Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses
Abstract
1. Introduction
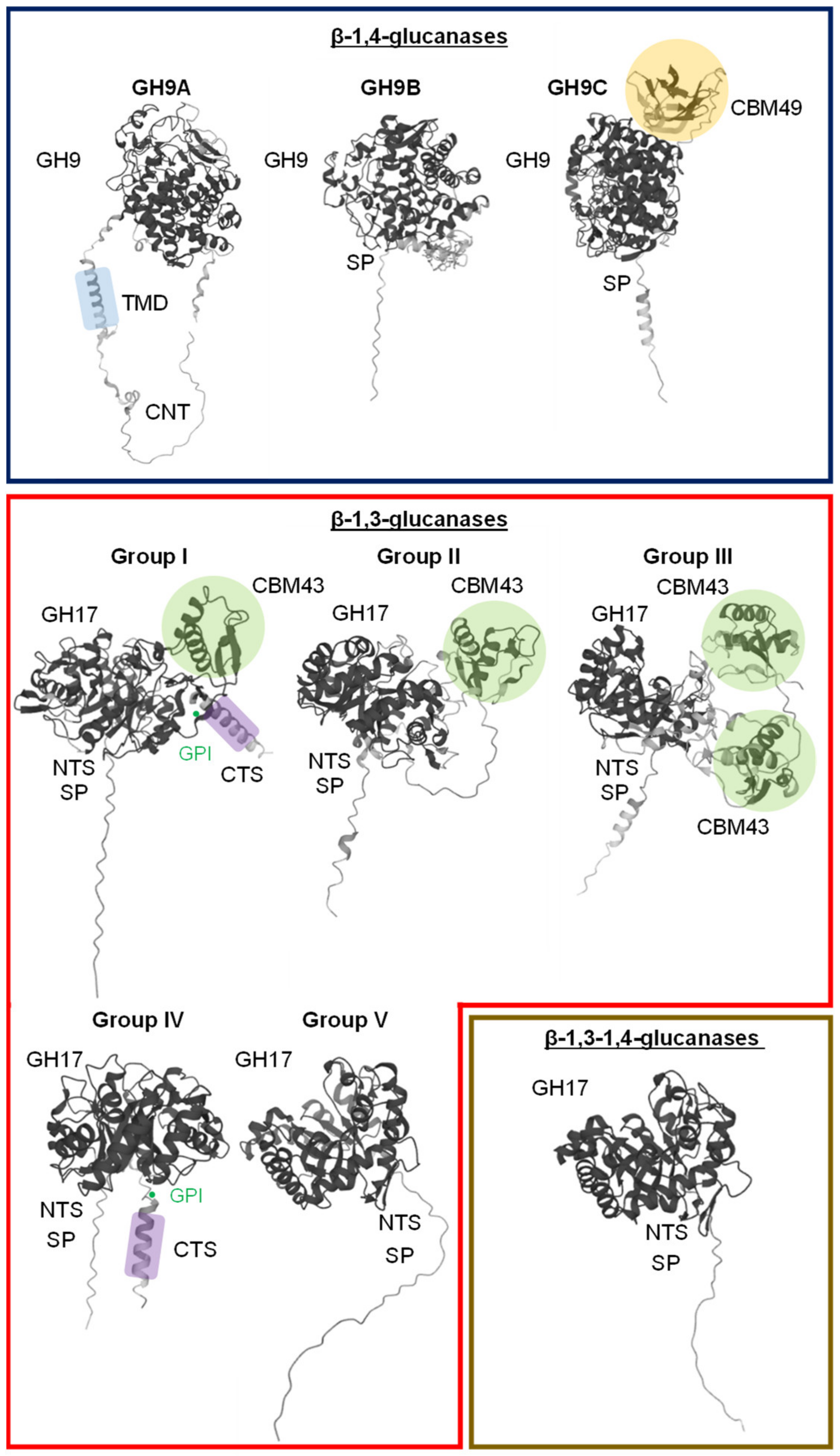
2. β-1,4-Glucanases
2.1. Classification and Evolutionary Origin
2.2. Biological Roles
3. β-1,3-Glucanases
3.1. Classification and Evolutionary Origin
3.2. Biological Roles
4. β-1,3-1,4-Glucanases
Biological Roles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The Plant Cell Wall: A Complex and Dynamic Structure As Revealed by the Responses of Genes under Stress Conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The Plant Cell Wall: Biosynthesis, Construction, and Functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Labavitch, J.M. Cell Wall Turnover in Plant Development. Annu. Rev. Plant Physiol. 1981, 32, 385–406. [Google Scholar] [CrossRef]
- Tucker, M.R.; Lou, H.; Aubert, M.K.; Wilkinson, L.G.; Little, A.; Houston, K.; Pinto, S.C.; Shirley, N.J. Exploring the Role of Cell Wall-Related Genes and Polysaccharides during Plant Development. Plants 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Schomburg, I.; Chang, A.; Schomburg, D. BRENDA, Enzyme Data and Metabolic Information. Nucleic Acids Res. 2002, 30, 47–49. [Google Scholar] [CrossRef]
- Simmons, C.R. The Physiology and Molecular Biology of Plant 1,3-β-D-Glucanases and 1,3;1,4-β-D-Glucanases. Crit. Rev. Plant Sci. 1994, 13, 325–387. [Google Scholar] [CrossRef]
- Høj, P.B.; Fincher, G.B. Molecular Evolution of Plant Beta-Glucan Endohydrolases. Plant J. 1995, 7, 367–379. [Google Scholar] [CrossRef]
- Del Campillo, E. Multiple Endo-1,4-Beta-D-Glucanase (Cellulase) Genes in Arabidopsis. Curr. Top. Dev. Biol. 1999, 46, 39–61. [Google Scholar] [CrossRef]
- Henrissat, B. A Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1991, 280 Pt 2, 309–316. [Google Scholar] [CrossRef]
- Wilson, D.; Urbanowicz, B. Glycoside Hydrolase Family 9/Plant Endoglucanases. Available online: https://www.cazypedia.org/index.php/Glycoside_Hydrolase_Family_9/Plant_endoglucanases (accessed on 22 March 2022).
