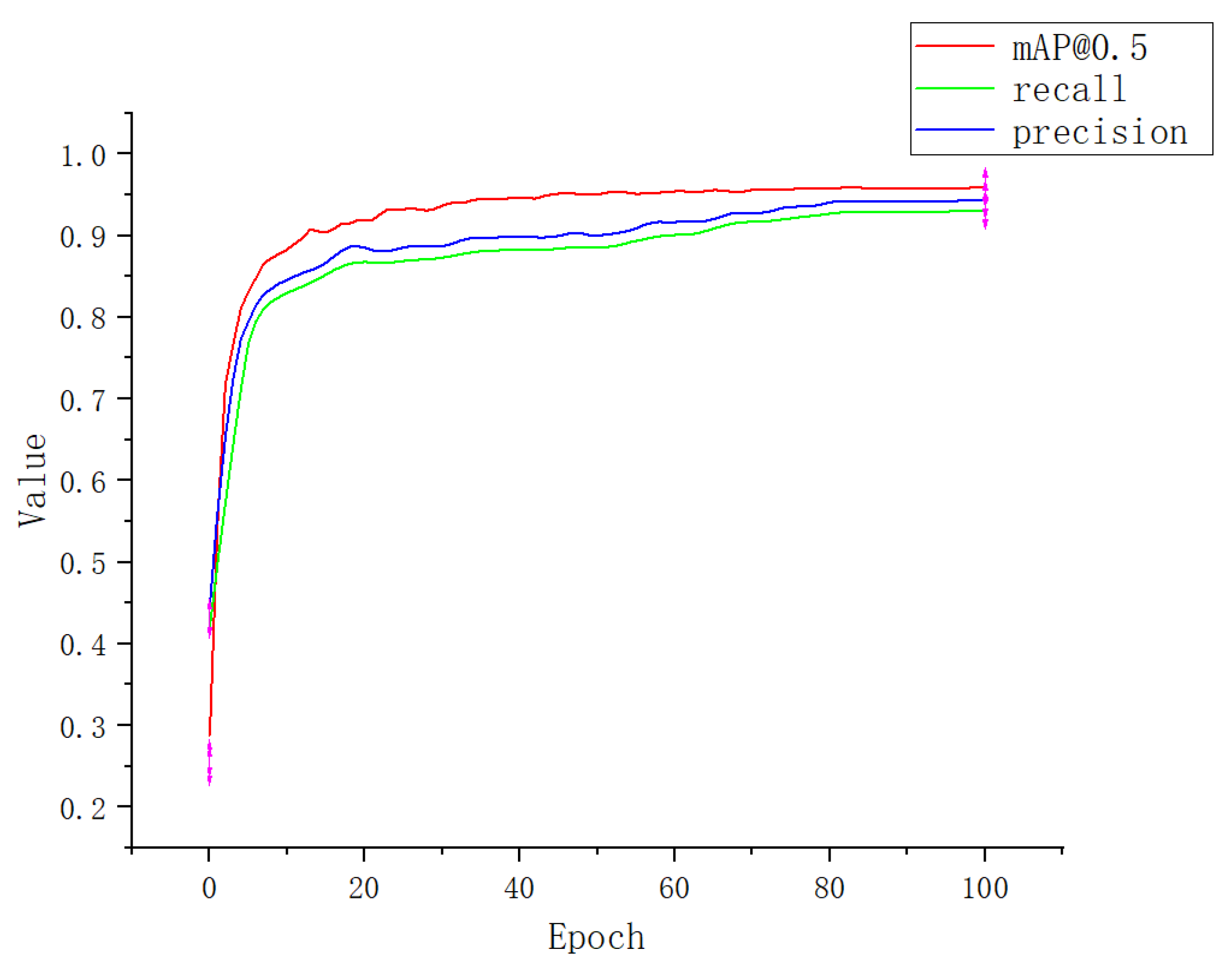
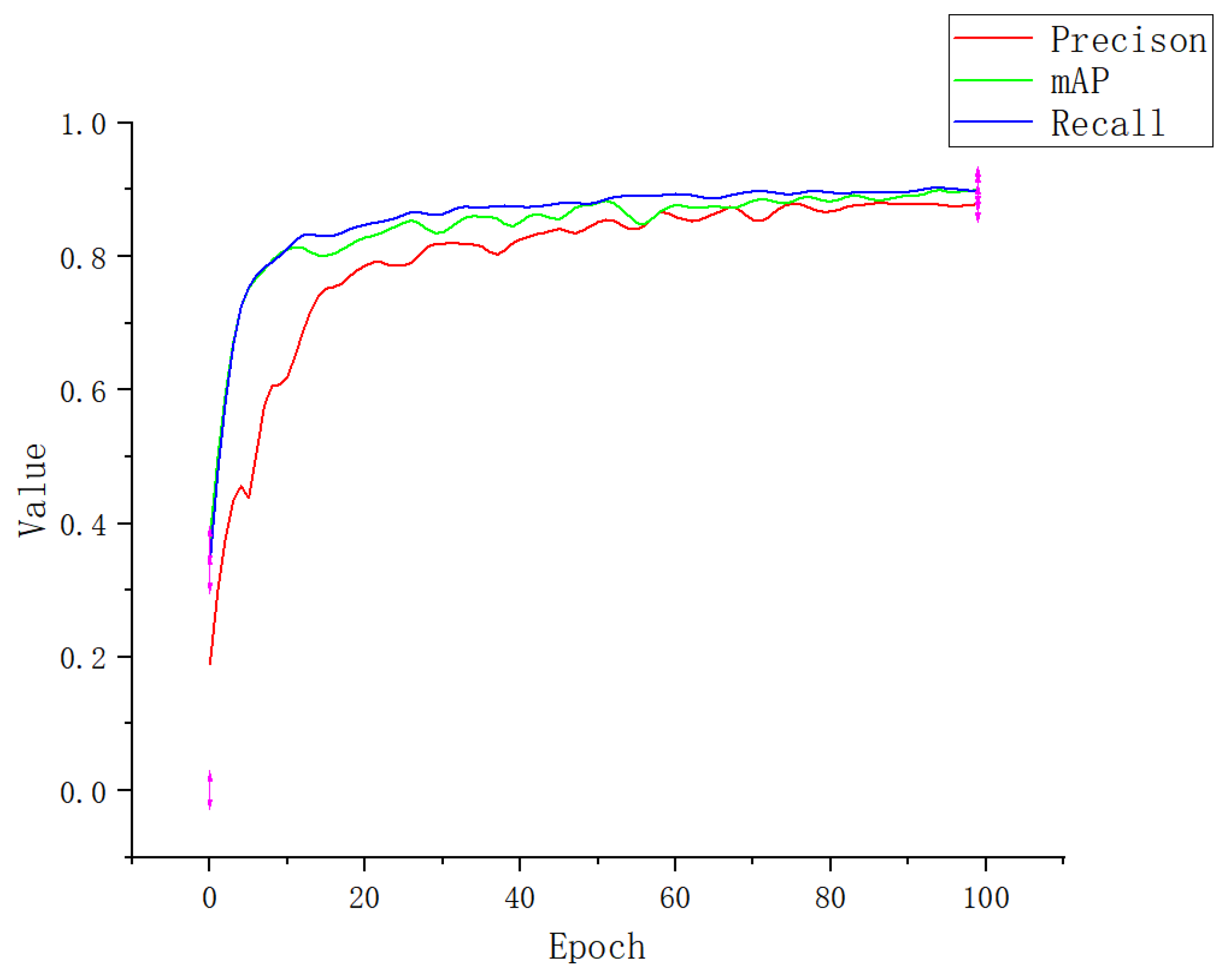
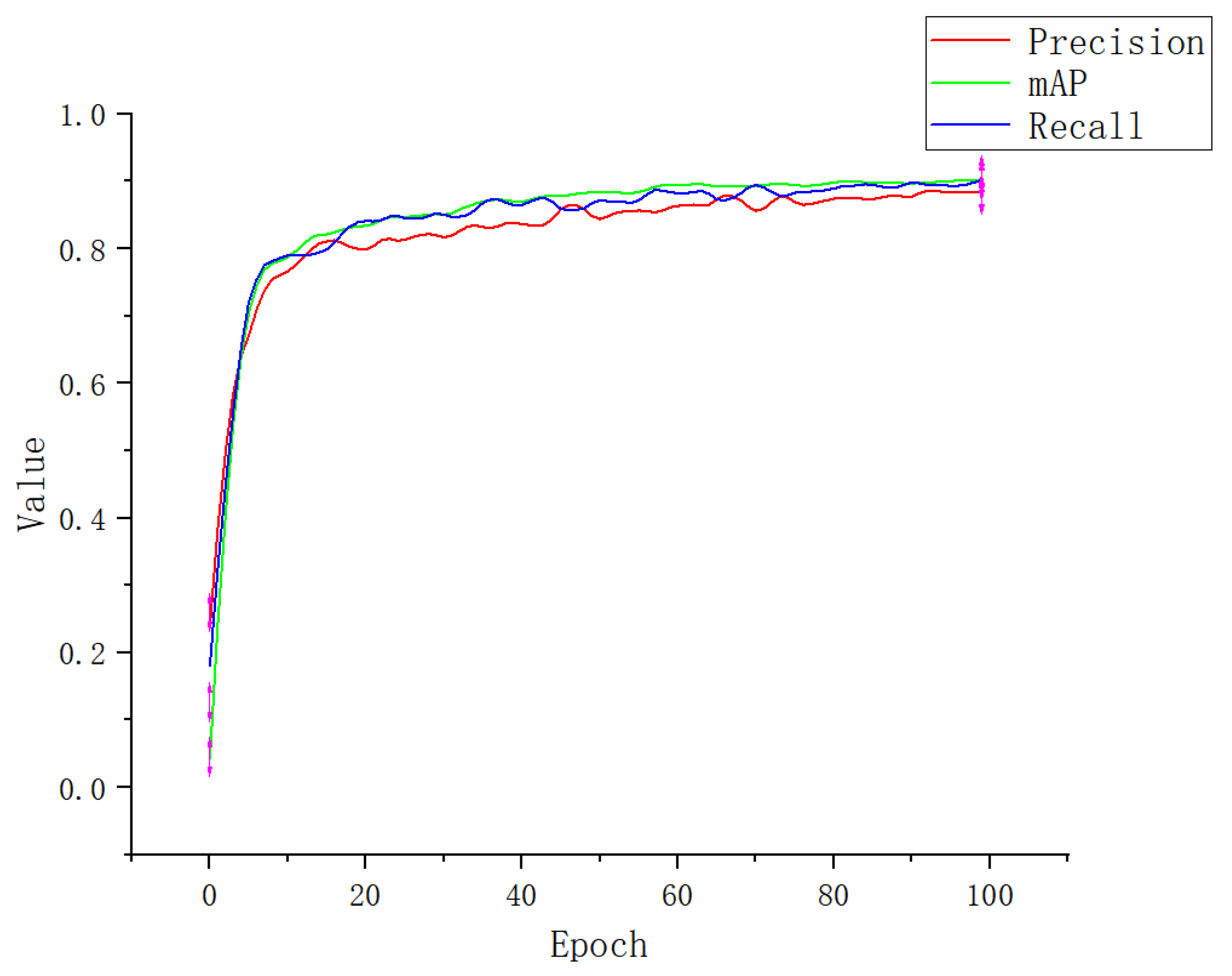
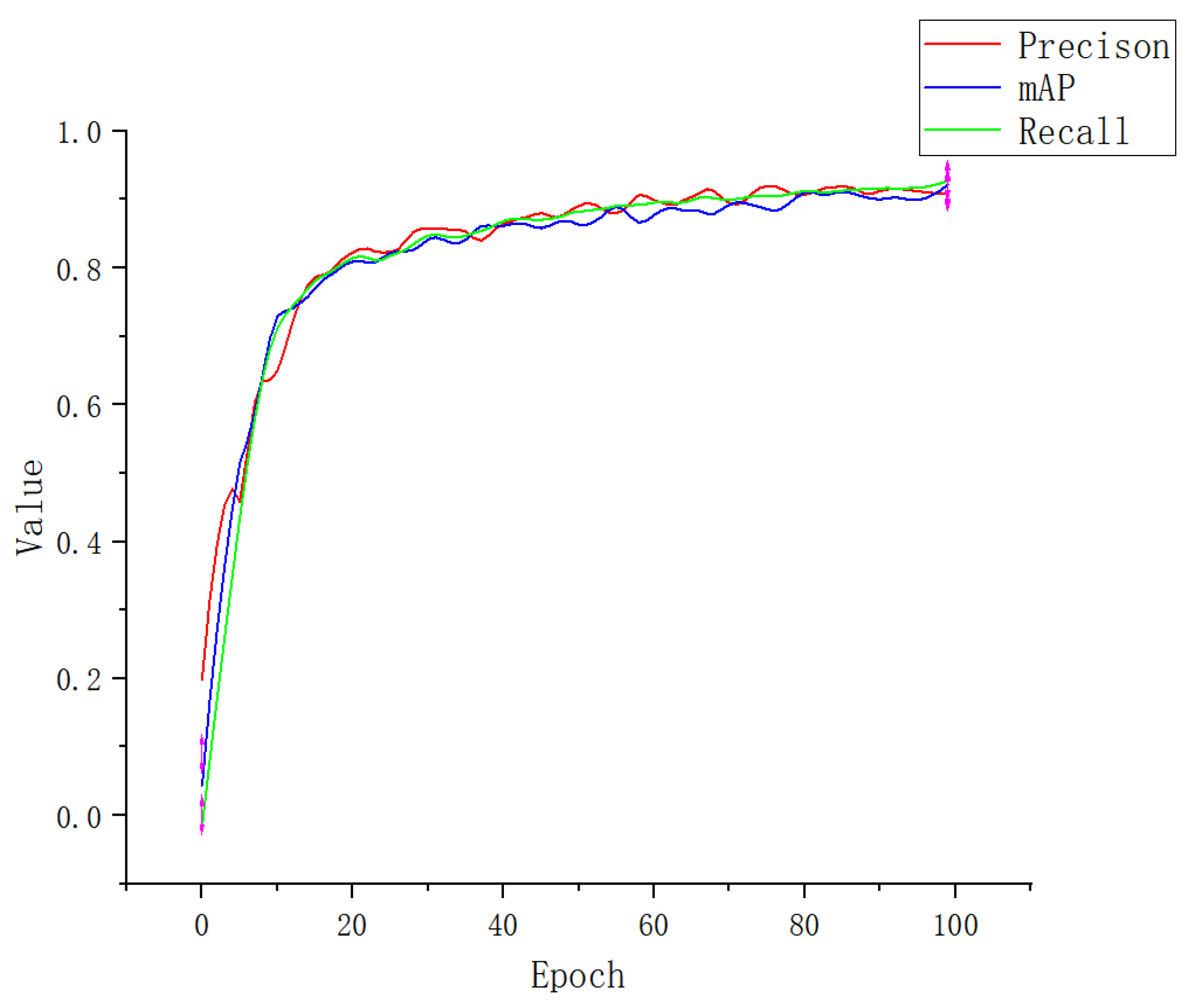
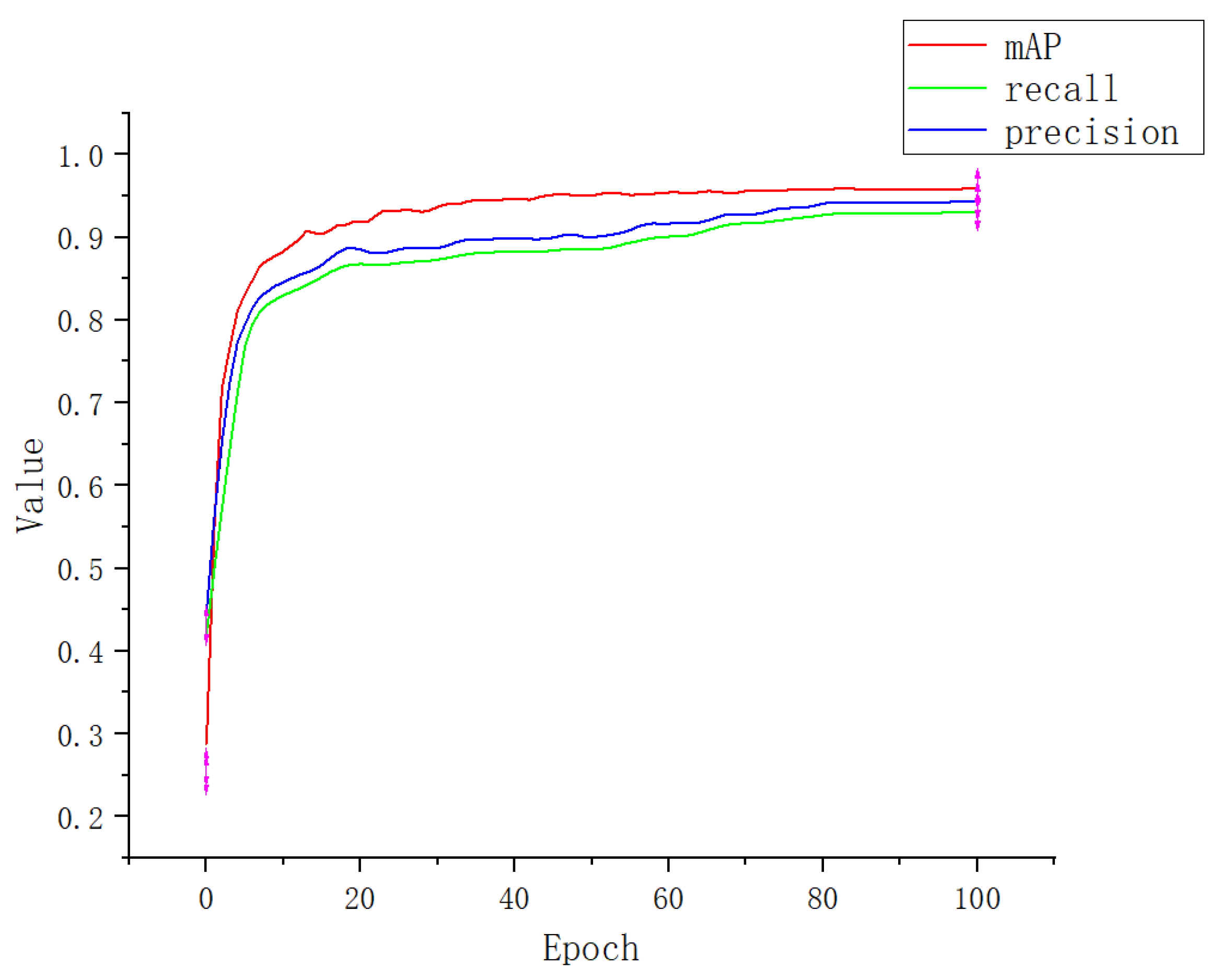
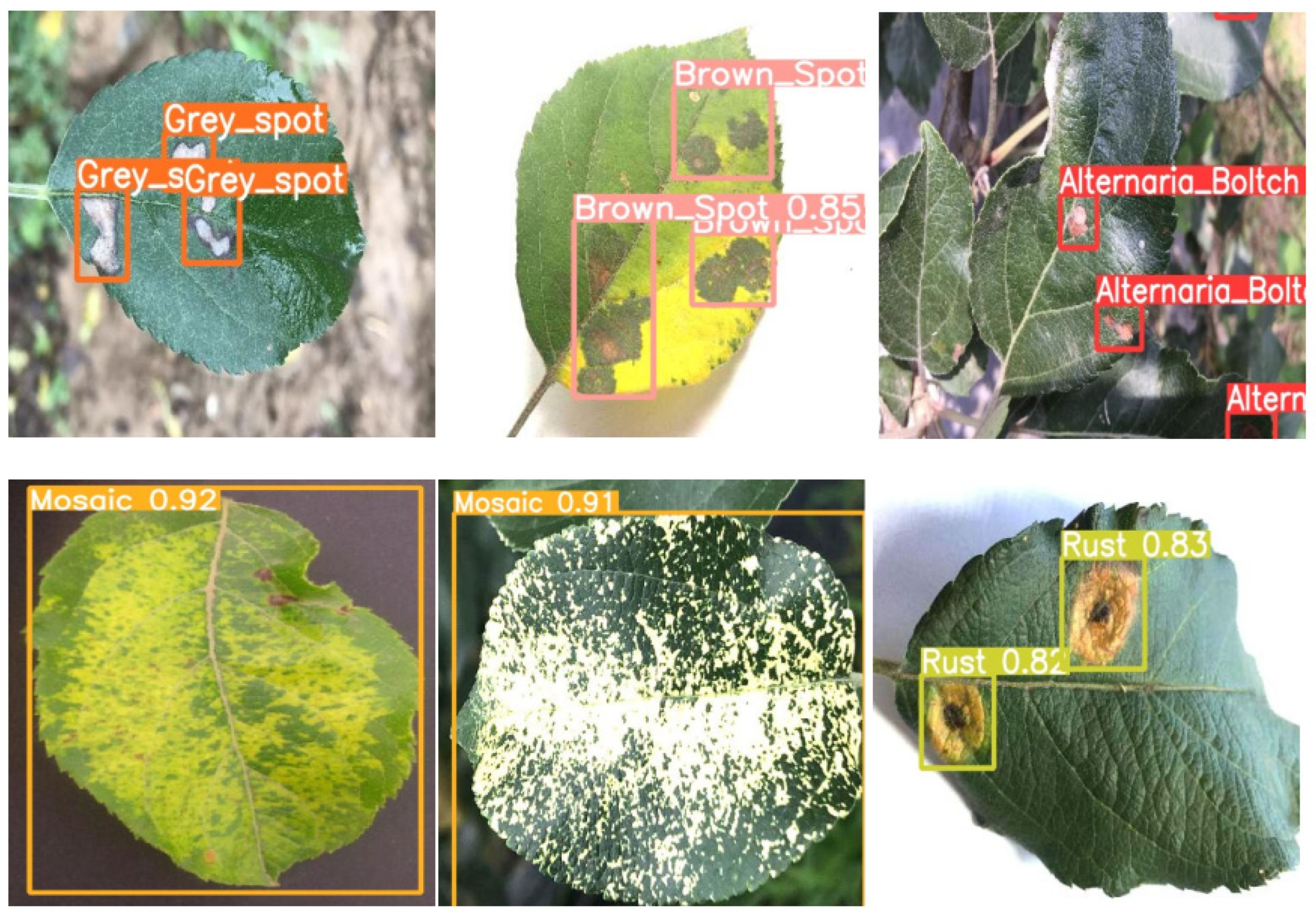
3.1. Experimental Result
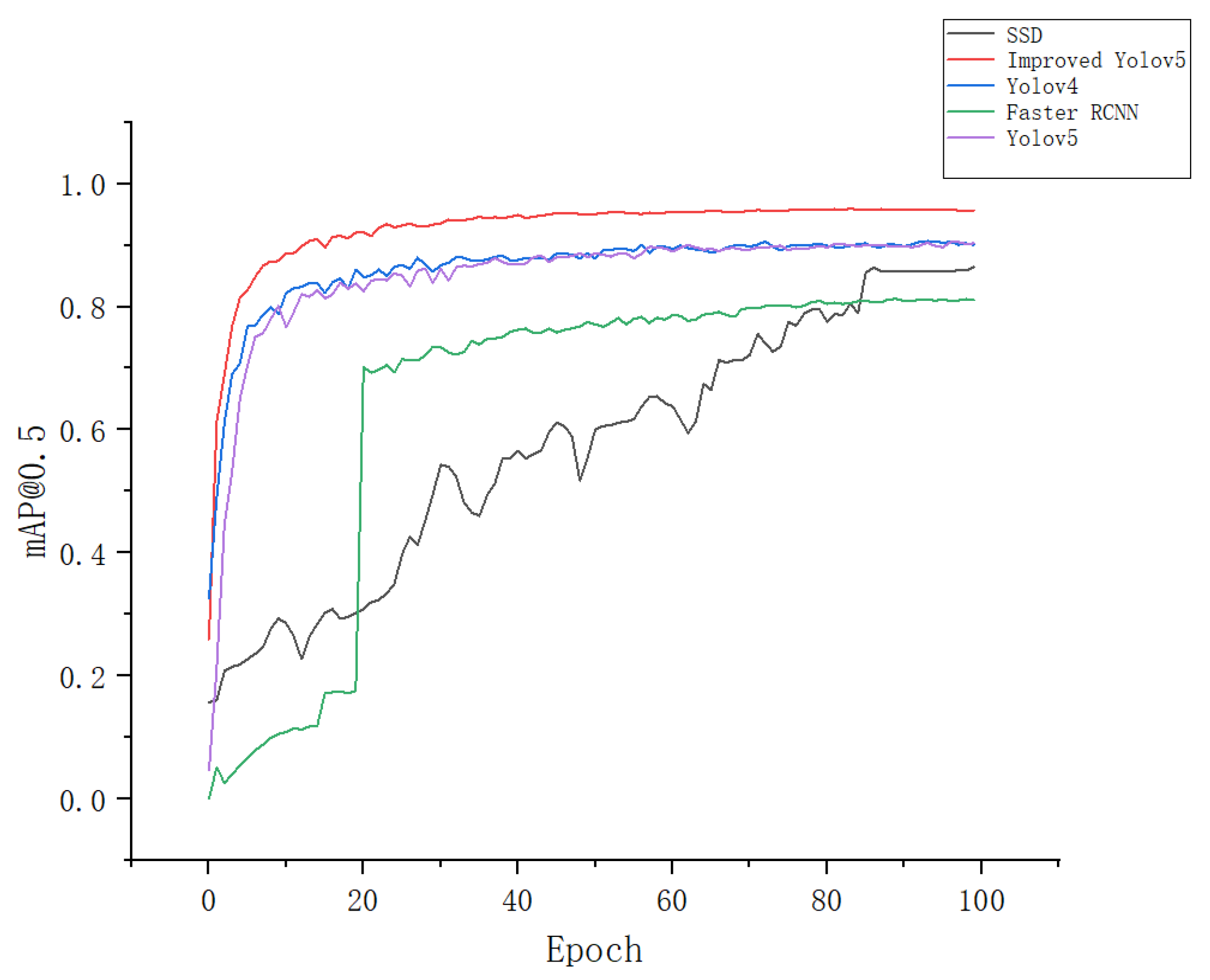
3.2. Comparison of the Accuracy of Different Network Models
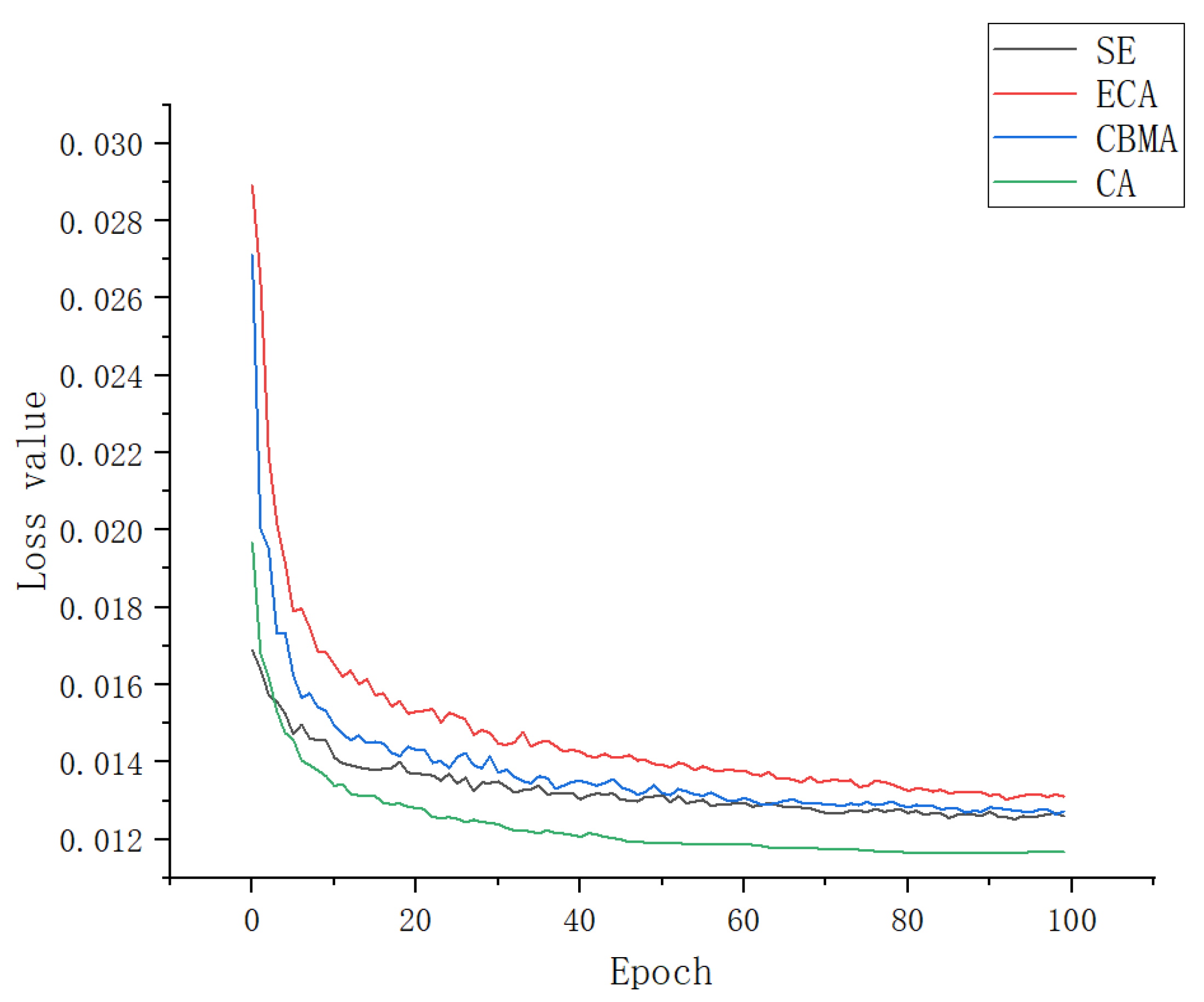
3.3. Ablation Experiment
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Khan, A.I.; Quadri, S.M.K.; Banday, S.; Shah, J.L. Deep diagnosis: A real-time apple leaf disease detection system based on deep learning. Comput. Electron. Agric. 2022, 198, 107093. [Google Scholar] [CrossRef]
- Ding, R.; Qiao, Y.; Yang, X.; Jiang, H.; Zhang, Y.; Huang, Z.; Wang, D.; Liu, H. Improved Res-Net Based Apple Leaf Diseases Identification. IFAC-Pap. 2022, 55, 78–82. [Google Scholar] [CrossRef]
- Hasan, S.; Jahan, S.; Islam, M.I. Disease detection of apple leaf with combination of color segmentation and modified DWT. J. King Saud Univ. - Comput. Inf. Sci. 2022, 34, 7212–7224. [Google Scholar] [CrossRef]
- Du Tot, M.; Nelson, L.M.; Tyson, R.C. Predicting the spread of postharvest disease in stored fruit, with application to apples. Postharvest Biol. Technol. 2013, 85, 45–56. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Zhang, Y.; Zhang, D.; Ouyang, H.; Chen, X. A Multiscale Lightweight and Efficient Model Based on YOLOv7: Applied to Citrus Orchard. Plants 2022, 11, 3260. [Google Scholar] [CrossRef]
- Qin, F.; Liu, D.; Sun, B.; Ruan, L.; Ma, Z.; Wang, H. Identification of Alfalfa Leaf Diseases Using Image Recognition Technology. PLoS ONE 2016, 11, e0168274. [Google Scholar] [CrossRef]
