Role of Nodulation-Enhancing Rhizobacteria in the Promotion of Medicago sativa Development in Nutrient-Poor Soils
Abstract
:1. Introduction
2. Results
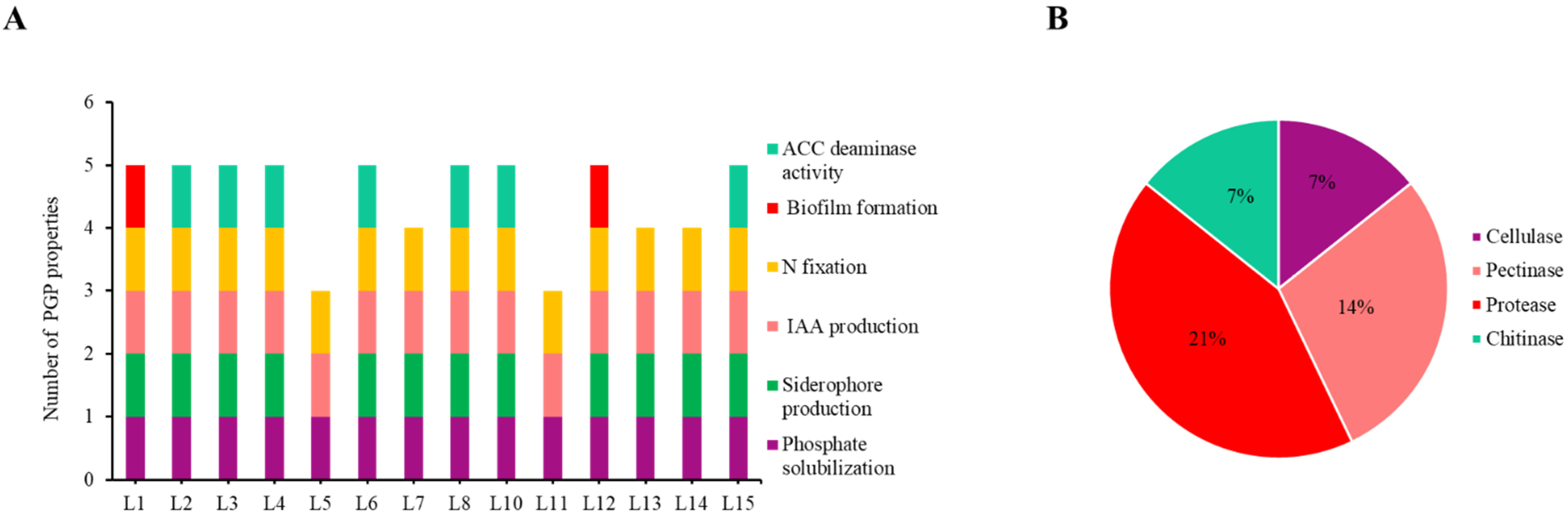
2.1. Isolation and Characterization of Rhizosphere Bacteria
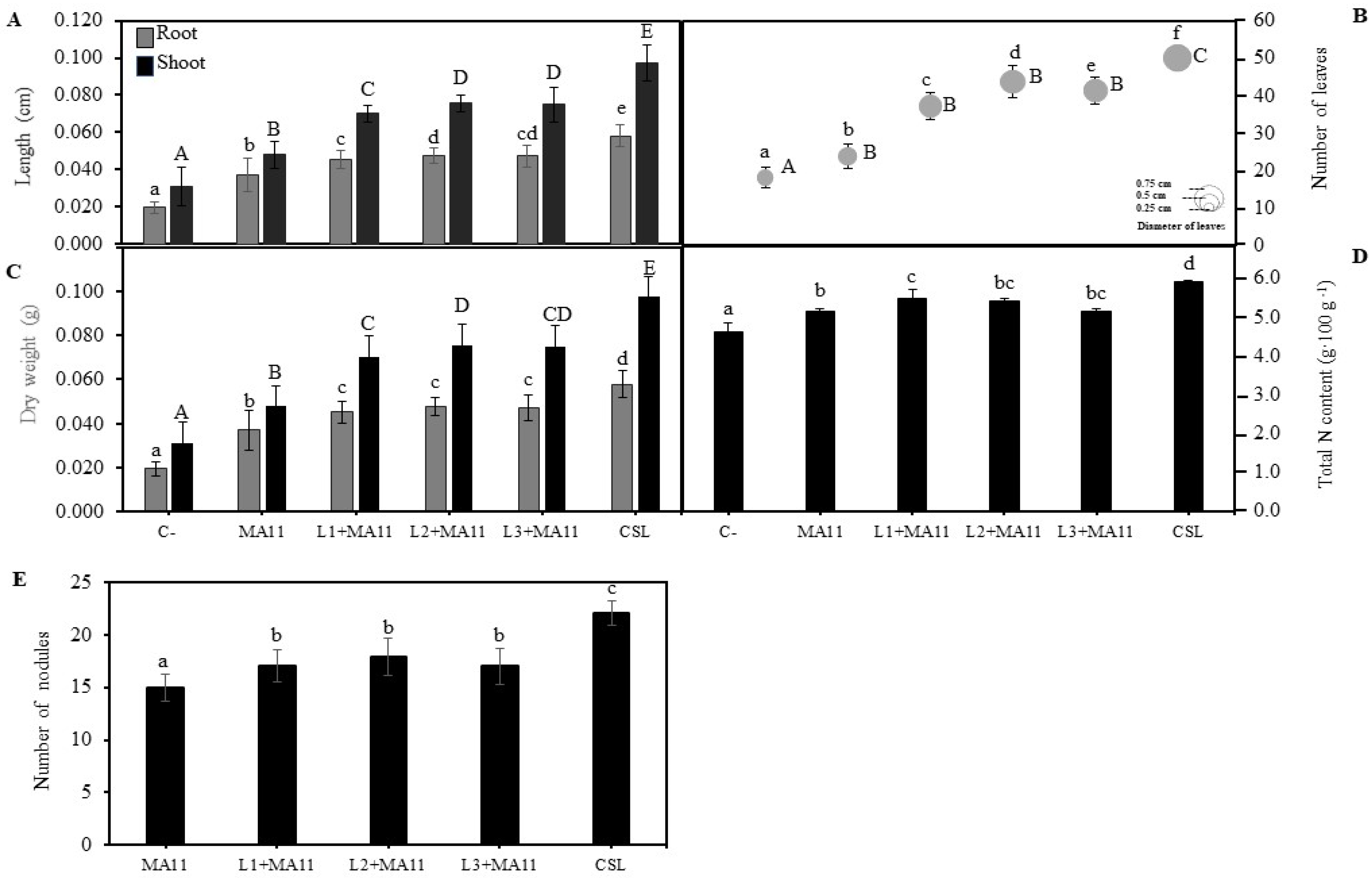
2.2. Effects of Selected PGPR in the Germination, Growth, and Nodulation of M. sativa Plants In Vitro
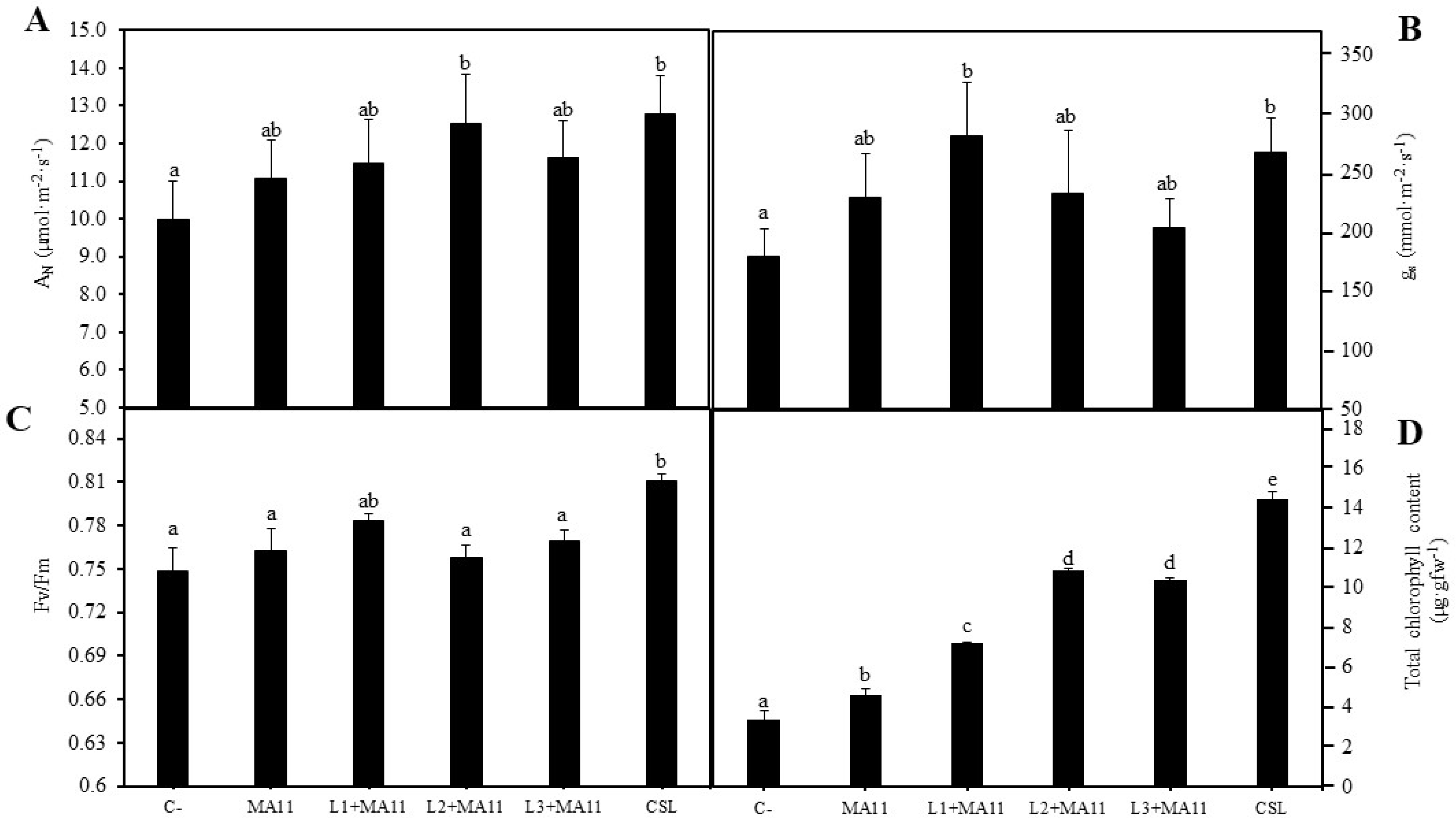
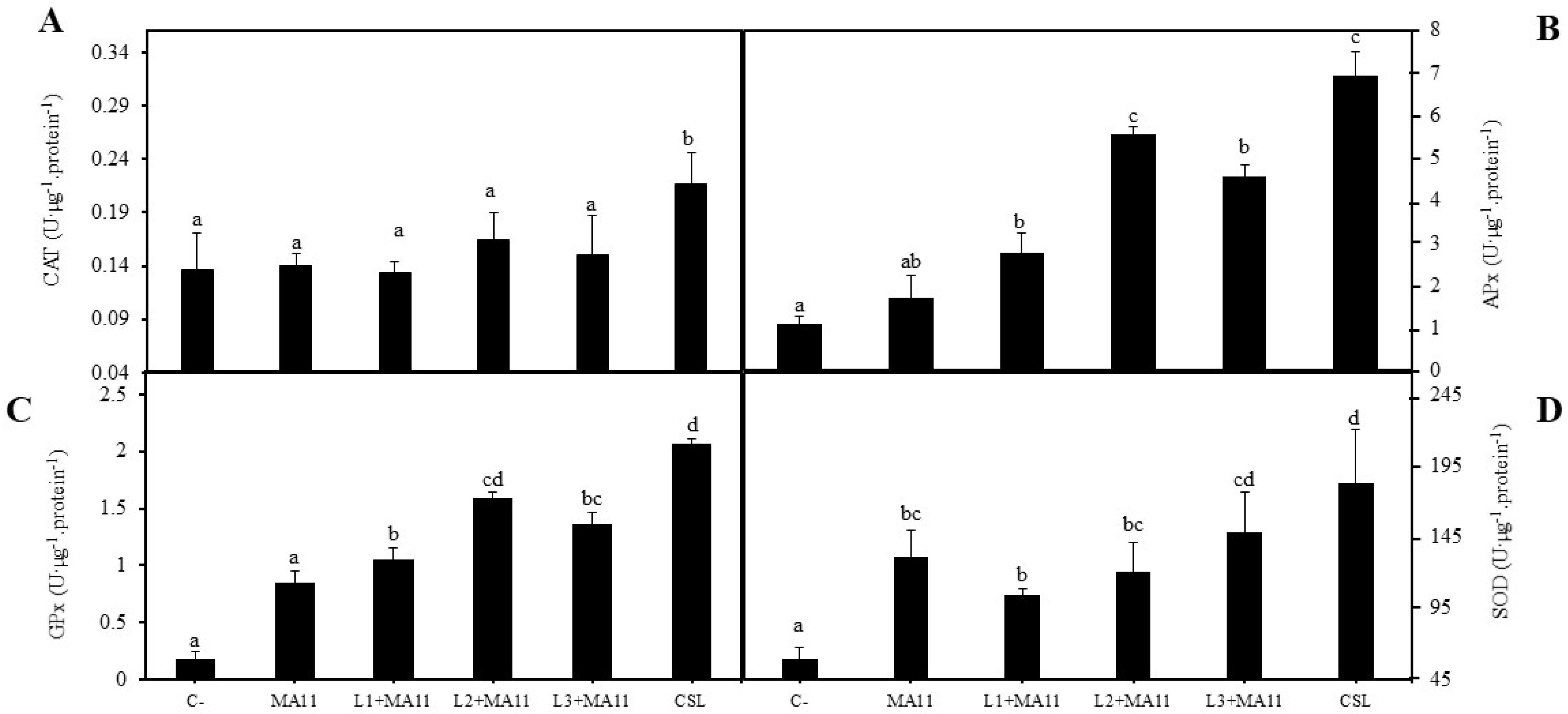
2.3. Effects of Selected PGPR in the Physiological Status and Nodulation of Plants under Nutrient-Poor Soil Stress
3. Discussion
4. Materials and Methods
4.1. Collection and Characterization of Soil
4.2. Isolation of Cultivable Rhizosphere Bacteria
4.3. Bacterial Screening Based on Phosphate Solubilization
4.4. Analysis of Diversity and Identification of Isolates
4.5. Bacteria Characterization
4.5.1. Study of Plant Growth Promoting Properties In Vitro
4.5.2. Study of Enzymatic Activities
4.6. Conditions and Inoculum Preparation
4.7. Seed Germination Assay
4.8. In Vitro Plant Cultivation
4.9. Greenhouse Experiments
4.9.1. Determination of Photosynthetic Parameters
4.9.2. Total Chlorophyll Content
4.9.3. Antioxidant Enzymes Measurement
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A.; et al. A new subfamily classification of the leguminosae based on a taxonomically comprehensive phylogeny: The Legume Phylogeny Working Group (LPWG). Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef] [Green Version]
- Lindström, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2019, 13, 1314–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markmann, K.; Parniske, M. Evolution of root endosymbiosis with bacteria: How novel are nodules? Trends Plant Sci. 2009, 14, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Chahboune, R.; Barrijal, S.; Moreno, S.; Bedmar, E.J. Characterization of Bradyrhizobium species isolated from root nodules of Cytisus villosus grown in Morocco. Syst. Appl. Microbiol. 2011, 34, 440–445. [Google Scholar] [CrossRef]
- Pucciariello, C.; Boscari, A.; Tagliani, A.; Brouquisse, R.; Perata, P. Exploring legume-rhizobia symbiotic models for waterlogging tolerance. Front. Plant Sci. 2019, 10, 578. [Google Scholar] [CrossRef] [PubMed]
- Sprent, J.I. Legume Nodulation: A Global Perspective; Wiley-Blackwell: Chichester, UK, 2009; p. 181. [Google Scholar] [CrossRef]
- Dai, J.; Qiu, W.; Wang, N.; Wang, T.; Nakanishi, H.; Zuo, Y. From Leguminosae/Gramineae Intercropping Systems to See Benefits of Intercropping on Iron Nutrition. Front. Plant Sci. 2019, 10, 605. [Google Scholar] [CrossRef] [Green Version]
- Raza, A.; Asghar, M.A.; Ahmad, B.; Bin, C.; Hussain, M.I.; Li, W.; Iqbal, T.; Yaseen, M.; Shafiq, I.; Yi, Z.; et al. Agro-Techniques for Lodging Stress Management in Maize-Soybean Intercropping System—A Review. Plants 2020, 9, 1592. [Google Scholar] [CrossRef]