- Nicol, F.; His, I.; Jauneau, A.; Vernhettes, S.; Canut, H.; Höfte, H. A Plasma Membrane-Bound Putative Endo-1,4-Beta-D-Glucanase Is Required for Normal Wall Assembly and Cell Elongation in Arabidopsis. EMBO J. 1998, 17, 5563–5576. [Google Scholar] [CrossRef] [PubMed]
- Szyjanowicz, P.M.J.; McKinnon, I.; Taylor, N.G.; Gardiner, J.; Jarvis, M.C.; Turner, S.R. The Irregular Xylem 2 Mutant Is an Allele of Korrigan That Affects the Secondary Cell Wall of Arabidopsis thaliana. Plant J. 2004, 37, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Vain, T.; Crowell, E.F.; Timpano, H.; Biot, E.; Desprez, T.; Mansoori, N.; Trindade, L.M.; Pagant, S.; Robert, S.; Höfte, H.; et al. The Cellulase KORRIGAN Is Part of the Cellulose Synthase Complex. Plant Physiol. 2014, 165, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, J.M.; Brumer, H. The XTH Gene Family: An Update on Enzyme Structure, Function, and Phylogeny in Xyloglucan Remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef]
- Smith, R.C.; Fry, S.C. Endotransglycosylation of Xyloglucans in Plant Cell Suspension Cultures. Biochem. J. 1991, 279, 529–535. [Google Scholar] [CrossRef]
- Nishitani, K.; Tominaga, R. Endo-Xyloglucan Transferase, a Novel Class of Glycosyltransferase That Catalyzes Transfer of a Segment of Xyloglucan Molecule to Another Xyloglucan Molecule. J. Biol. Chem. 1992, 267, 21058–21064. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH Family of Enzymes Involved in Xyloglucan Endotransglucosylation and Endohydrolysis: Current Perspectives and a New Unifying Nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Herburger, K.; Franková, L.; Pičmanová, M.; Loh, J.W.; Valenzuela-Ortega, M.; Meulewaeter, F.; Hudson, A.D.; French, C.E.; Fry, S.C. Hetero-Trans-β-Glucanase Produces Cellulose-Xyloglucan Covalent Bonds in the Cell Walls of Structural Plant Tissues and Is Stimulated by Expansin. Mol. Plant 2020, 13, 1047–1062. [Google Scholar] [CrossRef]
- Ishida, K.; Yokoyama, R. Reconsidering the Function of the Xyloglucan Endotransglucosylase/Hydrolase Family. J. Plant Res. 2022, 135, 145–156. [Google Scholar] [CrossRef]
- Hrmova, M.; Stratilová, B.; Stratilová, E. Broad Specific Xyloglucan:Xyloglucosyl Transferases Are Formidable Players in the Re-Modelling of Plant Cell Wall Structures. Int. J. Mol. Sci. 2022, 23, 1656. [Google Scholar] [CrossRef]
- Moore, A.E.; Stone, B.A. A β-1,3-Glucan Hydrolase from Nicotiana Glutinosa II. Specificity, Action Pattern and Inhibitor Studies. Biochim. Biophys. Acta BBA-Enzymol. 1972, 258, 248–264. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The Structure and Synthesis of the Fungal Cell Wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-Glucanases: Their Biological Functions and Transgenic Expression against Phytopathogenic Fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef]
- Abeles, F.B.; Bosshart, R.P.; Forrence, L.E.; Habig, W.H. Preparation and Purification of Glucanase and Chitinase from Bean Leaves 1. Plant Physiol. 1971, 47, 129–134. [Google Scholar] [CrossRef]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Parrish, F.W.; Perlin, A.S.; Reese, E.T. Selective Enzymolysis of Poly-β-D-glucans, and The Structure of The Polymers. Can. J. Chem. 1960, 38, 2094–2104. [Google Scholar] [CrossRef]
- McCleary, B.V. Purification of (1 → 3),(1 → 4)-β-d-Glucan from Barley Flour. In Methods in Enzymology; Biomass Part A: Cellulose and Hemicellulose; Academic Press: Cambridge, MA, USA, 1988; Volume 160, pp. 511–514. [Google Scholar]
- Perlin, A.S.; Suzuki, S. The Structure of Lichenin: Selective Enzymolysis Studies. Can. J. Chem. 1962, 40, 50–56. [Google Scholar] [CrossRef]
- Olafsdottir, E.S.; Ingólfsdottir, K. Polysaccharides from Lichens: Structural Characteristics and Biological Activity. Planta Med. 2001, 67, 199–208. [Google Scholar] [CrossRef]
- Stone, B.A. Chemistry of β-Glucans. In Chemistry, Biochemistry, and Biology of 1–3 Beta Glucans and Related Polysaccharides; Bacic, A., Fincher, G.B., Stone, B.A., Eds.; Academic Press: San Diego, CA, USA, 2009; Chapter 2.1; pp. 5–46. ISBN 978-0-12-373971-1. [Google Scholar]
- Varghese, J.N.; Garrett, T.P.; Colman, P.M.; Chen, L.; Høj, P.B.; Fincher, G.B. Three-Dimensional Structures of Two Plant Beta-Glucan Endohydrolases with Distinct Substrate Specificities. Proc. Natl. Acad. Sci. USA 1994, 91, 2785–2789. [Google Scholar] [CrossRef]
- Bulone, V.; Schwerdt, J.G.; Fincher, G.B. Co-Evolution of Enzymes Involved in Plant Cell Wall Metabolism in the Grasses. Front. Plant Sci. 2019, 10, 1009. [Google Scholar] [CrossRef]
- Libertini, E.; Li, Y.; McQueen-Mason, S.J. Phylogenetic Analysis of the Plant Endo-Beta-1,4-Glucanase Gene Family. J. Mol. Evol. 2004, 58, 506–515. [Google Scholar] [CrossRef]
- Urbanowicz, B.R.; Bennett, A.B.; del Campillo, E.; Catalá, C.; Hayashi, T.; Henrissat, B.; Höfte, H.; McQueen-Mason, S.J.; Patterson, S.E.; Shoseyov, O.; et al. Structural Organization and a Standardized Nomenclature for Plant Endo-1,4-β-Glucanases (Cellulases) of Glycosyl Hydrolase Family 9. Plant Physiol. 2007, 144, 1693–1696. [Google Scholar] [CrossRef]
- Buchanan, M.; Burton, R.A.; Dhugga, K.S.; Rafalski, A.J.; Tingey, S.V.; Shirley, N.J.; Fincher, G.B. Endo-(1,4)-β-Glucanase Gene Families in the Grasses: Temporal and Spatial Co-Transcription of Orthologous Genes1. BMC Plant Biol. 2012, 12, 235. [Google Scholar] [CrossRef]
- Du, Q.; Wang, L.; Yang, X.; Gong, C.; Zhang, D. Populus Endo-β-1,4-Glucanases Gene Family: Genomic Organization, Phylogenetic Analysis, Expression Profiles and Association Mapping. Planta 2015, 241, 1417–1434. [Google Scholar] [CrossRef]
- Henrissat, B.; Claeyssens, M.; Tomme, P.; Lemesle, L.; Mornon, J.-P. Cellulase Families Revealed by Hydrophobic Cluster Analysi. Gene 1989, 81, 83–95. [Google Scholar] [CrossRef]
- Davies, G.; Henrissat, B. Structures and Mechanisms of Glycosyl Hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Kundu, S.; Sharma, R. Origin, Evolution, and Divergence of Plant Class C GH9 Endoglucanases. BMC Evol. Biol. 2018, 18, 79. [Google Scholar] [CrossRef]
- Kundu, S. Insights into the Mechanism(s) of Digestion of Crystalline Cellulose by Plant Class C GH9 Endoglucanases. J. Mol. Model. 2019, 25, 240. [Google Scholar] [CrossRef]