- Rothe, P.; Kshirsagar, R. Cotton leaf disease identification using pattern recognition techniques. In Proceedings of the 2015 International Conference on Pervasive Computing (ICPC), Pune, India, 8–10 January 2015; pp. 1–6. [Google Scholar]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef]
- Wu, L.; Blackwell, M.; Dunham, S.; Hernández-Allica, J.; McGrath, S.P. Simulation of Phosphorus Chemistry, Uptake and Utilisation by Winter Wheat. Plants 2019, 8, 404. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Y.; Wang, J. Automatic image-based plant disease severity estimation using deep learning. Comput. Intell. Neuro Sci. 2017, 2017, 2917536. [Google Scholar] [CrossRef]
- Hang, J.; Zhang, D.; Chen, P.; Zhang, J.; Wang, B. Classification of Plant Leaf Diseases Based on Improved Convolutional Neural Network. Sensors 2019, 19, 4161. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xu, H.; Liu, B.; He, D.; He, J.; Zhang, H.; Geng, N. MEAN-SSD: A novel real-time detector for apple leaf diseases using improved light-weight convolutional neu-ral networks. Comput. Electron. Agric. 2021, 189, 106379. [Google Scholar] [CrossRef]
- Li, W.; Chen, P.; Wang, B.; Xie, C. Automatic localization and count of agricultural crop pests based on an improved deep learning pipeline. Sci. Rep. 2019, 9, 7024. [Google Scholar] [CrossRef]
- Yadav, D.; Yadav, A.K. A Novel Convolutional Neural Network Based Model for Recognition and Classification of Apple Leaf Diseases. Trait. Signal 2020, 37, 1093–1101. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Z.; Wu, J.; Tian, Y.; Tang, H.; Guo, X. Real-Time Vehicle Detection Based on Improved YOLO v5. Sustainability 2022, 14, 12274. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, J.; Fu, X.; Yu, T.; Guo, Y.; Wang, R. DC-SPP-YOLO: Dense connection and spatial pyramid pooling based YOLO for object detection. Inf. Sci. 2020, 522, 241–258. [Google Scholar] [CrossRef]
- Fang, S.; Wang, Y.; Zhou, G.; Chen, A.; Cai, W.; Wang, Q.; Hu, Y.; Li, L. Multi-channel feature fusion networks with hard coordinate attention mechanism for maize disease identification under complex backgrounds. Comput. Electron. Agric. 2022, 203, 107486. [Google Scholar] [CrossRef]
- Zhang, Y.; Lian, H.; Yang, G.; Zhao, S.; Ni, P.; Chen, H.; Li, C. Inaccurate-Supervised Learning with Generative Adversarial Nets. IEEE Trans. Cybern. 2021, 51, 1–15. [Google Scholar] [CrossRef]
- Putra, Y.C.; Wijayanto, A.W.; Chulafak, G.A. Oil palm trees detection and counting on Microsoft Bing Maps Very High Resolution (VHR) satellite imagery and Unmanned Aerial Vehicles (UAV) data using image processing thresholding approach. Ecol. Inform. 2022, 72, 101878. [Google Scholar] [CrossRef]
- Wu, D.; Lv, S.; Jiang, M.; Song, H. Using channel pruningbased YOLOv4 deep learning algorithm for the real-time and accurate detection of apple flowers in natural environments. J. Comput. Electron. Agricult. 2020, 178, 174–178. [Google Scholar] [CrossRef]
- Tan, S.; Bie, X.; Lu, G.; Tan, X. Real-time detection for mask-wearing of personnel based on YOLOv5 network model. J. Laser J. 2021, 42, 147–150. [Google Scholar]
- Obeso, A.M.; Benois-Pineau, J.; Vázquez, M.S.G.; Acosta, A.Á.R. Visual vs internal attention mechanisms in deep neu-ral networks for image classification and object detection. Pattern Recognit. 2022, 123, 108411. [Google Scholar] [CrossRef]