- Matamoros, M.A.; Becana, M. Molecular responses of legumes to abiotic stress: Post-translational modifications of proteins and redox signaling. J. Exp. Bot. 2021, 72, 5876–5892. [Google Scholar] [CrossRef]
- Sharma, M.P.; Grover, M.; Chourasiya, D.; Bharti, A.; Agnihotri, R.; Maheshwari, H.S.; Pareek, A.; Buyer, J.S.; Sharma, S.K.; Schütz, L.; et al. Deciphering the Role of Trehalose in Tripartite Symbiosis Among Rhizobia, Arbuscular Mycorrhizal Fungi, and Legumes for Enhancing Abiotic Stress Tolerance in Crop Plants. Front. Microbiol. 2020, 11, 2219. [Google Scholar] [CrossRef]
- Alemneh, A.A.; Zhou, Y.; Ryder, M.H.; Denton, M.D. Mechanisms in plant growth-promoting rhizobacteria that enhance legume-rhizobial symbioses. J. Appl. Microbiol. 2020, 129, 1133–1156. [Google Scholar] [CrossRef]
- Babalola, O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010, 32, 1559–1570. [Google Scholar] [CrossRef] [PubMed]
- Carvalhais, L.C.; Dennis, P.G.; Badri, D.V.; Kidd, B.; Vivanco, J.M.; Schenk, P. Linking Jasmonic Acid Signaling, Root Exudates, and Rhizosphere Microbiomes. Mol. Plant-Microbe Interact. 2015, 28, 1049–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro-Torre, S.; Bessadok, K.; Flores-Duarte, N.J.; Rodríguez-Llorente, I.D.; Caviedes, M.A.; Pajuelo, E. Helping Legumes under Stress Situations: Inoculation with Beneficial Microorganisms. In Legume Crops—Prospects, Production and Uses; Hasanuzzaman, H., Ed.; IntechOpen: London, UK, 2020; pp. 115–135. [Google Scholar] [CrossRef]
- Chamkhi, I.; El Omari, N.; Balahbib, A.; El Menyiy, N.; Benali, T.; Ghoulam, C. Is the rhizosphere a source of applicable multi-beneficial microorganisms for plant enhancement? Saudi J. Biol. Sci. 2021, 29, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [Green Version]
- Chandwani, S.; Amaresan, N. Role of ACC deaminase producing bacteria for abiotic stress management and sustainable agriculture production. Environ. Sci. Pollut. Res. 2022, 29, 22843–22859. [Google Scholar] [CrossRef]
- Jones, K.; Kobayashi, H.; Davies, B.; Taga, M.E.; Walker, G.C. How rhizobial symbionts invade plants: The Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 2007, 5, 619–633. [Google Scholar] [CrossRef] [Green Version]
- Pajuelo, E.; Rodríguez-Llorente, I.D.; Dary, M.; Palomares, A.J. Toxic effects of arsenic on Sinorhizobium-Medicago sativa symbiotic interaction. Environ. Pollut. 2008, 154, 203–211. [Google Scholar] [CrossRef]
- Zhu, Q.; Wang, B.; Tan, J.; Liu, T.; Li, L.; Liu, Y.-G. Plant Synthetic Metabolic Engineering for Enhancing Crop Nutritional Quality. Plant Commun. 2020, 1, 100017. [Google Scholar] [CrossRef]