- Mølhøj, M.; Johansen, B.; Ulvskov, P.; Borkhardt, B. Expression of a Membrane-Anchored Endo-1,4-β-Glucanase from Brassica napus, Orthologous to KOR from Arabidopsis thaliana, Is Inversely Correlated to Elongation in Light-Grown Plants. Plant Mol. Biol. 2001, 45, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Mølhøj, M.; Pagant, S.; Höfte, H. Towards Understanding the Role of Membrane-Bound Endo-β-1,4-Glucanases in Cellulose Biosynthesis. Plant Cell Physiol. 2002, 43, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Rasmussen, C.G. Cell Biology of Primary Cell Wall Synthesis in Plants. Plant Cell 2022, 34, 103–128. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, N.; Timmers, J.; Desprez, T.; Kamei, C.L.A.; Dees, D.C.T.; Vincken, J.-P.; Visser, R.G.F.; Höfte, H.; Vernhettes, S.; Trindade, L.M. KORRIGAN1 Interacts Specifically with Integral Components of the Cellulose Synthase Machinery. PLoS ONE 2014, 9, e112387. [Google Scholar] [CrossRef]
- Worden, N.; Wilkop, T.E.; Esteve, V.E.; Jeannotte, R.; Lathe, R.; Vernhettes, S.; Weimer, B.; Hicks, G.; Alonso, J.; Labavitch, J.; et al. CESA TRAFFICKING INHIBITOR Inhibits Cellulose Deposition and Interferes with the Trafficking of Cellulose Synthase Complexes and Their Associated Proteins KORRIGAN1 and POM2/CELLULOSE SYNTHASE INTERACTIVE PROTEIN1. Plant Physiol. 2015, 167, 381–393. [Google Scholar] [CrossRef]
- Zhang, Y.; Nikolovski, N.; Sorieul, M.; Vellosillo, T.; McFarlane, H.E.; Dupree, R.; Kesten, C.; Schneider, R.; Driemeier, C.; Lathe, R.; et al. Golgi-Localized STELLO Proteins Regulate the Assembly and Trafficking of Cellulose Synthase Complexes in Arabidopsis. Nat. Commun. 2016, 7, 11656. [Google Scholar] [CrossRef]
- Takahashi, J.; Rudsander, U.J.; Hedenström, M.; Banasiak, A.; Harholt, J.; Amelot, N.; Immerzeel, P.; Ryden, P.; Endo, S.; Ibatullin, F.M.; et al. KORRIGAN1 and Its Aspen Homolog PttCel9A1 Decrease Cellulose Crystallinity in Arabidopsis Stems. Plant Cell Physiol. 2009, 50, 1099–1115. [Google Scholar] [CrossRef]
- Peng, L.; Kawagoe, Y.; Hogan, P.; Delmer, D. Sitosterol-β-Glucoside as Primer for Cellulose Synthesis in Plants. Science 2002, 295, 147–150. [Google Scholar] [CrossRef]
- Somerville, C. Cellulose Synthesis in Higher Plants. Annu. Rev. Cell Dev. Biol. 2006, 22, 53–78. [Google Scholar] [CrossRef]
- Bhandari, S.; Fujino, T.; Thammanagowda, S.; Zhang, D.; Xu, F.; Joshi, C.P. Xylem-Specific and Tension Stress-Responsive Coexpression of KORRIGAN Endoglucanase and Three Secondary Wall-Associated Cellulose Synthase Genes in Aspen Trees. Planta 2006, 224, 828–837. [Google Scholar] [CrossRef]
- Zhou, H.-L.; He, S.-J.; Cao, Y.-R.; Chen, T.; Du, B.-X.; Chu, C.-C.; Zhang, J.-S.; Chen, S.-Y. OsGLU1, a Putative Membrane-Bound Endo-1,4-Beta-D-Glucanase from Rice, Affects Plant Internode Elongation. Plant Mol. Biol. 2006, 60, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, U.C.; Payyavula, R.S.; Labbé, J.L.; Engle, N.; Bali, G.; Jawdy, S.S.; Sykes, R.W.; Davis, M.; Ragauskas, A.; Tuskan, G.A.; et al. Down-Regulation of KORRIGAN-Like Endo-β-1,4-Glucanase Genes Impacts Carbon Partitioning, Mycorrhizal Colonization and Biomass Production in Populus. Front. Plant Sci. 2016, 7, 1455. [Google Scholar] [CrossRef] [PubMed]
- Brummell, D.A.; Catala, C.; Lashbrook, C.C.; Bennett, A.B. A Membrane-Anchored E-Type Endo-1,4-β-Glucanase Is Localized on Golgi and Plasma Membranes of Higher Plants. Proc. Natl. Acad. Sci. USA 1997, 94, 4794–4799. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Sun, J.; Li, L. PtrCel9A6, an Endo-1,4-β-Glucanase, Is Required for Cell Wall Formation during Xylem Differentiation in Populus. Mol. Plant 2013, 6, 1904–1917. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, H.; Sun, J.; Li, L. PtrKOR1 Is Required for Secondary Cell Wall Cellulose Biosynthesis in Populus. Tree Physiol. 2014, 34, 1289–1300. [Google Scholar] [CrossRef]
- López-Cruz, J.; Finiti, I.; Fernández-Crespo, E.; Crespo-Salvador, O.; García-Agustín, P.; González-Bosch, C. Absence of Endo-1,4-β-Glucanase KOR1 Alters the Jasmonate-Dependent Defence Response to Pseudomonas Syringae in Arabidopsis. J. Plant Physiol. 2014, 171, 1524–1532. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Bennett, A.B. Cooperative Disassembly of the Cellulose-Xyloglucan Network of Plant Cell Walls: Parallels between Cell Expansion and Fruit Ripening. Trends Plant Sci. 1999, 4, 176–183. [Google Scholar] [CrossRef]
- Brummell, D.A.; Hall, B.D.; Bennett, A.B. Antisense Suppression of Tomato Endo-1,4-Beta-Glucanase Cel2 MRNA Accumulation Increases the Force Required to Break Fruit Abscission Zones but Does Not Affect Fruit Softening. Plant Mol. Biol. 1999, 40, 615–622. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the Plant Cell Wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Del Campillo, E.; Abdel-Aziz, A.; Crawford, D.; Patterson, S.E. Root Cap Specific Expression of an Endo-β-1,4-Glucanase (Cellulase): A New Marker to Study Root Development in Arabidopsis. Plant Mol. Biol. 2004, 56, 309–323. [Google Scholar] [CrossRef]
- Lewis, D.R.; Olex, A.L.; Lundy, S.R.; Turkett, W.H.; Fetrow, J.S.; Muday, G.K. A Kinetic Analysis of the Auxin Transcriptome Reveals Cell Wall Remodeling Proteins That Modulate Lateral Root Development in Arabidopsis. Plant Cell 2013, 25, 3329–3346. [Google Scholar] [CrossRef] [PubMed]
- Karve, R.; Suárez-Román, F.; Iyer-Pascuzzi, A.S. The Transcription Factor NIN-LIKE PROTEIN7 Controls Border-Like Cell Release. Plant Physiol. 2016, 171, 2101–2111. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.L.; Bennett, A.B. Role of Cell Wall Hydrolases in Fruit Ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 675–703. [Google Scholar] [CrossRef]
- Harpster, M.H.; Lee, K.Y.; Dunsmuir, P. Isolation and Characterization of a Gene Encoding Endo-β-1,4-Glucanase from Pepper (Capsicum annuum L.). Plant Mol. Biol. 1997, 33, 47–59. [Google Scholar] [CrossRef]
- Manning, K. Isolation of a Set of Ripening-Related Genes from Strawberry: Their Identification and Possible Relationship to Fruit Quality Traits. Planta 1998, 205, 622–631. [Google Scholar] [CrossRef]
- Jara, K.; Castro, R.I.; Ramos, P.; Parra-Palma, C.; Valenzuela-Riffo, F.; Morales-Quintana, L. Molecular Insights into FaEG1, a Strawberry Endoglucanase Enzyme Expressed during Strawberry Fruit Ripening. Plants 2019, 8, 140. [Google Scholar] [CrossRef]
- Ohmiya, Y.; Takeda, T.; Nakamura, S.; Sakai, F.; Hayashi, T. Purification and Properties of a Wall-Bound Endo-1,4-β-Glucanase from Suspension-Cultured Poplar Cells1. Plant Cell Physiol. 1995, 36, 607–614. [Google Scholar] [CrossRef]
- Ohmiya, Y.; Nakai, T.; Park, Y.W.; Aoyama, T.; Oka, A.; Sakai, F.; Hayashi, T. The Role of PopCel1 and PopCel2 in Poplar Leaf Growth and Cellulose Biosynthesis. Plant J. 2003, 33, 1087–1097. [Google Scholar] [CrossRef]
