Sweet Cherry Diversity and Relationships in Modern and Local Varieties Based on SNP Markers
Abstract
1. Introduction
2. Results
2.1. Phenotypic Characterization
2.2. GBS and SNP Identification
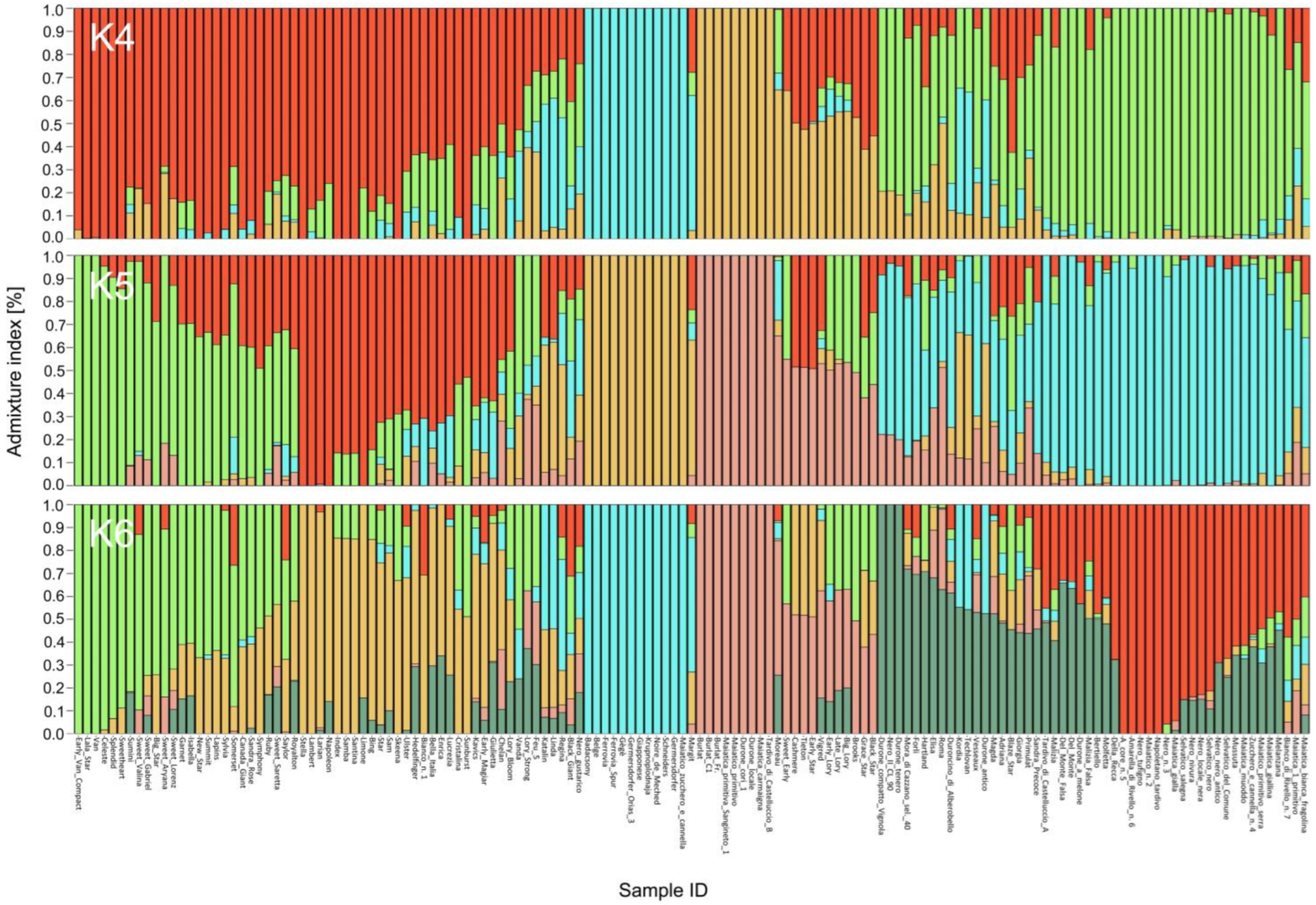
2.3. Allele Frequency and Ancestry Estimation