- Chodak, M.; Gołębiewski, M.; Morawska-Płoskonka, J.; Kuduk, K.; Niklińska, M. Soil chemical properties affect the reaction of forest soil bacteria to drought and rewetting stress. Ann. Microbiol. 2015, 65, 1627–1637. [Google Scholar] [CrossRef] [Green Version]
- Paul, D.; Lade, H. Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: A review. Agron. Sustain. Dev. 2014, 34, 737–752. [Google Scholar] [CrossRef]
- Vanlauwe, B.; Descheemaeker, K.; Giller, K.E.; Huising, J.; Merckx, R.; Nziguheba, G.; Wendt, J.; Zingore, S. Integrated soil fertility management in sub-Saharan Africa: Unravelling local adaptation. Soil 2015, 1, 491–508. [Google Scholar] [CrossRef] [Green Version]
- Chasek, P.; Akhtar-Schuster, M.; Orr, B.J.; Luise, A.; Ratsimba, H.R.; Safriel, U. Land degradation neutrality: The science-policy interface from the UNCCD to national implementation. Environ. Sci. Policy 2019, 92, 182–190. [Google Scholar] [CrossRef]
- Sá, J.C.D.M.; Lal, R.; Cerri, C.C.; Lorenz, K.; Hungria, M.; de Faccio Carvalho, P.C. Low-carbon agriculture in South America to mitigate global climate change and advance food security. Environ. Int. 2017, 98, 102–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanlauwe, B.; Hungria, M.; Kanampiu, F.; Giller, K. The role of legumes in the sustainable intensification of African smallholder agriculture: Lessons learnt and challenges for the future. Agric. Ecosyst. Environ. 2019, 284, 106583. [Google Scholar] [CrossRef]
- Abdalla, M.; Hastings, A.; Cheng, K.; Yue, Q.; Chadwick, D.; Espenberg, M.; Truu, J.; Rees, R.M.; Smith, P. A critical review of the impacts of cover crops on nitrogen leaching, net greenhouse gas balance and crop productivity. Glob. Chang. Biol. 2019, 25, 2530–2543. [Google Scholar] [CrossRef] [Green Version]
- Noori, F.; Etesami, H.; Zarini, H.N.; Khoshkholgh-Sima, N.A.; Salekdeh, G.H.; Alishahi, F. Mining alfalfa (Medicago sativa L.) nodules for salinity tolerant non-rhizobial bacteria to improve growth of alfalfa under salinity stress. Ecotoxicol. Environ. Saf. 2018, 162, 129–138. [Google Scholar] [CrossRef]
- Garrido Valero, M.S. Interpretación de Análisis de Suelos; Ministerio de Agricultura, Pesca y Alimentación: Madrid, Spain, 1993; p. 40.
- Moreira, M.U. Práticas de Solos; Publindústria: Porto, Portugal, 2012; p. 142. [Google Scholar]
- Food and Agriculture Organization (FAO). Status of the World’s Soil Resources (SWSR)-Main Report; Food and Agriculture Organization of the United Nations and Intergovernmental Technical Panel on Soils: Rome, Italy, 2015; p. 650. [Google Scholar]
- Odelade, K.A.; Babalola, O.O. Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity. Int. J. Environ. Res. Public Health 2019, 16, 3873. [Google Scholar] [CrossRef] [Green Version]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [Green Version]
- Hartman, K.; Tringe, S.G. Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef] [Green Version]