- Park, Y.W.; Tominaga, R.; Sugiyama, J.; Furuta, Y.; Tanimoto, E.; Samejima, M.; Sakai, F.; Hayashi, T. Enhancement of Growth by Expression of Poplar Cellulase in Arabidopsis thaliana. Plant J. 2003, 33, 1099–1106. [Google Scholar] [CrossRef]
- Huang, J.; Xia, T.; Li, G.; Li, X.; Li, Y.; Wang, Y.; Wang, Y.; Chen, Y.; Xie, G.; Bai, F.-W.; et al. Overproduction of Native Endo-β-1,4-Glucanases Leads to Largely Enhanced Biomass Saccharification and Bioethanol Production by Specific Modification of Cellulose Features in Transgenic Rice. Biotechnol. Biofuels 2019, 12, 11. [Google Scholar] [CrossRef]
- Glass, M.; Barkwill, S.; Unda, F.; Mansfield, S.D. Endo-β-1,4-Glucanases Impact Plant Cell Wall Development by Influencing Cellulose Crystallization. J. Integr. Plant Biol. 2015, 57, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Schwacke, R.; Schneider, A.; van der Graaff, E.; Fischer, K.; Catoni, E.; Desimone, M.; Frommer, W.B.; Flügge, U.-I.; Kunze, R. ARAMEMNON, a Novel Database for Arabidopsis Integral Membrane Proteins. Plant Physiol. 2003, 131, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Xi, W.; Shen, J.; Bi, T.; Li, L. Characterization of the Plasma Membrane Proteins and Receptor-like Kinases Associated with Secondary Vascular Differentiation in Poplar. Plant Mol. Biol. 2011, 76, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.; Bernfur, K.; Gustavsson, N.; Bygdell, J.; Wingsle, G.; Larsson, C. Proteomics of Plasma Membranes from Poplar Trees Reveals Tissue Distribution of Transporters, Receptors, and Proteins in Cell Wall Formation *. Mol. Cell. Proteom. 2010, 9, 368–387. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, L.; Wu, R. Plant Grafting: How Genetic Exchange Promotes Vascular Reconnection. New Phytol. 2017, 214, 56–65. [Google Scholar] [CrossRef]
- Notaguchi, M.; Kurotani, K.; Sato, Y.; Tabata, R.; Kawakatsu, Y.; Okayasu, K.; Sawai, Y.; Okada, R.; Asahina, M.; Ichihashi, Y.; et al. Cell-Cell Adhesion in Plant Grafting Is Facilitated by β-1,4-Glucanases. Science 2020, 369, 698–702. [Google Scholar] [CrossRef]
- Kurotani, K.; Wakatake, T.; Ichihashi, Y.; Okayasu, K.; Sawai, Y.; Ogawa, S.; Cui, S.; Suzuki, T.; Shirasu, K.; Notaguchi, M. Host-Parasite Tissue Adhesion by a Secreted Type of β-1,4-Glucanase in the Parasitic Plant Phtheirospermum Japonicum. Commun. Biol. 2020, 3, 407. [Google Scholar] [CrossRef]
- Finiti, I.; Leyva, M.O.; López-Cruz, J.; Rodrigues, B.C.; Vicedo, B.; Angulo, C.; Bennett, A.B.; Grant, M.; García-Agustín, P.; González-Bosch, C. Functional Analysis of Endo-1,4-β-Glucanases in Response to Botrytis Cinerea and Pseudomonas Syringae Reveals Their Involvement in Plant–Pathogen Interactions. Plant Biol. 2013, 15, 819–831. [Google Scholar] [CrossRef]
- Flors, V.; Leyva, M.D.L.O.; Vicedo, B.; Finiti, I.; Real, M.D.; García-Agustín, P.; Bennett, A.B.; González-Bosch, C. Absence of the Endo-β-1,4-Glucanases Cel1 and Cel2 Reduces Susceptibility to Botrytis Cinerea in Tomato. Plant J. 2007, 52, 1027–1040. [Google Scholar] [CrossRef]
- Woo, M.-O.; Beard, H.; MacDonald, M.H.; Brewer, E.P.; Youssef, R.M.; Kim, H.; Matthews, B.F. Manipulation of Two α-Endo-β-1,4-Glucanase Genes, AtCel6 and GmCel7, Reduces Susceptibility to Heterodera Glycines in Soybean Roots. Mol. Plant Pathol. 2014, 15, 927–939. [Google Scholar] [CrossRef]
- Goellner, M.; Wang, X.; Davis, E.L. Endo-β-1,4-Glucanase Expression in Compatible Plant-Nematode Interactions. Plant Cell 2001, 13, 2241–2256. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Endo, B.Y. Histology and Ultrastructural Modification Induced By Cyst Nematodes. In Cyst Nematodes; Lamberti, F., Taylor, C.E., Eds.; Nato ASI Series; Springer: Boston, MA, USA, 1986; pp. 133–146. ISBN 978-1-4613-2251-1. [Google Scholar]
- Hussey, R.; Grundler, F. Nematode Parasitism of Plants. In Physiology and Biochemistry of Free-Living and Plant Parasitic Nematodes; CAB International Press: Oxfordshire, UK, 1998; pp. 213–243. [Google Scholar]
- Tucker, M.L.; Burke, A.; Murphy, C.A.; Thai, V.K.; Ehrenfried, M.L. Gene Expression Profiles for Cell Wall-Modifying Proteins Associated with Soybean Cyst Nematode Infection, Petiole Abscission, Root Tips, Flowers, Apical Buds, and Leaves. J. Exp. Bot. 2007, 58, 3395–3406. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, K.; Hofmann, J.; Blöchl, A.; Szakasits, D.; Bohlmann, H.; Grundler, F.M.W. Arabidopsis Endo-1,4-β-Glucanases Are Involved in the Formation of Root Syncytia Induced by Heterodera Schachtii. Plant J. 2008, 53, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.; Ibrahim, H.; Klink, V. Changes in the Expression of Genes in Soybean Roots Infected by Nematodes. In Soybean-Genetics and Novel Techniques for Yield Enhancement; InTech: London, UK, 2011; ISBN 978-953-307-721-5. [Google Scholar]
- Trainotti, L.; Spolaore, S.; Pavanello, A.; Baldan, B.; Casadoro, G. A Novel E-Type Endo-β-1,4-Glucanase with a Putative Cellulose-Binding Domain Is Highly Expressed in Ripening Strawberry Fruits. Plant Mol. Biol. 1999, 40, 323–332. [Google Scholar] [CrossRef]
- Del Campillo, E.; Gaddam, S.; Mettle-Amuah, D.; Heneks, J. A Tale of Two Tissues: AtGH9C1 Is an Endo-β-1,4-Glucanase Involved in Root Hair and Endosperm Development in Arabidopsis. PLoS ONE 2012, 7, e49363. [Google Scholar] [CrossRef]
- Urbanowicz, B.R.; Catalá, C.; Irwin, D.; Wilson, D.B.; Ripoll, D.R.; Rose, J.K.C. A Tomato Endo-β-1,4-Glucanase, SlCel9C1, Represents a Distinct Subclass with a New Family of Carbohydrate Binding Modules (CBM49) *. J. Biol. Chem. 2007, 282, 12066–12074. [Google Scholar] [CrossRef]
- McLean, B.W.; Bray, M.R.; Boraston, A.B.; Gilkes, N.R.; Haynes, C.A.; Kilburn, D.G. Analysis of Binding of the Family 2a Carbohydrate-Binding Module from Cellulomonas Fimi Xylanase 10A to Cellulose: Specificity and Identification of Functionally Important Amino Acid Residues. Protein Eng. 2000, 13, 801–809. [Google Scholar] [CrossRef]
- Duan, C.-J.; Feng, Y.-L.; Cao, Q.-L.; Huang, M.-Y.; Feng, J.-X. Identification of a Novel Family of Carbohydrate-Binding Modules with Broad Ligand Specificity. Sci. Rep. 2016, 6, 19392. [Google Scholar] [CrossRef]
- Boraston, A.B.; Nurizzo, D.; Notenboom, V.; Ducros, V.; Rose, D.R.; Kilburn, D.G.; Davies, G.J. Differential Oligosaccharide Recognition by Evolutionarily-Related β-1,4 and β-1,3 Glucan-Binding Modules. J. Mol. Biol. 2002, 319, 1143–1156. [Google Scholar] [CrossRef]
- Charnock, S.J.; Bolam, D.N.; Nurizzo, D.; Szabó, L.; McKie, V.A.; Gilbert, H.J.; Davies, G.J. Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module, CBM29-2, in Complex with Cello- and Mannohexaose. Proc. Natl. Acad. Sci. USA 2002, 99, 14077–14082. [Google Scholar] [CrossRef]