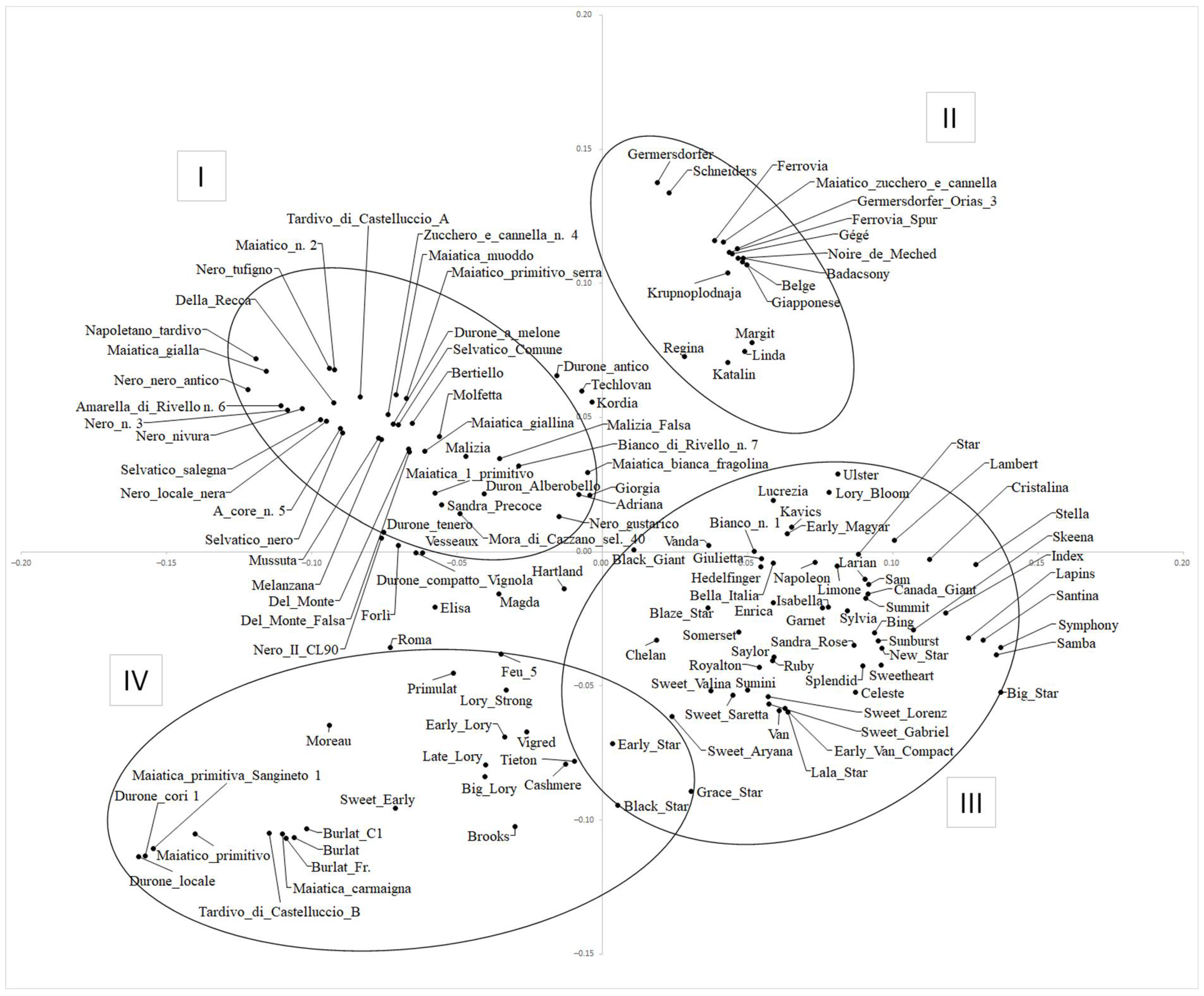
2.4. Principal Component Analysis
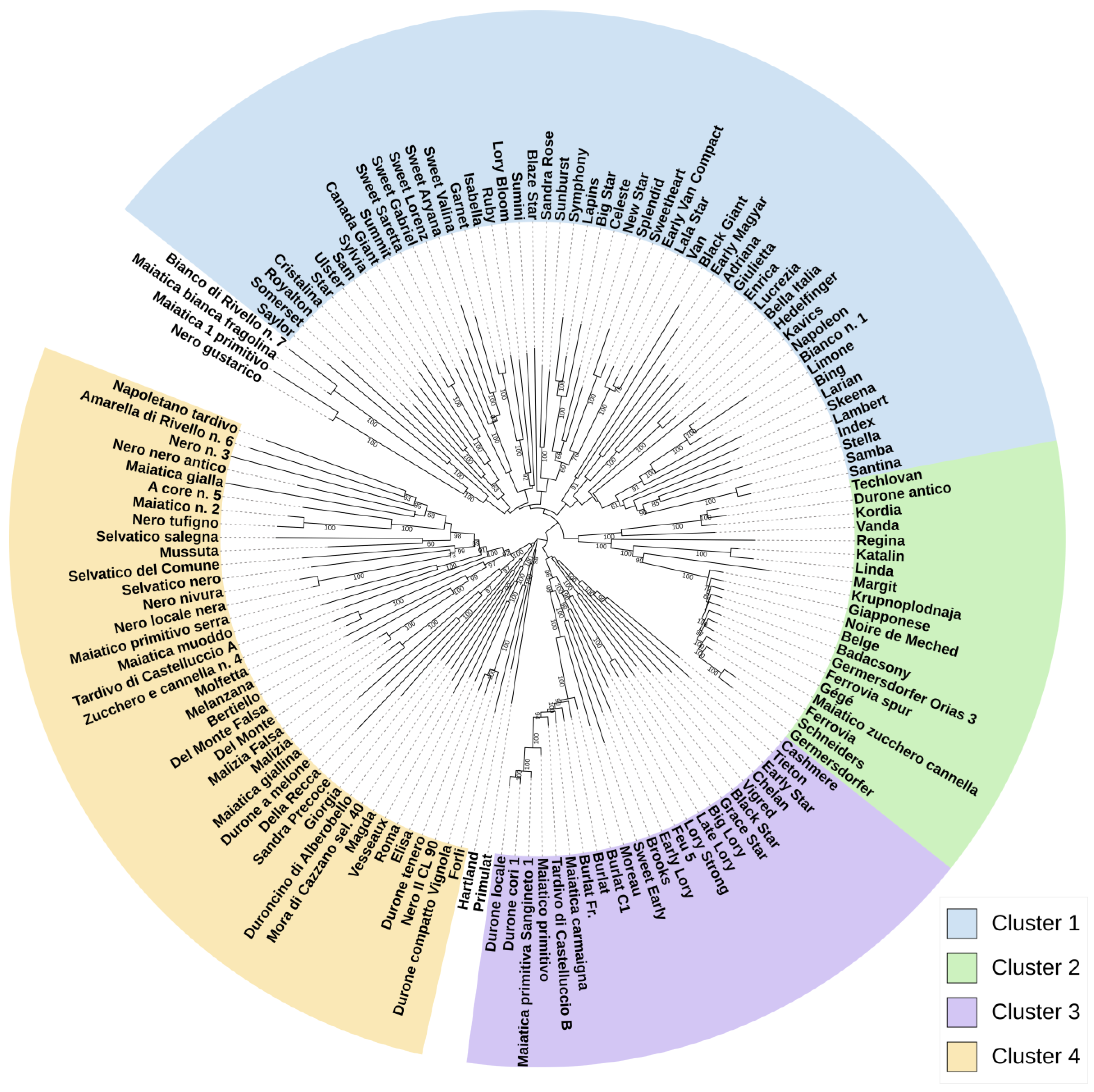
2.5. Genetic Relationships
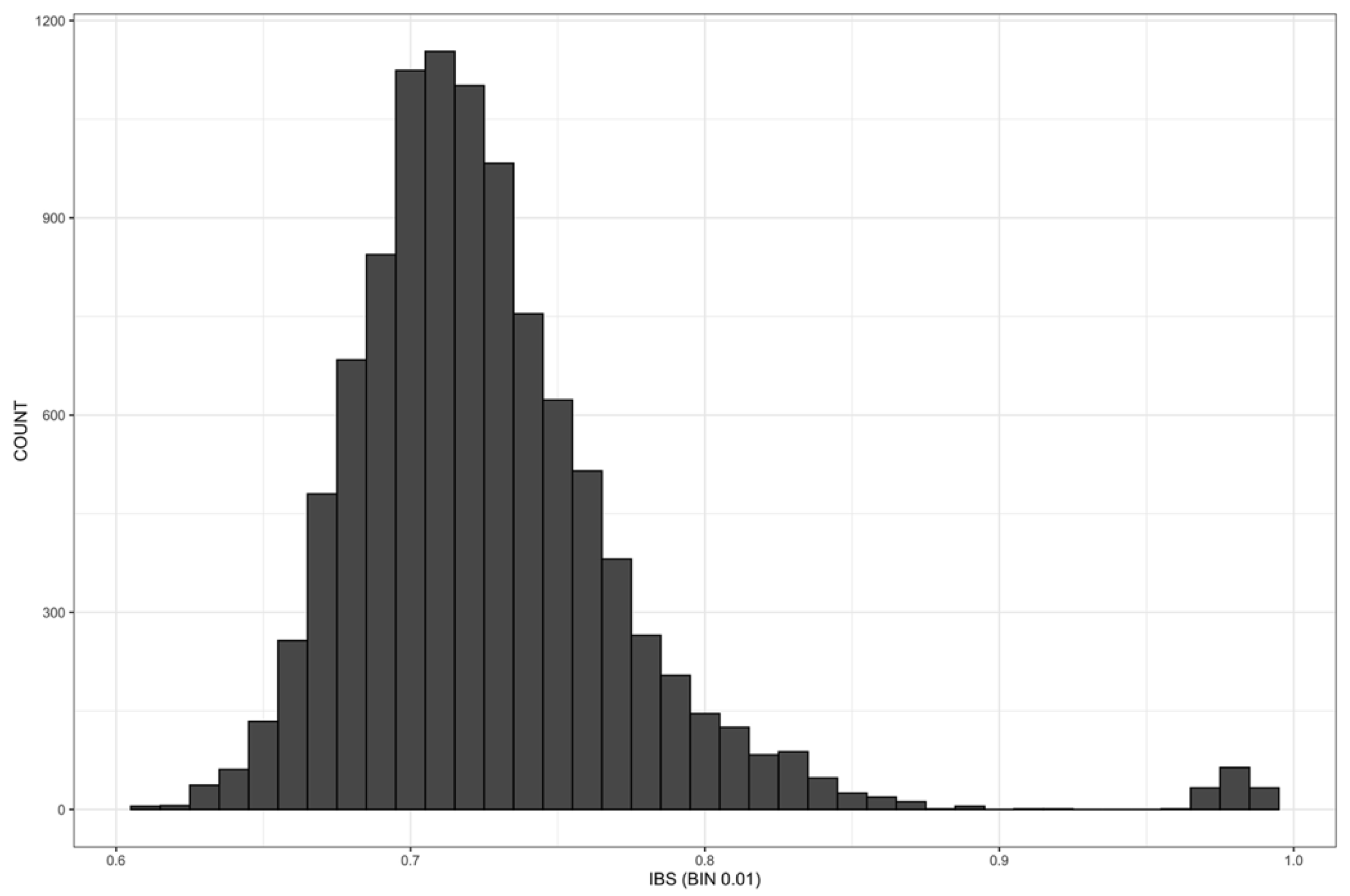
2.6. Identity by State
3. Discussion
4. Materials and Methods
4.1. Plant Material, Phenotypic Characterization, and Genotyping-by-Sequencing
4.2. Preprocessing of GBS Tags, SNP Calling, and Filtering
4.3. Population Structure and Genetic Relationships
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCune, L.M.; Kubota, C.; Stendell-Hollis, N.R.; Thomson, C.A. Cherries and health: A review. Crit. Rev. Food Sci. Nutr. 2011, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT 2022. Food and Agriculture Organization of the United Nations (FAO). Available online: https://www.fao.org/faostat/en/#home (accessed on 20 July 2022).