- Qiu, L.; Zhang, Q.; Zhu, H.; Reich, P.B.; Banerjee, S.; van der Heijden, M.G.A.; Sadowsky, M.J.; Ishii, S.; Jia, X.; Shao, M.; et al. Erosion reduces soil microbial diversity, network complexity and multifunctionality. ISME J. 2021, 15, 2474–2489. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Feng, J.; Shi, Z.; Zhou, X.; Yuan, M.; Tao, X.; Hale, L.; Yuan, T.; Wang, J.; Qin, Y.; et al. Climate warming leads to divergent succession of grassland microbial communities. Nat. Clim. Chang. 2018, 8, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Ochoa-Hueso, R.; Collins, S.L.; Delgado-Baquerizo, M.; Hamonts, K.; Pockman, W.T.; Sinsabaugh, R.L.; Smith, M.D.; Knapp, A.K.; Power, S.A. Drought consistently alters the composition of soil fungal and bacterial communities in grasslands from two continents. Glob. Chang. Biol. 2018, 24, 2818–2827. [Google Scholar] [CrossRef] [PubMed]
- Araújo, A.S.F.; Borges, C.D.; Tsai, S.M.; Cesarz, S.; Eisenhauer, N. Soil bacterial diversity in degraded and restored lands of Northeast Brazil. Antonie van Leeuwenhoek 2014, 106, 891–899. [Google Scholar] [CrossRef]
- Sah, S.; Singh, R. Phylogenetical coherence of Pseudomonas in unexplored soils of Himalayan region. 3 Biotech 2016, 6, 170. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Wang, X.; Deng, P.; Ma, L.; Baird, S.M.; Li, X.; Lu, S. Pseudomonas glycinae sp. nov. isolated from the soybean rhizosphere. MicrobiologyOpen 2020, 9, e1101. [Google Scholar] [CrossRef]
- Zhao, H.; Ma, Y.; Wu, X.; Zhang, L. Pseudomonas viciae sp. nov., isolated from rhizosphere of broad bean. Int. J. Syst. Evol. Microbiol. 2020, 70, 5012–5018. [Google Scholar] [CrossRef]
- Wang, M.-Q.; Wang, Z.; Yu, L.-N.; Zhang, C.-S.; Bi, J.; Sun, J. Pseudomonas qingdaonensis sp. nov., an aflatoxin-degrading bacterium, isolated from peanut rhizospheric soil. Arch. Microbiol. 2019, 201, 673–678. [Google Scholar] [CrossRef]
- Wang, M.Q.; Zhang, C.S.; Yu, L.N.; Yang, W.Q.; Jiao, K.; Gong, K.J.; Chi, X.Y.; Bi, J.; Song, Y.; Yang, Q.L.; et al. Pseudomonas laoshanensis sp. nov., isolated from peanut field soil. Arch. Microbiol. 2021, 203, 829–834. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, X.; Kahaer, M.; Tian, T.; Sun, Y. Chryseobacterium endalhagicum sp. nov., isolated from seed of leguminous plant. Int. J. Syst. Evol. Microbiol. 2021, 71, 005077. [Google Scholar] [CrossRef]
- Korir, H.; Mungai, N.W.; Thuita, M.; Hamba, Y.; Masso, C.; Tola, Y.H. Co-inoculation Effect of Rhizobia and Plant Growth Promoting Rhizobacteria on Common Bean Growth in a Low Phosphorus Soil. Front. Plant Sci. 2017, 8, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, N.; Zhao, S.; Tian, C.-Y. Effect of halotolerant rhizobacteria isolated from halophytes on the growth of sugar beet (Beta vulgaris L.) under salt stress. FEMS Microbiol. Lett. 2017, 364, fnx091. [Google Scholar] [CrossRef] [PubMed]