- Kim, D.Y.; Lee, M.J.; Cho, H.-Y.; Lee, J.S.; Lee, M.-H.; Chung, C.W.; Shin, D.-H.; Rhee, Y.H.; Son, K.-H.; Park, H.-Y. Genetic and Functional Characterization of an Extracellular Modular GH6 Endo-β-1,4-Glucanase from an Earthworm Symbiont, Cellulosimicrobium Funkei HY-13. Antonie Van Leeuwenhoek 2016, 109, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Komae, K. A Rice Family 9 Glycoside Hydrolase Isozyme with Broad Substrate Specificity for Hemicelluloses in Type II Cell Walls. Plant Cell Physiol. 2006, 47, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Bairoch, A. New Families in the Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1993, 293, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Linthorst, H.J.; Melchers, L.S.; Mayer, A.; van Roekel, J.S.; Cornelissen, B.J.; Bol, J.F. Analysis of Gene Families Encoding Acidic and Basic Beta-1,3-Glucanases of Tobacco. Proc. Natl. Acad. Sci. USA 1990, 87, 8756–8760. [Google Scholar] [CrossRef]
- Hudspeth, R.L.; Hobbs, S.L.; Anderson, D.M.; Grula, J.W. Characterization and Expression of Chitinase and 1,3-β-Glucanase Genes in Cotton. Plant Mol. Biol. 1996, 31, 911–916. [Google Scholar] [CrossRef]
- Jin, W.; Horner, H.T.; Palmer, R.G.; Shoemaker, R.C. Analysis and Mapping of Gene Families Encoding Beta-1,3-Glucanases of Soybean. Genetics 1999, 153, 445–452. [Google Scholar] [CrossRef]
- Romero, G.O.; Simmons, C.; Yaneshita, M.; Doan, M.; Thomas, B.R.; Rodriguez, R.L. Characterization of Rice Endo-Beta-Glucanase Genes (Gns2–Gns14) Defines a New Subgroup within the Gene Family. Gene 1998, 223, 311–320. [Google Scholar] [CrossRef]
- Hwang, D.H.; Kim, S.T.; Kim, S.G.; Kang, K.Y. Comprehensive Analysis of the Expression of Twenty-Seven Beta-1,3-Glucanase Genes in Rice (Oryza sativa L.). Mol. Cells 2007, 23, 207–214. [Google Scholar]
- Doxey, A.C.; Yaish, M.W.F.; Moffatt, B.A.; Griffith, M.; McConkey, B.J. Functional Divergence in the Arabidopsis β-1,3-Glucanase Gene Family Inferred by Phylogenetic Reconstruction of Expression States. Mol. Biol. Evol. 2007, 24, 1045–1055. [Google Scholar] [CrossRef]
- Xu, X.; Feng, Y.; Fang, S.; Xu, J.; Wang, X.; Guo, W. Genome-Wide Characterization of the β-1,3-Glucanase Gene Family in Gossypium by Comparative Analysis. Sci. Rep. 2016, 6, 29044. [Google Scholar] [CrossRef]
- Barral, P.; Suárez, C.; Batanero, E.; Alfonso, C.; de Dios Alché, J.; Rodríguez-García, M.I.; Villalba, M.; Rivas, G.; Rodríguez, R. An Olive Pollen Protein with Allergenic Activity, Ole e 10, Defines a Novel Family of Carbohydrate-Binding Modules and Is Potentially Implicated in Pollen Germination. Biochem. J. 2005, 390, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Sticher, L.; Hinz, U.; Meyer, A.D.; Meins, F. Intracellular Transport and Processing of a Tobacco Vacuolar β-1,3-Glucanase. Planta 1992, 188, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Davies, G.J. Glycoside Hydrolases and Glycosyltransferases. Families, Modules, and Implications for Genomics1. Plant Physiol. 2000, 124, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Leubner-Metzger, G.; Meins, F. Functions and Regulation of Plant SS-1,3-Glucanases (PR-2): Pathogenesis-Related Proteins in Plants. In Pathogenesis-Related Proteins in Plants; Datta, S.K., Muthukrishnan, S., Eds.; CRC Press, LLC: Boca Raton, FL, USA, 1999; pp. 49–76. [Google Scholar]
- Bachman, E.S.; McClay, D.R. Molecular Cloning of the First Metazoan Beta-1,3 Glucanase from Eggs of the Sea Urchin Strongylocentrotus Purpuratus. Proc. Natl. Acad. Sci. USA 1996, 93, 6808–6813. [Google Scholar] [CrossRef]
- Sun, L.; Gurnon, J.R.; Adams, B.J.; Graves, M.V.; Van Etten, J.L. Characterization of a β-1,3-Glucanase Encoded by Chlorella Virus PBCV-1. Virology 2000, 276, 27–36. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Ortiz-Castellanos, L. Cell Wall Glucans of Fungi. A Review. Cell Surf. 2019, 5, 100022. [Google Scholar] [CrossRef]
- Sela-Buurlage, M.B.; Ponstein, A.S.; Bres-Vloemans, S.A.; Melchers, L.S.; Van Den Elzen, P.J.M.; Cornelissen, B.J.C. Only Specific Tobacco (Nicotiana tabacum) Chitinases and [Beta]-1,3-Glucanases Exhibit Antifungal Activity. Plant Physiol. 1993, 101, 857–863. [Google Scholar] [CrossRef]
- Amian, A.; Papenbrock, J.; Jacobsen, H.-J.; Hassan, F. Enhancing Transgenic Pea (Pisum sativum L.) Resistance against Fungal Diseases through Stacking of Two Antifungal Genes (Chitinase and Glucanase). GM Crops 2011, 2, 104–109. [Google Scholar] [CrossRef]
- Zhang, S.-B.; Zhang, W.-J.; Zhai, H.-C.; Lv, Y.-Y.; Cai, J.-P.; Jia, F.; Wang, J.-S.; Hu, Y.-S. Expression of a Wheat β-1,3-Glucanase in Pichia pastoris and Its Inhibitory Effect on Fungi Commonly Associated with Wheat Kernel. Protein Expr. Purif. 2019, 154, 134–139. [Google Scholar] [CrossRef]
- Liu, B.; Lu, Y.; Xin, Z.; Zhang, Z. Identification and Antifungal Assay of a Wheat β-1,3-Glucanase. Biotechnol. Lett. 2009, 31, 1005–1010. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Wang, X.; Zhong, L.; Shi, G.; Xu, Y.; Li, Y.; Li, R.; Huang, Y.; Ye, X.; et al. A Novel β-1,3-Glucanase Gns6 from Rice Possesses Antifungal Activity against Magnaporthe oryzae. J. Plant Physiol. 2021, 265, 153493. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Ravi, I.; Sharma, V. Induction of β-1,3-Glucanase and Chitinase Activity in the Defense Response of Eruca sativa Plants against the Fungal Pathogen Alternaria brassicicola. J. Plant Interact. 2013, 8, 155–161. [Google Scholar] [CrossRef]
- Salim, A.P.; Saminaidu, K.; Marimuthu, M.; Perumal, Y.; Rethinasamy, V.; Palanisami, J.R.; Vadivel, K. Defense Responses in Tomato Landrace and Wild Genotypes to Early Blight Pathogen Alternaria solani Infection and Accumulation of Pathogenesis-Related Proteins. Arch. Phytopathol. Plant Prot. 2011, 44, 1147–1164. [Google Scholar] [CrossRef]
- Aggarwal, R.; Purwar, S.; Kharbikar, L.; Gupta, S. Induction of a Wheat β-1,3-Glucanase Gene during the Defense Response to Bipolaris sorokiniana. Acta Phytopathol. Entomol. Hung. 2011, 46, 39–47. [Google Scholar] [CrossRef]
- Sundaresha, S.; Kumar, A.M.; Rohini, S.; Math, S.A.; Keshamma, E.; Chandrashekar, S.C.; Udayakumar, M. Enhanced Protection against Two Major Fungal Pathogens of Groundnut, Cercospora arachidicola and Aspergillus flavus in Transgenic Groundnut over-Expressing a Tobacco β 1–3 Glucanase. Eur. J. Plant Pathol. 2010, 126, 497–508. [Google Scholar] [CrossRef]
- Wróbel-Kwiatkowska, M.; Lorenc-Kukula, K.; Starzycki, M.; Oszmiański, J.; Kepczyńska, E.; Szopa, J. Expression of β-1,3-Glucanase in Flax Causes Increased Resistance to Fungi. Physiol. Mol. Plant Pathol. 2004, 65, 245–256. [Google Scholar] [CrossRef]