- Yu, W.; Xiang, Q.; Hu, Y.; Du, Y.; Kang, X.; Zheng, D.; Shi, H.; Xu, Q.; Li, Z.; Niu, Y.; et al. An improved automated diatom detection method based on YOLOv5 framework and its preliminary study for taxonomy recognition in the forensic diatom test. Front. Microbiol. 2022, 13, 963059. [Google Scholar] [CrossRef] [PubMed]
- Jubayer, F.; Soeb, J.A.; Mojumder, A.N.; Paul, M.K.; Barua, P.; Kayshar, S.; Akter, S.S.; Rahman, M.; Islam, A. Detection of mold on the food surface using YOLOv5. Curr. Res. Food Sci. 2021, 4, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gu, X.; Xu, X.; Xu, D.; Zhang, T.; Liu, Z.; Dong, Q. Detection of concealed cracks from ground penetrating radar images based on deep learning algorithm. Constr. Build. Mater. 2021, 273, 121949. [Google Scholar] [CrossRef]
- Huu, P.N.; Xuan, K.D. Proposing Algorithm Using YOLOV4 and VGG-16 for Smart-Education. Appl. Comput. Intell. Soft Comput. 2021, 2021, 1682395. [Google Scholar] [CrossRef]
- Wang, G.; Rao, Z.; Sun, H.; Zhu, C.; Liu, Z. A belt tearing detection method of YOLOv4-BELT for multi-source interference environment. Measurement 2022, 189, 110469. [Google Scholar] [CrossRef]
- Feng, L.; Zhao, Y.; Sun, Y.; Zhao, W.; Tang, J. Action Recognition Using a Spatial-Temporal Network for Wild Felines. Animals 2021, 11, 485. [Google Scholar] [CrossRef]
- Wang, P.; Niu, T.; Mao, Y.; Zhang, Z.; Liu, B.; He, D. Identification of Apple Leaf Diseases by Improved Deep Convolutional Neural Networks with an Attention Mechanism. Front. Plant Sci. 2021, 12, 723294. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the International Conference on Neural Information Processing System, Lake Tahoe, NV, USA, 3 December 2012. [Google Scholar]