- Webster, A.D. The taxonomic classification of sweet and sour cherries and a brief history of their cultivation. In Cherries: Crop Physiology, Production and Uses; Webster, A.D., Looney, N.E., Eds.; CAB International: Wallingford, UK, 1996; pp. 3–24. [Google Scholar]
- Watkins, R. Cherry, plum, peach, apricot and almond. In Evolution of Crop Plants; Simmonds, N.W., Ed.; Longman: London, UK; New York, NY, USA, 1976; pp. 242–247. [Google Scholar]
- Roselli, G.; Mariotti, P. Il germoplasma del Ciliegio. Vol 1: Provincia di Pisa; ARSIA: Florence, Italy, 1999; ISBN 88-8295-009-3. [Google Scholar]
- Giovannini, D.; Leone, A.; Liverani, A.; Sirri, S.; Tellarini, S. Studi di caratterizzazione molecolare di vecchie varietà di ciliegio della tradizione romagnola. Frutticoltura 2013, 4, 62–65. (In Italian) [Google Scholar]
- Marchese, A.; Giovannini, D.; Leone, A.; Mafrica, R.; Palasciano, M.; Cantini, C.; Di Vaio, C.; De Salvador, F.R.; Giacalone, G.; Caruso, T.; et al. S-genotype identification, genetic diversity and structure analysis of Italian sweet cherry germplasm. Tree Genet. Genomes 2017, 13, 93. [Google Scholar] [CrossRef]
- ISTAT. Available online: http://dati.istat.it/Index.aspx?QueryId=33705# (accessed on 30 July 2022).
- Bargioni, G. Sweet cherry scions: Characteristics of the principal commercial cultivars, breeding objectives and methods. In Cherries, Crop Physiology, Production and Uses; Webster, A.D., Looney, N.E., Eds.; CAB International: Wallingford, UK, 1996; pp. 73–125. [Google Scholar]
- Mariette, S.; Tavaud, M.; Arunyawat, U.; Capdeville, G.; Millan, M.; Salin, F. Population structure and genetic bottleneck in sweet cherry estimated with SSRs and the gametophytic self-incompatibility locus. BMC Genet. 2010, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Ganopoulos, I.V.; Kazantzis, K.; Chatzicharisis, I.; Karayiannis, I.; Tsaftaris, A.S. Genetic diversity, structure and fruit trait associations in Greek sweet cherry cultivars using microsatellite based (SSR/ISSR) and morpho-physiological markers. Euphytica 2011, 181, 237–251. [Google Scholar] [CrossRef]
- Sharma, K.; Xuan, H.; Sedlák, P. Assessment of genetic diversity of Czech sweet cherry cultivars using microsatellite markers. Biochem. Syst. Ecol. 2015, 63, 6–12. [Google Scholar] [CrossRef]
- Campoy, J.A.; Lerigoleur-Balsemin, E.; Christmann, H.; Beauvieux, R.; Girollet, N.; Quero-García, J.; Dirlewanger, E.; Barreneche, T. Genetic diversity, linkage disequilibrium, population structure and construction of a core collection of Prunus avium L. landraces and bred cultivars. BMC Plant Biol. 2016, 16, 49. [Google Scholar] [CrossRef]
- Krmpot, T.; Radoš, L.; Vokurka, A. Genetic characterisation of autochthonous sweet cherry genotypes (Prunus avium L.) using SSR markers. Genetika 2020, 52, 43–53. [Google Scholar] [CrossRef]