- Mackintosh, C.A.; Lewis, J.; Radmer, L.E.; Shin, S.; Heinen, S.J.; Smith, L.A.; Wyckoff, M.N.; Dill-Macky, R.; Evans, C.K.; Kravchenko, S.; et al. Overexpression of Defense Response Genes in Transgenic Wheat Enhances Resistance to Fusarium Head Blight. Plant Cell Rep. 2007, 26, 479–488. [Google Scholar] [CrossRef]
- Maziah, M.; Sreeramanan, S.; Puad, A.; Meon, S. Production of Transgenic Banana Cultivar, Rastali (AAB) via Agrobacterium-Mediated Transformation with a Rice Chitinase Gene. J. Plant Sci. 2007, 2, 504–517. [Google Scholar] [CrossRef]
- Fesel, P.H.; Zuccaro, A. β-Glucan: Crucial Component of the Fungal Cell Wall and Elusive MAMP in Plants. Fungal Genet. Biol. 2016, 90, 53–60. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Keen, N.T.; Wang, M.C. A Receptor on Soybean Membranes for a Fungal Elicitor of Phytoalexin Accumulation. Plant Physiol. 1983, 73, 497–506. [Google Scholar] [CrossRef]
- Sharp, J.K.; Valent, B.; Albersheim, P. Purification and Partial Characterization of a Beta-Glucan Fragment That Elicits Phytoalexin Accumulation in Soybean. J. Biol. Chem. 1984, 259, 11312–11320. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Yamada, A.; Hong, N.; Ogawa, T.; Ishii, T.; Shibuya, N. Differences in the Recognition of Glucan Elicitor Signals between Rice and Soybean: β-Glucan Fragments from the Rice Blast Disease Fungus Pyricularia Oryzae That Elicit Phytoalexin Biosynthesis in Suspension-Cultured Rice Cells. Plant Cell 2000, 12, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Cosio, E.G.; Feger, M.; Miller, C.J.; Antelo, L.; Ebel, J. High-Affinity Binding of Fungal β-Glucan Elicitors to Cell Membranes of Species of the Plant Family Fabaceae. Planta 1996, 200, 92–99. [Google Scholar] [CrossRef]
- Côté, F.; Roberts, K.A.; Hahn, M.G. Identification of High-Affinity Binding Sites for the Hepta-Beta-Glucoside Elicitor in Membranes of the Model Legumes Medicago truncatula and Lotus japonicus. Planta 2000, 211, 596–605. [Google Scholar] [CrossRef] [PubMed]
- Wanke, A.; Malisic, M.; Wawra, S.; Zuccaro, A. Unraveling the Sugar Code: The Role of Microbial Extracellular Glycans in Plant-Microbe Interactions. J. Exp. Bot. 2021, 72, 15–35. [Google Scholar] [CrossRef] [PubMed]
- Mélida, H.; Sopeña-Torres, S.; Bacete, L.; Garrido-Arandia, M.; Jordá, L.; López, G.; Muñoz-Barrios, A.; Pacios, L.F.; Molina, A. Non-Branched β-1,3-Glucan Oligosaccharides Trigger Immune Responses in Arabidopsis. Plant J. 2018, 93, 34–49. [Google Scholar] [CrossRef]
- Wanke, A.; Rovenich, H.; Schwanke, F.; Velte, S.; Becker, S.; Hehemann, J.-H.; Wawra, S.; Zuccaro, A. Plant Species-Specific Recognition of Long and Short β-1,3-Linked Glucans Is Mediated by Different Receptor Systems. Plant J. 2020, 102, 1142–1156. [Google Scholar] [CrossRef]
- Chandrasekar, B.; Wanke, A.; Wawra, S.; Saake, P.; Mahdi, L.; Charura, N.; Neidert, M.; Malisic, M.; Thiele, M.; Dama, M.; et al. Fungi Hijack a Ubiquitous Plant Apoplastic Endoglucanase to Release a ROS Scavenging β-Glucan Decasaccharide to Subvert Immune Responses. Plant Cell, 2022; accepted. [Google Scholar] [CrossRef]
- Hird, D.L.; Worrall, D.; Hodge, R.; Smartt, S.; Paul, W.; Scott, R. The Anther-Specific Protein Encoded by the Brassica Napus and Arabidopsis thaliana A6 Gene Displays Similarity to β-1,3-Glucanases. Plant J. 1993, 4, 1023–1033. [Google Scholar] [CrossRef]
- Parveen, S.; Mazumder, M.; Bhattacharya, A.; Mukhopadhyay, S.; Saha, U.; Mukherjee, A.; Mondal, B.; Debnath, A.J.; Das, S.; Sikdar, S.; et al. Identification of Anther-Specific Genes from Sesame and Functional Assessment of the Upstream Region of a Tapetum-Specific β-1,3-Glucanase Gene. Plant Mol. Biol. Rep. 2018, 36, 149–161. [Google Scholar] [CrossRef]
- Zieliński, K.; Dubas, E.; Gerši, Z.; Krzewska, M.; Janas, A.; Nowicka, A.; Matušíková, I.; Żur, I.; Sakuda, S.; Moravčíková, J. β-1,3-Glucanases and Chitinases Participate in the Stress-Related Defence Mechanisms That Are Possibly Connected with Modulation of Arabinogalactan Proteins (AGP) Required for the Androgenesis Initiation in Rye (Secale cereale L.). Plant Sci. Int. J. Exp. Plant Biol. 2021, 302, 110700. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Zha, W.; Cheng, X.; Liu, C.; Lv, L.; Liu, C.; Wang, Z.; Du, B.; Chen, R.; Zhu, L.; et al. A Rice β-1,3-Glucanase Gene Osg1 Is Required for Callose Degradation in Pollen Development. Planta 2011, 233, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Delp, G.; Palva, E.T. A Novel Flower-Specific Arabidopsis Gene Related to Both Pathogen-Induced and Developmentally Regulated Plant Beta-1,3-Glucanase Genes. Plant Mol. Biol. 1999, 39, 565–575. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; Geelen, D. The Impact of Environmental Stress on Male Reproductive Development in Plants: Biological Processes and Molecular Mechanisms. Plant Cell Environ. 2014, 37, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Huang, L.-J.; Feng, D.; Jiang, W.; Miu, W.; Li, N. Plasmodesmata-Related Structural and Functional Proteins: The Long Sought-After Secrets of a Cytoplasmic Channel in Plant Cell Walls. Int. J. Mol. Sci. 2019, 20, 2946. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Erlanger, M.; Rosenthal, M.; Epel, B.L. A Plasmodesmata-Associated β-1,3-Glucanase in Arabidopsis. Plant J. 2007, 49, 669–682. [Google Scholar] [CrossRef]
- Wu, S.-W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.-Y. Callose Balancing at Plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef]
- Kim, I.; Zambryski, P.C. Cell-to-Cell Communication via Plasmodesmata during Arabidopsis Embryogenesis. Curr. Opin. Plant Biol. 2005, 8, 593–599. [Google Scholar] [CrossRef]
- Benitez-Alfonso, Y.; Faulkner, C.; Pendle, A.; Miyashima, S.; Helariutta, Y.; Maule, A. Symplastic Intercellular Connectivity Regulates Lateral Root Patterning. Dev. Cell 2013, 26, 136–147. [Google Scholar] [CrossRef]
- Guseman, J.M.; Lee, J.S.; Bogenschutz, N.L.; Peterson, K.M.; Virata, R.E.; Xie, B.; Kanaoka, M.M.; Hong, Z.; Torii, K.U. Dysregulation of Cell-to-Cell Connectivity and Stomatal Patterning by Loss-of-Function Mutation in Arabidopsis Chorus (Glucan Synthase-like 8). Development 2010, 137, 1731–1741. [Google Scholar] [CrossRef]
- Gisel, A.; Hempel, F.D.; Barella, S.; Zambryski, P. Leaf-to-Shoot Apex Movement of Symplastic Tracer Is Restricted Coincident with Flowering in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 1713–1717. [Google Scholar] [CrossRef]