- Hartmann, D.; Franzenm, D.; Brodehl, S. Studying the Evolution of Neural Activation Patterns During Training of Feed-Forward ReLU Networks. Front. Artif. Intell. 2021, 4, 642374. [Google Scholar] [CrossRef]
- Shi, C.; Wang, Y.; Xiao, H.; Li, H. Extended convolution model for computing the far-field directivity of an amplitude-modulated parametric loudspeaker. J. Phys. D Appl. Phys. 2022, 55, 244002. [Google Scholar] [CrossRef]
- Li, J.; Han, Y.; Zhang, M.; Li, G.; Zhang, B. Multi-scale residual network model combined with Global Average Pooling for action recognition. Multimed. Tools Appl. 2021, 81, 1375–1393. [Google Scholar] [CrossRef]
- Zhou, L.; Li, Y.; Rao, X.; Liu, C.; Zuo, X.; Liu, Y. Ship Target Detection in Optical Remote Sensing Images Based on Multiscale Feature Enhancement. Comput. Intell. Neurosci. 2022, 2022, 2605140. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Li, B.; Zhang, F.; Meng, X.; Zhang, L. A Lightweight YOLOv5-MNE Algorithm for SAR Ship Detection. Sensors 2022, 22, 7088. [Google Scholar] [CrossRef]
- Qin, F.; Zhu, Q.; Zhang, Y.; Wang, R.; Wang, X.; Zhou, M.; Yang, Y. Effect of substrates on lasing properties of GAN transferable membranes. Opt. Mater. 2021, 122, 111663. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, H.; Gao, J.; Li, N. 3D Model Retrieval Algorithm Based on DSP-SIFT Descriptor and Codebook Combination. Appl. Sci. 2022, 12, 11523. [Google Scholar] [CrossRef]
- Tsourounis, D.; Kastaniotis, D.; Theoharatos, C.; Kazantzidis, A.; Economou, G. SIFT-CNN: When Convolutional Neural Networks Meet Dense SIFT Descriptors for Image and Sequence Classification. J. Imaging 2022, 8, 256. [Google Scholar] [CrossRef]