- Höfer, M.; Braun-Lüllemann, A.; Schiffler, J.; Schuster, M.; Flachowsky, H. Pomological and molecular characterization of sweet cherry cultivars (Prunus avium L.) of the German Fruit Genebank. OpenAgrar Repos. 2021. Available online: https://www.openagrar.de/receive/openagrar_mods_00066782 (accessed on 1 July 2022). [CrossRef]
- Schüller, E.; Fernández, F.F.; Antanaviciute, L.; Anhalt-brüderl, U.; Spornberger, A.; Forneck, A. Autochthonous Austrian varieties of Prunus avium L. Represent a regional gene pool, assessed using SSR and AFLP markers. Genes 2021, 12, 322. [Google Scholar] [CrossRef]
- Guarino, C.; Santoro, S.; De Simone, L.; Cipriani, G. Prunus avium: Nuclear DNA study in wild populations and sweet cherry cultivars. Genome 2009, 52, 320–337. [Google Scholar] [CrossRef] [PubMed]
- Di Vaio, C.; Villano, C.; Marallo, N. Molecular analysis of native cultivars of sweet cherry in Southern Italy. Hortic. Sci. 2015, 42, 114–118. [Google Scholar] [CrossRef]
- Muccillo, L.; Colantuoni, V.; Sciarrillo, R.; Baiamonte, G.; Salerno, G.; Marziano, M.; Sabatino, L.; Guarino, C. Molecular and environmental analysis of Campania (Italy) sweet cherry (Prunus avium L.) cultivars for biocultural refugia identification and conservation. Sci. Rep. 2019, 9, 6796. [Google Scholar] [CrossRef]
- Fernandez i Marti, A.; Athanson, B.; Koepke, T.; Font i Forcada, C.; Dhingra, A.; Oraguzie, N. Genetic diversity and relatedness of sweet cherry (Prunus avium L.) cultivars based on single nucleotide polymorphic markers. Front. Plant Sci. 2012, 3, 116. [Google Scholar] [CrossRef] [PubMed]
- Barreneche, T.; de la Concepción, M.C.; Blouin-Delmas, M.; Ordidge, M.; Nybom, H.; Lacis, G.; Feldmane, D.; Sedlak, J.; Meland, M.; Kaldmäe, H.; et al. Ssr-based analysis of genetic diversity and structure of sweet cherry (Prunus avium L.) from 19 countries in Europe. Plants 2021, 10, 1983. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef]
- Pinosio, S.; Marroni, F.; Zuccolo, A.; Vitulo, N.; Mariette, S.; Sonnante, G.; Aravanopoulos, F.A.; Ganopoulos, I.; Palasciano, M.; Vidotto, M.; et al. A draft genome of sweet cherry (Prunus avium L.) reveals genome-wide and local effects of domestication. Plant J. 2020, 103, 1420–1432. [Google Scholar] [CrossRef]
- Xanthopoulou, A.; Manioudaki, M.; Bazakos, C.; Kissoudis, C.; Farsakoglou, A.M.; Karagiannis, E.; Michailidis, M.; Polychroniadou, C.; Zambounis, A.; Kazantzis, K.; et al. Whole genome re-sequencing of sweet cherry (Prunus avium L.) yields insights into genomic diversity of a fruit species. Hortic. Res. 2020, 7, 60. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-bysequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Deschamps, S.; Llaca, V.; May, G.D. Genotyping-by-sequencing in plants. Biology 2012, 1, 460–483. [Google Scholar] [CrossRef]
- Poland, J.A.; Rife, T.W. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef]
- Pavan, S.; Curci, P.L.; Zuluaga, D.L.; Blanco, E.; Sonnante, G. Genotyping-by-sequencing highlights patterns of genetic structure and domestication in artichoke and cardoon. PLoS ONE 2018, 13, e0205988. [Google Scholar] [CrossRef] [PubMed]