- Paterlini, A.; Dorussen, D.; Fichtner, F.; van Rongen, M.; Delacruz, R.; Vojnović, A.; Helariutta, Y.; Leyser, O. Callose Accumulation in Specific Phloem Cell Types Reduces Axillary Bud Growth in Arabidopsis thaliana. New Phytol. 2021, 231, 516–523. [Google Scholar] [CrossRef]
- Rinne, P.L.H.; Kaikuranta, P.M.; Van Der Schoot, C. The Shoot Apical Meristem Restores Its Symplasmic Organization during Chilling-Induced Release from Dormancy. Plant J. 2001, 26, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yuan, Y.; Liu, Z.; Liu, C.; Xin, H.; Zhang, Y.; Gai, S. Chilling and Gibberellin Acids Hyperinduce β-1,3-Glucanases to Reopen Transport Corridor and Break Endodormancy in Tree Peony (Paeonia suffruticosa). Plant Physiol. Biochem. 2021, 167, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Leubner-Metzger, G. Seed After-Ripening and over-Expression of Class I β-1,3-Glucanase Confer Maternal Effects on Tobacco Testa Rupture and Dormancy Release. Planta 2002, 215, 959–968. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Roy, S.; Sengupta, D.N. Characterization of Cultivar Differences in Beta-1,3 Glucanase Gene Expression, Glucanase Activity and Fruit Pulp Softening Rates during Fruit Ripening in Three Naturally Occurring Banana Cultivars. Plant Cell Rep. 2009, 28, 1641–1653. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Perry, L.; Benitez-Alfonso, Y. A Phylogenetic and Transcriptomic Study of the β-1,3-Glucanase Family in Tomato Identifies Candidate Targets for Fruit Improvement. bioRxiv, 2021; preprint. [Google Scholar] [CrossRef]
- Beffa, R.S.; Hofer, R.M.; Thomas, M.; Meins, F. Decreased Susceptibility to Viral Disease of [Beta]-1,3-Glucanase-Deficient Plants Generated by Antisense Transformation. Plant Cell 1996, 8, 1001–1011. [Google Scholar] [CrossRef]
- Iglesias, V.A.; Meins, F. Movement of Plant Viruses Is Delayed in a Beta-1,3-Glucanase-Deficient Mutant Showing a Reduced Plasmodesmatal Size Exclusion Limit and Enhanced Callose Deposition. Plant J. 2000, 21, 157–166. [Google Scholar] [CrossRef]
- Zavaliev, R.; Levy, A.; Gera, A.; Epel, B.L. Subcellular Dynamics and Role of Arabidopsis β-1,3-Glucanases in Cell-to-Cell Movement of Tobamoviruses. Mol. Plant-Microbe Interact. MPMI 2013, 26, 1016–1030. [Google Scholar] [CrossRef]
- Li, Z.; Variz, H.; Chen, Y.; Liu, S.-L.; Aung, K. Plasmodesmata-Dependent Intercellular Movement of Bacterial Effectors. Front. Plant Sci. 2021, 12, 464. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Wang, Y.; Zhang, F.; Yuan, X.; Chen, H.; Chen, X.; Chen, X.; Cui, X. Soybean Endo-1,3-Beta-Glucanase (GmGLU) Interaction with Soybean mosaic Virus-Encoded P3 Protein May Contribute to the Intercelluar Movement. Front. Genet. 2020, 11, 536771. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.N.; Lasky, D.; Karki, H.; Zhang, Z.; Goyer, A.; Halterman, D.; Rakotondrafara, A.M. HCPro Suppression of Callose Deposition Contributes to Strain-Specific Resistance Against Potato Virus Y. Phytopathology 2020, 110, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Tseng, K.-C.; Chang, W.-C.; Seo, J.-K.; Kim, K.-H. Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses 2018, 10, 581. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B.E.L. Plant Cell Wall Dynamics in Compatible and Incompatible Potato Response to Infection Caused by Potato Virus Y (PVYNTN). Int. J. Mol. Sci. 2018, 19, 862. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Regulation and Function of Defense-Related Callose Deposition in Plants. Int. J. Mol. Sci. 2021, 22, 2393. [Google Scholar] [CrossRef]
- Griffith, M.; Yaish, M.W.F. Antifreeze Proteins in Overwintering Plants: A Tale of Two Activities. Trends Plant Sci. 2004, 9, 399–405. [Google Scholar] [CrossRef]
- Yaish, M.W.F.; Doxey, A.C.; McConkey, B.J.; Moffatt, B.A.; Griffith, M. Cold-Active Winter Rye Glucanases with Ice-Binding Capacity. Plant Physiol. 2006, 141, 1459–1472. [Google Scholar] [CrossRef]
- Romero, I.; Fernandez-Caballero, C.; Goñi, O.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. Functionality of a Class I Beta-1,3-Glucanase from Skin of Table Grapes Berries. Plant Sci. 2008, 174, 641–648. [Google Scholar] [CrossRef]
- Ding, C.-K.; Wang, C.; Gross, K.C.; Smith, D.L. Jasmonate and Salicylate Induce the Expression of Pathogenesis-Related-Protein Genes and Increase Resistance to Chilling Injury in Tomato Fruit. Planta 2002, 214, 895–901. [Google Scholar] [CrossRef]
- Sanchez-Ballesta, M.T.; Gosalbes, M.J.; Rodrigo, M.J.; Granell, A.; Zacarias, L.; Lafuente, M.T. Characterization of a β-1,3-Glucanase from Citrus Fruit as Related to Chilling-Induced Injury and Ethylene Production. Postharvest Biol. Technol. 2006, 40, 133–140. [Google Scholar] [CrossRef]
- Fincher, G.B. Exploring the Evolution of (1,3;1,4)-β-d-Glucans in Plant Cell Walls: Comparative Genomics Can Help! Curr. Opin. Plant Biol. 2009, 12, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Fincher, G.B.; Lock, P.A.; Morgan, M.M.; Lingelbach, K.; Wettenhall, R.E.; Mercer, J.F.; Brandt, A.; Thomsen, K.K. Primary Structure of the (1→3,1→4)-Beta-D-Glucan 4-Glucohydrolase from Barley Aleurone. Proc. Natl. Acad. Sci. USA 1986, 83, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Simmons, C.R.; Litts, J.C.; Huang, N.; Rodriguez, R.L. Structure of a Rice β-Glucanase Gene Regulated by Ethylene, Cytokinin, Wounding, Salicylic Acid and Fungal Elicitors. Plant Mol. Biol. 1992, 18, 33–45. [Google Scholar] [CrossRef]
- Lai, D.M.; Høj, P.B.; Fincher, G.B. Purification and Characterization of (1→3,1→4)-Beta-Glucan Endohydrolases from Germinated Wheat (Triticum aestivum). Plant Mol. Biol. 1993, 22, 847–859. [Google Scholar] [CrossRef]
- Yun, S.J.; Martin, D.J.; Gengenbach, B.G.; Rines, H.W.; Somers, D.A. Sequence of a (1-3,1-4)-Beta-Glucanase CDNA from Oat. Plant Physiol. 1993, 103, 295–296. [Google Scholar] [CrossRef]
- Akiyama, T.; Jin, S.; Yoshida, M.; Hoshino, T.; Opassiri, R.; Ketudat Cairns, J.R. Expression of an Endo-(1,3;1,4)-β-Glucanase in Response to Wounding, Methyl Jasmonate, Abscisic Acid and Ethephon in Rice Seedlings. J. Plant Physiol. 2009, 166, 1814–1825. [Google Scholar] [CrossRef]
- Kraemer, F.J.; Lunde, C.; Koch, M.; Kuhn, B.M.; Ruehl, C.; Brown, P.J.; Hoffmann, P.; Göhre, V.; Hake, S.; Pauly, M.; et al. A Mixed-Linkage (1,3;1,4)-β-D-Glucan Specific Hydrolase Mediates Dark-Triggered Degradation of This Plant Cell Wall Polysaccharide. Plant Physiol. 2021, 185, 1559–1573. [Google Scholar] [CrossRef]
- Fan, M.; Jensen, J.K.; Zemelis-Durfee, S.; Kim, S.-J.; Chan, J.-Y.; Beaudry, C.M.; Brandizzi, F.; Wilkerson, C.G. Disruption of Brachypodium Lichenase Alters Metabolism of Mixed-Linkage Glucan and Starch. Plant J. 2022, 109, 927–939. [Google Scholar] [CrossRef]