- Benidire, L.; El Khalloufi, F.; Oufdou, K.; Barakat, M.; Tulumello, J.; Ortet, P.; Heulin, T.; Achouak, W. Phytobeneficial bacteria improve saline stress tolerance in Vicia faba and modulate microbial interaction network. Sci. Total Environ. 2020, 729, 139020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-L.; Jia, F.; Li, M.; Yu, F.; Zhou, B.; Hao, Q.-H.; Wang, X.-L. Endophytic Bacillus strains isolated from alfalfa (Medicago sativa L.) seeds: Enhancing the lifespan of Caenorhabditis elegans. Lett. Appl. Microbiol. 2018, 68, 226–233. [Google Scholar] [CrossRef] [PubMed]
- de Meyer, S.E.; de Beuf, K.; Vekeman, B.; Willems, A. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol. Biochem. 2015, 83, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Turan, M.; Kitir, N.; Alkaya, Ü.; Günes, A.; Tüfenkçi, S.; Yildirim, E.; Nikerel, E. Making Soil More Accessible to Plants: The Case of Plant Growth Promoting Rhizobacteria. In Plant Growth; Rigobelo, E.C., Ed.; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Alori, E.T.; Babalola, O.O. Microbial Inoculants for Improving Crop Quality and Human Health in Africa. Front. Microbiol. 2018, 9, 2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, C.K.; Saraf, M. Plant growth promoting rhizobacteria (PGPR): A review. E3 J. Agric. Res. Dev. 2015, 5, 108–0119. [Google Scholar]
- Bashan, Y.; De-Bashan, L.E. Inoculant Preparation and Formulations for Azospirillum spp. In Handbook for Azospirillum: Technical Issues and Protocols; Cassán, F., Okon, Y., Creus, C., Eds.; Springer: Cham, Switzerland; Edinburgh, UK, 2015; pp. 469–485. [Google Scholar] [CrossRef]
- Gupta, G.; Parihar, S.S.; Ahirwar, N.K.; Snehi, S.K.; Singh, V. Plant growth promoting rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 2015, 7, 96–102. [Google Scholar] [CrossRef]
- Selim, S.M.; Zayed, M.S. Role of Biofertilizers in Sustainable Agriculture Under Abiotic Stresses. In Microorganisms for Green Revolution. Microorganisms for Sustainability; Panpatte, D., Jhala, Y., Vyas, R., Shelat, H., Eds.; Springer: Singapore, 2017; Volume 6, pp. 281–301. [Google Scholar] [CrossRef]
- Kang, B.G.; Kim, W.T.; Yun, H.S.; Chang, S.C. Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol. Rep. 2010, 4, 179–183. [Google Scholar] [CrossRef]
- Guinel, F.C. Ethylene, a Hormone at the Center-Stage of Nodulation. Front. Plant Sci. 2015, 6, 1121. [Google Scholar] [CrossRef] [Green Version]
- Benito, P.; Alonso-Vega, P.; Aguado, C.; Luján, R.; Anzai, Y.; Hirsch, A.M.; Trujillo, M.E. Monitoring the colonization and infection of legume nodules by Micromonospora in co-inoculation experiments with rhizobia. Sci. Rep. 2017, 7, 11051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Hidalgo, P.; Hirsch, A.M. The Nodule Microbiome: N2-Fixing Rhizobia Do Not Live Alone. Phytobiomes J. 2017, 1, 70–82. [Google Scholar] [CrossRef] [Green Version]