- Lioi, L.; Zuluaga, D.L.; Pavan, S.; Sonnante, G. Genotyping-by-sequencing reveals molecular genetic diversity in Italian common bean landraces. Sci. Hortic. 2019, 11, 154. [Google Scholar] [CrossRef]
- Albertini, A.; della Strada, G. Monografia di Cultivar di Ciliegio Dolce; Tipografia Interstampa s.r.l: Rome, Italy, 1996. (In Italian) [Google Scholar]
- Albertini, A.; della Strada, G. Monografia di cultivar di ciliegio dolce e acido. In Progetto Finalizzato MiPAF Liste di Orientamento Varietale dei Fruttiferi; Tipografia Arti Grafiche Ciampino s.r.l: Rome, Italy, 2001; Volume 175. (In Italian) [Google Scholar]
- Choi, C.; Kappel, F. Inbreeding, coancestry, and founding clones of sweet cherries from North America. J. Am. Soc. Hortic. Sci. 2004, 129, 535–543. [Google Scholar] [CrossRef]
- Kappel, F.; Granger, A.; Hrotkó, K.; Schuster, M. Cherry. In Fruit Breed, Handbook of Plant Breeding; Springer: Boston, MA, USA, 2012; pp. 459–504. [Google Scholar] [CrossRef]
- Schuster, M.M. Self-incompatibility (S) genotypes of cultivated sweet cherries–An overview update 2020. OpenAgrar Repos. 2020. Available online: https://www.openagrar.de/receive/openagrar_mods_00064311 (accessed on 30 May 2022). [CrossRef]
- Quero-García, J.; Schuster, M.; López-Ortega, G.; Charlot, G. Sweet cherry varieties and improvement. In Cherries: Botany, Production and Uses; Quero-García, J., Iezzoni, A., Puławska, J., Lang, G., Eds.; CABI: Oxfordshire, UK; Boston, MA, USA, 2017; pp. 60–94. ISBN 9781780648378. [Google Scholar]
- Rosyara, U.R.; Sebolt, A.M.; Peace, C.; Iezzoni, A.F. Identification of the paternal parent of “Bing” sweet cherry and confirmation of descendants using single nucleotide polymorphism markers. J. Am. Soc. Hortic. Sci. 2014, 139, 148–156. [Google Scholar] [CrossRef]
- Palasciano, M.A.; Fanizza, G.; Resta, P. Le varietà locali non sono sempre autoctone: L’esempio della ciliegia “Ferrovia” (Prunus avium L.). Ital. J. Agron. 2009, 4, 699–704. (In Italian) [Google Scholar]
- Godini, A.; De Palma, L.; Palasciano, M. New and old sweet cherry cultivars suitable for Apulia (Southern Italy). Acta Hortic. 1996, 410, 75–80. [Google Scholar] [CrossRef]
- Corona, M.G.; Resta, P.; Fanizza, G.; Palasciano, M.; Godini, A. Prima indagine su varietà pugliesi di ciliegio dolce (Prunus avium L.) mediante lo studio dei polimorfismi del DNA (RAPDs). In Proceedings of the Atti Convegno Nazionale del Ciliegio, Valenzano, Bari, Italy, 19–21 June 1997; pp. 201–214. (In Italian). [Google Scholar]
- Lichou, J.; Edin, M.; Tronel, C.; Saunier, R. Le Cerisier: Les Variétés de Cerise; C.T.I.F.L.: Paris, France, 1990. (In French) [Google Scholar]
- Baldini, E. Indagine Sulle Cultivar di Ciliegio Diffuse in Italia; Minigraf: Bologna, Italy, 1973. (In Italian) [Google Scholar]