- Zhuang, L.; Yu, J.; Song, Y. Panoramic image mosaic method based on image segmentation and Improved SIFT algorithm. J. Phys. Conf. Ser. 2021, 2113, 012066. [Google Scholar] [CrossRef]
- Jiao, Y. Optimization and Innovation of SIFT Feature Matching Algorithm in Static Image Stitching Scheme. J. Phys. Conf. Ser. 2021, 1881, 032017. [Google Scholar] [CrossRef]
- Liu, Y.; Qiu, T.; Wang, J.; Qi, W. A Nighttime Vehicle Detection Method with Attentive GAN for Accurate Classification and Regression. Entropy 2021, 23, 1490. [Google Scholar] [CrossRef]
- Yan, Q.; Yang, B.; Wang, W.; Wang, B.; Chen, P.; Zhang, J. Apple leaf diseases recognition based on an improved convolutional neural network. Sensors 2020, 20, 3535. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xue, Z.P.; Guo, Y. Research on Plant Leaf Disease Identification Based on Transfer Learning Algorithm. In Proceedings of the 4th International Conference on Artificial Intelligence, Automation and Control Technologies (AIACT 2020), Hangzhou, China, 24–26 April 2020; Volume 1576, p. 012023. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Reddy, V.K.; Av, R.K. Multi-channel neuro signal classification using Adam-based coyote optimization enabled deep belief network. Biomed. Signal Process. Control 2022, 77, 103774. [Google Scholar] [CrossRef]
- Huang, Z.; Yin, Z.; Ma, Y.; Fan, C.; Chai, A. Mobile phone component object detection algorithm based on improved SSD. Procedia Comput. Sci. 2021, 183, 107–114. [Google Scholar] [CrossRef]
- Chen, K.; Xuan, Y.; Lin, A.; Guo, S. Esophageal cancer detection based on classification of gastrointestinal CT images using improved Faster RCNN. Comput. Methods Programs Biomed. 2021, 207, 106172. [Google Scholar] [CrossRef]