- Rossi, F. Beneficial biofilms for land rehabilitation and fertilization. FEMS Microbiol. Lett. 2020, 367, fnaa184. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, E.; Robledo, M.; Jiménez-Zurdo, J.I.; Velázquez, E.; Rivas, R.; Murray, J.D.; Mateos, P.F. Legumes display common and host-specific responses to the rhizobial cellulase CelC2 during primary symbiotic infection. Sci. Rep. 2019, 9, 13907. [Google Scholar] [CrossRef] [Green Version]
- Bhadrecha, P.; Bala, M.; Khasa, Y.P.; Arshi, A.; Singh, J.; Kumar, M. Hippophae rhamnoides L. rhizobacteria exhibit diversified cellulase and pectinase activities. Physiol. Mol. Biol. Plants 2020, 26, 1075–1085. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, H.; Leng, J.; Niu, H.; Chen, X.; Liu, D.; Chen, Y.; Gao, N.; Ying, H. Isolation and characterization of plant growth-promoting rhizobacteria and their effects on the growth of Medicago sativa L. under salinity conditions. Antonie van Leeuwenhoek 2020, 113, 1263–1278. [Google Scholar] [CrossRef]
- Pajuelo, E.; Arjona, S.; Rodríguez-Llorente, I.D.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Merchán, F.; Navarro-Torre, S. Coastal Ecosystems as Sources of Biofertilizers in Agriculture: From Genomics to Application in an Urban Orchard. Front. Mar. Sci. 2021, 8, 685076. [Google Scholar] [CrossRef]
- Liu, J.; Liu, X.; Zhang, Q.; Li, S.; Sun, Y.; Lu, W.; Ma, C. Response of alfalfa growth to arbuscular mycorrhizal fungi and phosphate-solubilizing bacteria under different phosphorus application levels. AMB Express 2020, 10, 200. [Google Scholar] [CrossRef]
- Yao, X.; Chen, P.; Cheng, T.; Sun, K.; Megharaj, M.; He, W. Inoculation of Bacillus megaterium strain A14 alleviates cadmium accumulation in peanut: Effects and underlying mechanisms. J. Appl. Microbiol. 2021, 131, 819–832. [Google Scholar] [CrossRef]
- Raklami, A.; Oufdou, K.; Tahiri, A.-I.; Mateos-Naranjo, E.; Navarro-Torre, S.; Rodríguez-Llorente, I.D.; Meddich, A.; Redondo-Gómez, S.; Pajuelo, E. Safe Cultivation of Medicago sativa in Metal-Polluted Soils from Semi-Arid Regions Assisted by Heat- and Metallo-Resistant PGPR. Microorganisms 2019, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Saidi, S.; Cherif-Silini, H.; Bouket, A.C.; Silini, A.; Eshelli, M.; Luptakova, L.; Alenezi, F.N.; Belbahri, L. Improvement of Medicago sativa Crops Productivity by the Co-inoculation of Sinorhizobium meliloti-Actinobacteria under Salt Stress. Curr. Microbiol. 2021, 78, 1344–1357. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: Description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Marín, J.; Naranjo, E.M.; Caviedes, M.; Redondo-Gómez, S.; Pajuelo, E.; Rodríguez-Llorente, I. Scouting contaminated estuaries: Heavy metal resistant and plant growth promoting rhizobacteria in the native metal rhizoaccumulator Spartina maritima. Mar. Pollut. Bull. 2015, 90, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Bouyoucos, G.J. Directions for making mechanical analyses of soils by the hydrometer method. Soil Sci. 1936, 42, 225–230. [Google Scholar] [CrossRef]