- Wünsch, A.; Hormaza, J.I. Molecular characterisation of sweet cherry (Prunus avium l.) genotypes using peach [Prunus persica (L.) Batsch] SSR sequences. Heredity 2002, 89, 56–63. [Google Scholar] [CrossRef]
- Palasciano, M. Ciliegio dolce. In Atlante dei Fruttiferi Autoctoni Italiani; Fideghelli, C., Ed.; BetMultimedia: Florence, Italy, 2017; pp. 706–884. (In Italian) [Google Scholar]
- Hedrick, U.P. The history of cultivated cherries. In The Cherries of New York; J.B. Lyon: Albany, NY, USA, 1915; pp. 39–64. [Google Scholar]
- Bargioni, G. Il Ciliegio Dolce; Edagricole: Bologna, Italy, 1982. (In Italian) [Google Scholar]
- Petriccione, M.; Rega, P.; Di Vaio, C. Campania. In Atlante dei Fruttiferi Autoctoni Italiani; Fideghelli, C., Ed.; BetMultimedia: Florence, Italy, 2016; pp. 287–304. [Google Scholar]
- UPOV. Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability: Cherry; Union Internationale Pour la Protection des Obtentions Végétales: Geneva, Switzerland, 1995; p. 16.
- Guajardo, V.; Solís, S.; Sagredo, B.; Gainza, F.; Muñoz, C.; Gasic, K.; Hinrichsen, P. Construction of high density sweet cherry (Prunus avium L.) linkage maps using microsatellite markers and SNPs detected by genotyping-by-sequencing (GBS). PLoS ONE 2015, 10, e0127750. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chaichoompu, K.; Abegaz, F.; Tongsima, S.; Shaw, P.J.; Sakuntabhai, A.; Pereira, L.; Van Steen, K. KRIS: Keen and Reliable Interface Subroutines for Bioinformatic Analysis. 2018. Available online: https://CRAN.R-project.org/package=KRIS (accessed on 3 January 2022).
- Hudson, R.R.; Slatkin, M.; Maddison, W.P. Estimation of levels of gene flow from DNA sequence data. Genetics 1992, 132, 583–589. [Google Scholar] [CrossRef]
- Bhatia, G.; Patterson, N.; Sankararaman, S.; Price, A.L. Estimating and interpreting FST: The impact of rare variants. Genome Res. 2013, 23, 1514–1521. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palasciano, M.; Zuluaga, D.L.; Cerbino, D.; Blanco, E.; Aufiero, G.; D’Agostino, N.; Sonnante, G. Sweet Cherry Diversity and Relationships in Modern and Local Varieties Based on SNP Markers. Plants 2023, 12, 136. https://doi.org/10.3390/plants12010136
Palasciano M, Zuluaga DL, Cerbino D, Blanco E, Aufiero G, D’Agostino N, Sonnante G. Sweet Cherry Diversity and Relationships in Modern and Local Varieties Based on SNP Markers. Plants. 2023; 12(1):136. https://doi.org/10.3390/plants12010136
Chicago/Turabian StylePalasciano, Marino, Diana L. Zuluaga, Domenico Cerbino, Emanuela Blanco, Gaetano Aufiero, Nunzio D’Agostino, and Gabriella Sonnante. 2023. "Sweet Cherry Diversity and Relationships in Modern and Local Varieties Based on SNP Markers" Plants 12, no. 1: 136. https://doi.org/10.3390/plants12010136
APA StylePalasciano, M., Zuluaga, D. L., Cerbino, D., Blanco, E., Aufiero, G., D’Agostino, N., & Sonnante, G. (2023). Sweet Cherry Diversity and Relationships in Modern and Local Varieties Based on SNP Markers. Plants, 12(1), 136. https://doi.org/10.3390/plants12010136