- Litts, J.C.; Simmons, C.R.; Karrer, E.E.; Huang, N.; Rodriguez, R.L. The Isolation and Characterization of a Barley 1,3-1,4-β-Glucanase Gene. Eur. J. Biochem. 1990, 194, 831–838. [Google Scholar] [CrossRef]
- Slakeski, N.; Fincher, G.B. Developmental Regulation of (1→3, 1→4)-β-Glucanase Gene Expression in Barley. Plant Physiol. 1992, 99, 1226–1231. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Slakeski, N.; Baulcombe, D.C.; Devos, K.M.; Ahluwalia, B.; Doan, D.N.; Fincher, G.B. Structure and Tissue-Specific Regulation of Genes Encoding Barley (1→3,1→4)-β-Glucan Endohydrolases. Mol. Gen. Genet. MGG 1990, 224, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Collins, H.M.; Betts, N.S.; Dockter, C.; Berkowitz, O.; Braumann, I.; Cuesta-Seijo, J.A.; Skadhauge, B.; Whelan, J.; Bulone, V.; Fincher, G.B. Genes That Mediate Starch Metabolism in Developing and Germinated Barley Grain. Front. Plant Sci. 2021, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- Fincher, G.B. Molecular and Cellular Biology Associated with Endosperm Mobilization in Germinating Cereal Grains. Annu. Rev. Plant Biol. 1989, 40, 305–346. [Google Scholar] [CrossRef]
- Carpita, N.C.; Defernez, M.; Findlay, K.; Wells, B.; Shoue, D.A.; Catchpole, G.; Wilson, R.H.; McCann, M.C. Cell Wall Architecture of the Elongating Maize Coleoptile. Plant Physiol. 2001, 127, 551–565. [Google Scholar] [CrossRef]
- Kiemle, S.N.; Zhang, X.; Esker, A.R.; Toriz, G.; Gatenholm, P.; Cosgrove, D.J. Role of (1,3)(1,4)-β-Glucan in Cell Walls: Interaction with Cellulose. Biomacromolecules 2014, 15, 1727–1736. [Google Scholar] [CrossRef]
- Smith-Moritz, A.M.; Hao, Z.; Fernández-Niño, S.G.; Fangel, J.U.; Verhertbruggen, Y.; Holman, H.-Y.N.; Willats, W.G.T.; Ronald, P.C.; Scheller, H.V.; Heazlewood, J.L.; et al. Structural Characterization of a Mixed-Linkage Glucan Deficient Mutant Reveals Alteration in Cellulose Microfibril Orientation in Rice Coleoptile Mesophyll Cell Walls. Front. Plant Sci. 2015, 6, 628. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural Models of Primary Cell Walls in Flowering Plants: Consistency of Molecular Structure with the Physical Properties of the Walls during Growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Enzymes and Other Agents That Enhance Cell Wall Extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 391–417. [Google Scholar] [CrossRef]
- Roulin, S.; Buchala, A.J.; Fincher, G.B. Induction of (1→3,1→4)-β-D-Glucan Hydrolases in Leaves of Dark-Incubated Barley Seedlings. Planta 2002, 215, 51–59. [Google Scholar] [CrossRef][Green Version]
- Nishizawa, Y.; Saruta, M.; Nakazono, K.; Nishio, Z.; Soma, M.; Yoshida, T.; Nakajima, E.; Hibi, T. Characterization of Transgenic Rice Plants Over-Expressing the Stress-Inducible β-Glucanase Gene Gns1. Plant Mol. Biol. 2003, 51, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Barghahn, S.; Arnal, G.; Jain, N.; Petutschnig, E.; Brumer, H.; Lipka, V. Mixed Linkage β-1,3/1,4-Glucan Oligosaccharides Induce Defense Responses in Hordeum vulgare and Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 1201. [Google Scholar] [CrossRef] [PubMed]
- Rebaque, D.; del Hierro, I.; López, G.; Bacete, L.; Vilaplana, F.; Dallabernardina, P.; Pfrengle, F.; Jordá, L.; Sánchez-Vallet, A.; Pérez, R.; et al. Cell Wall-Derived Mixed-Linked β-1,3/1,4-Glucans Trigger Immune Responses and Disease Resistance in Plants. Plant J. 2021, 106, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Pettolino, F.; Sasaki, I.; Turbic, A.; Wilson, S.M.; Bacic, A.; Hrmova, M.; Fincher, G.B. Hyphal Cell Walls from the Plant Pathogen Rhynchosporium secalis Contain (1,3/1,6)-β-D-Glucans, Galacto- and Rhamnomannans, (1,3;1,4)-β-D-Glucans and Chitin. FEBS J. 2009, 276, 3698–3709. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mendoza, D.; Rodríguez-Carvajal, M.Á.; Romero-Jiménez, L.; de Araujo Farias, G.; Lloret, J.; Gallegos, M.T.; Sanjuán, J. Novel Mixed-Linkage β-Glucan Activated by c-Di-GMP in Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 2015, 112, E757–E765. [Google Scholar] [CrossRef]
- Samar, D.; Kieler, J.B.; Klutts, J.S. Identification and Deletion of Tft1, a Predicted Glycosyltransferase Necessary for Cell Wall β-1,3;1,4-Glucan Synthesis in Aspergillus fumigatus. PLoS ONE 2015, 10, e0117336. [Google Scholar] [CrossRef]
- Ao, J.; Free, S.J. Genetic and Biochemical Characterization of the GH72 Family of Cell Wall Transglycosylases in Neurospora crassa. Fungal Genet. Biol. 2017, 101, 46–54. [Google Scholar] [CrossRef]
- Woodward, J.R.; Fincher, G.B. Substrate Specificities and Kinetic Properties of Two (1→3),(1→4)-β-d-Glucan Endo-Hydrolases from Germinating Barley (Hordeum vulgare). Carbohydr. Res. 1982, 106, 111–122. [Google Scholar] [CrossRef]
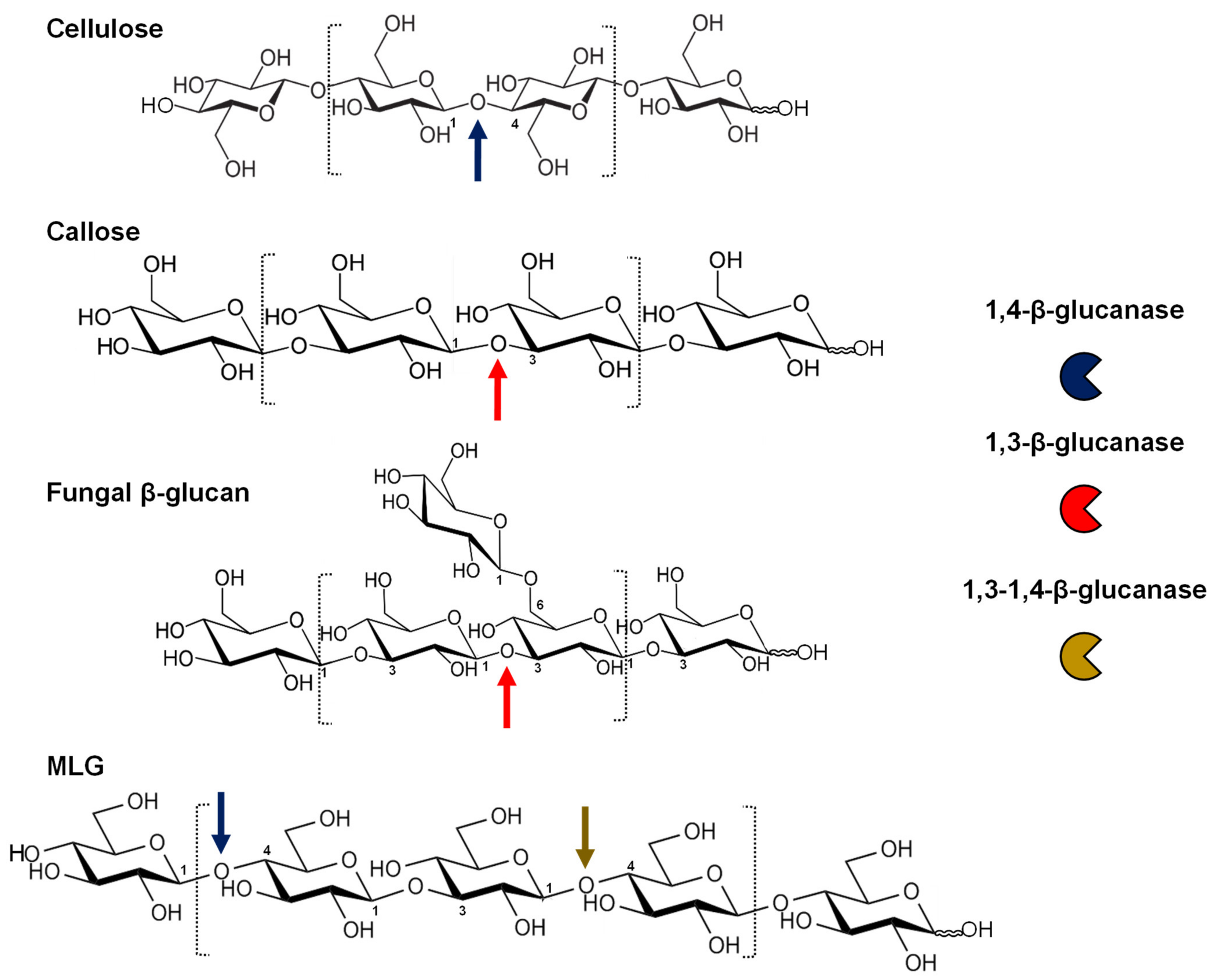
| Type | Substrate | Physiological Roles |
|---|---|---|
| 1,4-β-glucanases | Crystalline cellulose | Irreversible wall disassembly: root hair emergence, endosperm breakdown, fruit ripening |
| Amorphous cellulose | Secondary wall mechanical strength Wall remodelling: cell expansion, fruit ripening, nematode attack Cell-cell adhesion: grafting, plant parasitism Cellulose biosynthesis (plasma membrane-associated) | |
| MLG 1 (Fungal wall) | Antifungal activity Elicitor release: MAMP 2 | |
| 1,3-β-glucanases | Callose | Plasmodesmata and symplastic transport: dormancy release, fruit development, cell-to-cell communication Reproductive organs: pollen, style and stigma development |
| 1,3-β-glucan (Fungal wall) | Antifungal activity Elicitor release Fungal effector release | |
| 1,3-1,4-β-glucanases | MLG 1 | Cell wall loosening during germination Energy source in the dark |
| MLG 1 (Fungal wall) | Antifungal activity Elicitor release |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perrot, T.; Pauly, M.; Ramírez, V. Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses. Plants 2022, 11, 1119. https://doi.org/10.3390/plants11091119
Perrot T, Pauly M, Ramírez V. Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses. Plants. 2022; 11(9):1119. https://doi.org/10.3390/plants11091119
Chicago/Turabian StylePerrot, Thomas, Markus Pauly, and Vicente Ramírez. 2022. "Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses" Plants 11, no. 9: 1119. https://doi.org/10.3390/plants11091119
APA StylePerrot, T., Pauly, M., & Ramírez, V. (2022). Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses. Plants, 11(9), 1119. https://doi.org/10.3390/plants11091119