- Navarro-Torre, S.; Naranjo, E.M.; Caviedes, M.; Pajuelo, E.; Rodríguez-Llorente, I. Isolation of plant-growth-promoting and metal-resistant cultivable bacteria from Arthrocnemum macrostachyum in the Odiel marshes with potential use in phytoremediation. Mar. Pollut. Bull. 2016, 110, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Ji, S.H.; Gururani, M.; Chun, S.-C. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef]
- del Castillo, I.; Hernández, P.; Lafuente, A.; Rodríguez-Llorente, I.; Caviedes, M.; Pajuelo, E. Self-bioremediation of cork-processing wastewaters by (chloro)phenol-degrading bacteria immobilised onto residual cork particles. Water Res. 2012, 46, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Harley, J.P.; Prescott, L.M. Laboratory Exercises in Microbiology, 5th ed.; McGraw-Hill: New York, NY, USA, 2002; p. 320. [Google Scholar]
- Mesa-Marín, J.; Naranjo, E.M.; Caviedes, M.A.; Redondo-Gómez, S.; Pajuelo, E.; Rodríguez-Llorente, I.D. Endophytic Cultivable Bacteria of the Metal Bioaccumulator Spartina maritima Improve Plant Growth but Not Metal Uptake in Polluted Marshes Soils. Front. Microbiol. 2015, 6, 1450. [Google Scholar] [CrossRef]
- Elbeltagy, A.; Nishioka, K.; Suzuki, H.; Sato, T.; Sato, Y.-I.; Morisaki, H.; Mitsui, H.; Minamisawa, K. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci. Plant Nutr. 2000, 46, 617–629. [Google Scholar] [CrossRef]
- Ehrhardt, D.; Atkinson, E.; Long, S.R. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 1992, 256, 998–1000. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Torre, S.; Rodríguez-Llorente, I.D.; Doukkali, B.; Caviedes, M.A.; Pajuelo, E. Competition for alfalfa nodulation under metal stress by the metal-tolerant strain Ochrobactrum cytisi Azn6. Ann. Appl. Biol. 2019, 175, 184–192. [Google Scholar] [CrossRef]
- Carrasco, J.A.; Armario, P.; Pajuelo, E.; Burgos, A.; Caviedes, M.A.; Lopez, J.A.C.; Chamber, M.A.; Palomares, A.J. Isolation and characterisation of symbiotically effective Rhizobium resistant to arsenic and heavy metals after the toxic spill at the Aznalcóllar pyrite mine. Soil Biol. Biochem. 2005, 37, 1131–1140. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Schreiber, U.; Schliwa, U.; Bilger, W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 1986, 10, 51–62. [Google Scholar] [CrossRef]
- Krall, J.P.; Edwards, G.E. Relationship between photosystem II activity and CO2 fixation in leaves. Physiol. Plant. 1992, 86, 180–187. [Google Scholar] [CrossRef]