- Zhou, T.; Canu, S.; Ruan, S. Fusion based on attention mechanism and context constraint for multi-modal brain tumor segmentation. Comput. Med. Imaging Graph. 2020, 86, 101811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, P.; Gao, R.X. Attention Mechanism-Incorporated Deep Learning for AM Part Quality Prediction. Procedia CIRP93 2020, 93, 96–101. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, S.; Tanta, A.; Gupta, R. Extraction of Multiple Diseases in Apple Leaf Using Machine Learning. Int. J. Image Graph. 2021, 21, 2140009. [Google Scholar] [CrossRef]
- Pradhan, P.; Kumar, B.; Mohan, S. Comparison of various deep convolutional neural network models to discriminate apple leaf diseases using transfer learning. J. Plant Dis. Prot. 2022, 129, 1461–1473. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.; Liu, B.I.N. Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Zhao, J. MGA-YOLO: A lightweight one-stage network for apple leaf disease detection. Front. Plant Sci. 2022, 13, 927424. [Google Scholar] [CrossRef]
- Yu, H.; Cheng, X.; Chen, C.; Heidari, A.A.; Liu, J.; Cai, Z.; Chen, H. Apple leaf disease recognition method with improved residual network. Multimed. Tools Appl. 2022, 81, 7759–7782. [Google Scholar] [CrossRef]


| Classes | Number of Training Data | Number of Test Data | Label Number |
|---|---|---|---|
| Alternaria blotch | 2000 | 500 | 0 |
| Brown spot | 2000 | 500 | 1 |
| Gray spot | 2000 | 500 | 2 |
| Mosaic | 2000 | 500 | 3 |
| Rust | 2000 | 500 | 4 |
| Model | Precision/% | Recall/% | mAP@0.5/% |
|---|---|---|---|
| SSD | 86.2 | 88.7 | 86.5 |