- Hiscox, J.D.; Israelstam, G.F. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979, 57, 1332–1334. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte, B.; Goessling, J.; Marques, J.C.; Caçador, I. Ecophysiological constraints of Aster tripolium under extreme thermal events impacts: Merging biophysical, biochemical and genetic insights. Plant Physiol. Biochem. 2015, 97, 217–228. [Google Scholar] [CrossRef] [PubMed]

| Physicochemical Properties | |||||
|---|---|---|---|---|---|
| Texture (%) * | Organic Material (%) | Conductivity (mS·cm−1) | pH | ||
| 92/4/4 | 0.98 ± 0.01 | 1.245 ± 0.009 | 8.5 ± 0.014 | ||
| Nutrient concentration (mg·kg−1) | |||||
| Ca | K | Mg | Mn | Na | P |
| 3.162 ± 0.448 | 0.253 ± 0.021 | 0.277 ± 0.029 | 189.822 ± 2.860 | 0.159 ± 0.006 | 0.014 ± 0.002 |
| Strain | Sequence Size (bp) | Related Species | % ID | Accession Number |
|---|---|---|---|---|
| L1 | 469 | Pseudomonas kitaguniensis | 98.01 | OM397092 |
| L2 | 1272 | Chryseobacterium soli | 99.76 | OM397093 |
| L3 | 1366 | Priestia megaterium | 100 | OM397094 |
| L6 | 1298 | Pseudomonas canadensis | 100 | OM397096 |
| L7 | 1270 | Bacillus paramycoides | 100 | OM397095 |
| L10 | 1293 | Pseudomonas baetica | 99.77 | OM397097 |
| L12 | 1277 | Pseudomonas moorei | 99.76 | OM397098 |
| L13 | 1301 | Pseudomonas moorei | 99.69 | OM397099 |
| L14 | 937 | Pseudomonas moraviensis | 99.89 | OM442986 |
| L15 | 1317 | Buttiauxella noackiae | 99.92 | OM397100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores-Duarte, N.J.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Pajuelo, E.; Rodriguez-Llorente, I.D.; Navarro-Torre, S. Role of Nodulation-Enhancing Rhizobacteria in the Promotion of Medicago sativa Development in Nutrient-Poor Soils. Plants 2022, 11, 1164. https://doi.org/10.3390/plants11091164
Flores-Duarte NJ, Mateos-Naranjo E, Redondo-Gómez S, Pajuelo E, Rodriguez-Llorente ID, Navarro-Torre S. Role of Nodulation-Enhancing Rhizobacteria in the Promotion of Medicago sativa Development in Nutrient-Poor Soils. Plants. 2022; 11(9):1164. https://doi.org/10.3390/plants11091164
Chicago/Turabian StyleFlores-Duarte, Noris J., Enrique Mateos-Naranjo, Susana Redondo-Gómez, Eloísa Pajuelo, Ignacio D. Rodriguez-Llorente, and Salvadora Navarro-Torre. 2022. "Role of Nodulation-Enhancing Rhizobacteria in the Promotion of Medicago sativa Development in Nutrient-Poor Soils" Plants 11, no. 9: 1164. https://doi.org/10.3390/plants11091164
APA StyleFlores-Duarte, N. J., Mateos-Naranjo, E., Redondo-Gómez, S., Pajuelo, E., Rodriguez-Llorente, I. D., & Navarro-Torre, S. (2022). Role of Nodulation-Enhancing Rhizobacteria in the Promotion of Medicago sativa Development in Nutrient-Poor Soils. Plants, 11(9), 1164. https://doi.org/10.3390/plants11091164