| Faster RCNN | 82.1 | 84.9 | 81.2 |
| YOLOv4 | 84.5 | 86.7 | 90.3 |
| YOLOv5 | 87.6 | 90.3 | 89.8 |
| Apple-Net | 93.1 | 94.4 | 95.9 |
| Module | Precision/% | Recall/% | mAP/% |
|---|---|---|---|
| YOLOv5s + SE | 87.9 | 90.4 | 90.1 |
| YOLOv5s + ECA | 91.2 | 91.6 | 92.8 |
| YOLOv5s + CBAM | 89.5 | 90.9 | 90.3 |
| YOLOv5s + CA | 93.1 | 94.4 | 95.9 |
| References | Methods/Models | Categories | Images | mAP@0.5 | Accuracy |
|---|---|---|---|---|---|
| [6] | SVM | 4 | 899 | / | 92.49 |
| [14] | MEAN-SSD | 5 | 26,767 | 83.12 | 97.07 |
| [43] | VGG-Net | 4 | 2446 | / | 99.01 |
| [44] | Res-Net | 4 | 4174 | / | 83.75 |
| [51] | KNN | 2 | 744 | / | 96.40 |
| [52] | DenseNet-201 | 4 | 2537 | / | 98.75 |
| [53] | DP-CNNS | 5 | 26,377 | 78.8 | 97.14 |
| [54] | MGA-YOLO | 4 | 8838 | 89.3 | 94.80 |
| [55] | MSO Res-Net | 5 | 11,397 | 89.6 | 95.70 |
| Proposed | Apple-Net | 5 | 15,000 | 95.9 | 96.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, R.; Zou, H.; Li, Z.; Ni, R. Apple-Net: A Model Based on Improved YOLOv5 to Detect the Apple Leaf Diseases. Plants 2023, 12, 169. https://doi.org/10.3390/plants12010169
Zhu R, Zou H, Li Z, Ni R. Apple-Net: A Model Based on Improved YOLOv5 to Detect the Apple Leaf Diseases. Plants. 2023; 12(1):169. https://doi.org/10.3390/plants12010169
Chicago/Turabian StyleZhu, Ruilin, Hongyan Zou, Zhenye Li, and Ruitao Ni. 2023. "Apple-Net: A Model Based on Improved YOLOv5 to Detect the Apple Leaf Diseases" Plants 12, no. 1: 169. https://doi.org/10.3390/plants12010169
APA StyleZhu, R., Zou, H., Li, Z., & Ni, R. (2023). Apple-Net: A Model Based on Improved YOLOv5 to Detect the Apple Leaf Diseases. Plants, 12(1), 169. https://doi.org/10.3390/plants12010169






